Mechanisms of Bone Fragility: From Osteogenesis Imperfecta to Secondary Osteoporosis
Abstract
1. Introduction
2. Genetic Causes of Bone Fragility
2.1. Primary Osteoporosis Affecting Collagen (Osteogenesis Imperfecta)
2.1.1. Clinical Symptoms and Classification
2.1.2. Genetic Classification and Protein Function in OI
2.1.3. Pathway-Specific Therapy
2.2. Primary Osteoporosis Caused by WNT-Signaling Pathway Defects
Pathway-Specific Treatment
2.3. Primary Osteoporosis Caused by Defects in the TGF-β Pathway
Pathway-Specific Treatment
2.4. Primary Osteoporosis Caused by RANKL/RANK/OPG Defects: TNFRSF11B (Juvenile Paget Disease) and TNFRSF11A (Familial Expansile Osteolysis)
Pathway-Specific Treatment
2.5. Bone Fragility in Hajdu Cheney Syndrome
3. Acquired Causes of Bone Fragility
3.1. Immobility-Induced Osteoporosis Caused by the Osteocyte Biomechanic Sensing Mechanism
Pathway-Specific Treatment
3.2. Cytokine-Induced Osteoporosis in Leukemia/Cancer or Chronic Inflammatory Conditions via RANKL Activation
3.3. Steroid-Induced Osteoporosis (Osteotoxic Glucocorticoid Medication)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Literature Search Strategy
References
- Florencio-Silva, R.; Sasso, G.R.; Sasso-Cerri, E.; Simoes, M.J.; Cerri, P.S. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. BioMed Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Econs, M.J.; DiMeglio, L.A.; Insogna, K.L.; Levine, M.A.; Orchard, P.J.; Miller, W.P.; Petryk, A.; Rush, E.T.; Shoback, D.M.; et al. Diagnosis and Management of Osteopetrosis: Consensus Guidelines From the Osteopetrosis Working Group. J. Clin. Endocrinol. Metab. 2017, 102, 3111–3123. [Google Scholar] [CrossRef] [PubMed]
- Uday, S.; Högler, W. Rickets and Osteomalacia. In Encyclopedia of Endocrine Diseases, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 5, pp. 339–354. [Google Scholar]
- Forlino, A.; Marini, J.C. Osteogenesis imperfecta. Lancet 2016, 387, 1657–1671. [Google Scholar] [CrossRef]
- Cho, T.J.; Lee, K.E.; Lee, S.K.; Song, S.J.; Kim, K.J.; Jeon, D.; Lee, G.; Kim, H.N.; Lee, H.R.; Eom, H.H.; et al. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am. J. Hum. Genet. 2012, 91, 343–348. [Google Scholar] [CrossRef]
- Semler, O.; Garbes, L.; Keupp, K.; Swan, D.; Zimmermann, K.; Becker, J.; Iden, S.; Wirth, B.; Eysel, P.; Koerber, F.; et al. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am. J. Hum. Genet. 2012, 91, 349–357. [Google Scholar] [CrossRef]
- Becker, J.; Semler, O.; Gilissen, C.; Li, Y.; Bolz, H.J.; Giunta, C.; Bergmann, C.; Rohrbach, M.; Koerber, F.; Zimmermann, K.; et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2011, 88, 362–371. [Google Scholar] [CrossRef]
- Morello, R.; Bertin, T.K.; Chen, Y.; Hicks, J.; Tonachini, L.; Monticone, M.; Castagnola, P.; Rauch, F.; Glorieux, F.H.; Vranka, J.; et al. CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 2006, 127, 291–304. [Google Scholar] [CrossRef]
- Caparros-Martin, J.A.; Valencia, M.; Pulido, V.; Martinez-Glez, V.; Rueda-Arenas, I.; Amr, K.; Farra, C.; Lapunzina, P.; Ruiz-Perez, V.L.; Temtamy, S.; et al. Clinical and molecular analysis in families with autosomal recessive osteogenesis imperfecta identifies mutations in five genes and suggests genotype-phenotype correlations. Am. J. Med. Genet. A 2013, 161A, 1354–1369. [Google Scholar] [CrossRef]
- Cabral, W.A.; Chang, W.; Barnes, A.M.; Weis, M.; Scott, M.A.; Leikin, S.; Makareeva, E.; Kuznetsova, N.V.; Rosenbaum, K.N.; Tifft, C.J.; et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet. 2007, 39, 359–365. [Google Scholar] [CrossRef]
- Van Dijk, F.S.; Nesbitt, I.M.; Zwikstra, E.H.; Nikkels, P.G.; Piersma, S.R.; Fratantoni, S.A.; Jimenez, C.R.; Huizer, M.; Morsman, A.C.; Cobben, J.M.; et al. PPIB mutations cause severe osteogenesis imperfecta. Am. J. Hum. Genet. 2009, 85, 521–527. [Google Scholar] [CrossRef]
- Christiansen, H.E.; Schwarze, U.; Pyott, S.M.; AlSwaid, A.; Al Balwi, M.; Alrasheed, S.; Pepin, M.G.; Weis, M.A.; Eyre, D.R.; Byers, P.H. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010, 86, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Alanay, Y.; Avaygan, H.; Camacho, N.; Utine, G.E.; Boduroglu, K.; Aktas, D.; Alikasifoglu, M.; Tuncbilek, E.; Orhan, D.; Bakar, F.T.; et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010, 86, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Lapunzina, P.; Aglan, M.; Temtamy, S.; Caparros-Martin, J.A.; Valencia, M.; Leton, R.; Martinez-Glez, V.; Elhossini, R.; Amr, K.; Vilaboa, N.; et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2010, 87, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Pihlajaniemi, T.; Dickson, L.A.; Pope, F.M.; Korhonen, V.R.; Nicholls, A.; Prockop, D.J.; Myers, J.C. Osteogenesis imperfecta: Cloning of a pro-alpha 2(I) collagen gene with a frameshift mutation. J. Biol. Chem. 1984, 259, 12941–12944. [Google Scholar] [CrossRef]
- Shaheen, R.; Alazami, A.M.; Alshammari, M.J.; Faqeih, E.; Alhashmi, N.; Mousa, N.; Alsinani, A.; Ansari, S.; Alzahrani, F.; Al-Owain, M.; et al. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. J. Med. Genet. 2012, 49, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Keupp, K.; Beleggia, F.; Kayserili, H.; Barnes, A.M.; Steiner, M.; Semler, O.; Fischer, B.; Yigit, G.; Janda, C.Y.; Becker, J.; et al. Mutations in WNT1 cause different forms of bone fragility. Am. J. Hum. Genet. 2013, 92, 565–574. [Google Scholar] [CrossRef]
- Pyott, S.M.; Tran, T.T.; Leistritz, D.F.; Pepin, M.G.; Mendelsohn, N.J.; Temme, R.T.; Fernandez, B.A.; Elsayed, S.M.; Elsobky, E.; Verma, I.; et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2013, 92, 590–597. [Google Scholar] [CrossRef]
- Symoens, S.; Malfait, F.; D’Hondt, S.; Callewaert, B.; Dheedene, A.; Steyaert, W.; Bachinger, H.P.; De Paepe, A.; Kayserili, H.; Coucke, P.J. Deficiency for the ER-stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet. J. Rare Dis. 2013, 8, 154. [Google Scholar] [CrossRef]
- Mendoza-Londono, R.; Fahiminiya, S.; Majewski, J.; Care4Rare Canada, C.; Tetreault, M.; Nadaf, J.; Kannu, P.; Sochett, E.; Howard, A.; Stimec, J.; et al. Recessive osteogenesis imperfecta caused by missense mutations in SPARC. Am. J. Hum. Genet. 2015, 96, 979–985. [Google Scholar] [CrossRef]
- Doyard, M.; Bacrot, S.; Huber, C.; Di Rocco, M.; Goldenberg, A.; Aglan, M.S.; Brunelle, P.; Temtamy, S.; Michot, C.; Otaify, G.A.; et al. FAM46A mutations are responsible for autosomal recessive osteogenesis imperfecta. J. Med. Genet. 2018, 55, 278–284. [Google Scholar] [CrossRef]
- Lindert, U.; Cabral, W.A.; Ausavarat, S.; Tongkobpetch, S.; Ludin, K.; Barnes, A.M.; Yeetong, P.; Weis, M.; Krabichler, B.; Srichomthong, C.; et al. MBTPS2 mutations cause defective regulated intramembrane proteolysis in X-linked osteogenesis imperfecta. Nat. Commun. 2016, 7, 11920. [Google Scholar] [CrossRef] [PubMed]
- Moosa, S.; Yamamoto, G.L.; Garbes, L.; Keupp, K.; Beleza-Meireles, A.; Moreno, C.A.; Valadares, E.R.; de Sousa, S.B.; Maia, S.; Saraiva, J.; et al. Autosomal-Recessive Mutations in MESD Cause Osteogenesis Imperfecta. Am. J. Hum. Genet. 2019, 105, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Garbes, L.; Kim, K.; Riess, A.; Hoyer-Kuhn, H.; Beleggia, F.; Bevot, A.; Kim, M.J.; Huh, Y.H.; Kweon, H.S.; Savarirayan, R.; et al. Mutations in SEC24D, encoding a component of the COPII machinery, cause a syndromic form of osteogenesis imperfecta. Am. J. Hum. Genet. 2015, 96, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Dubail, J.; Brunelle, P.; Baujat, G.; Huber, C.; Doyard, M.; Michot, C.; Chavassieux, P.; Khairouni, A.; Topouchian, V.; Monnot, S.; et al. Homozygous Loss-of-Function Mutations in CCDC134 Are Responsible for a Severe Form of Osteogenesis Imperfecta. J. Bone Miner. Res. 2020, 35, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, D.; Zheng, W.; Wang, O.; Jiang, Y.; Xia, W.; Xing, X.; Li, M. A novel missense mutation in P4HB causes mild osteogenesis imperfecta. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Ha-Vinh, R.; Alanay, Y.; Bank, R.A.; Campos-Xavier, A.B.; Zankl, A.; Superti-Furga, A.; Bonafe, L. Phenotypic and molecular characterization of Bruck syndrome (osteogenesis imperfecta with contractures of the large joints) caused by a recessive mutation in PLOD2. Am. J. Med. Genet. A 2004, 131, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wu, H.; Zhang, C.; Feng, J.; Chen, L.; Xie, R.; Wang, F.; Chen, X.; Zhou, H.; Sun, H.; et al. Clinical, Genetics, and Bioinformatic Characterization of Mutations Affecting an Essential Region of PLS3 in Patients with BMND18. Int. J. Endocrinol. 2018, 2018, 8953217. [Google Scholar] [CrossRef]
- Van Dijk, F.S.; Semler, O.; Etich, J.; Kohler, A.; Jimenez-Estrada, J.A.; Bravenboer, N.; Claeys, L.; Riesebos, E.; Gegic, S.; Piersma, S.R.; et al. Interaction between KDELR2 and HSP47 as a Key Determinant in Osteogenesis Imperfecta Caused by Bi-allelic Variants in KDELR2. Am. J. Hum. Genet. 2020, 107, 989–999. [Google Scholar] [CrossRef]
- Sillence, D.O.; Senn, A.; Danks, D.M. Genetic heterogeneity in osteogenesis imperfecta. J. Med. Genet. 1979, 16, 101–116. [Google Scholar] [CrossRef]
- Warman, M.L.; Cormier-Daire, V.; Hall, C.; Krakow, D.; Lachman, R.; LeMerrer, M.; Mortier, G.; Mundlos, S.; Nishimura, G.; Rimoin, D.L.; et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am. J. Med. Genet. A 2011, 155A, 943–968. [Google Scholar] [CrossRef]
- Manogari, C.; Imaan Amina, R.; Peter, B. The Evolution of the Nosology of Osteogenesis Imperfecta. Clin. Genet. 2021, 99, 42–52. [Google Scholar]
- Marini, J.C.; Forlino, A.; Bachinger, H.P.; Bishop, N.J.; Byers, P.H.; Paepe, A.; Fassier, F.; Fratzl-Zelman, N.; Kozloff, K.M.; Krakow, D.; et al. Osteogenesis imperfecta. Nat. Rev. Dis Primers 2017, 3, 17052. [Google Scholar] [CrossRef] [PubMed]
- Alten, E.D.; Chaturvedi, A.; Cullimore, M.; Fallon, A.A.; Habben, L.; Hughes, I.; O’Malley, N.T.; Rahimi, H.; Renodin-Mead, D.; Schmidt, B.L.; et al. No longer a historical ailment: Two cases of childhood scurvy with recommendations for bone health providers. Osteoporos Int. 2020, 31, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Pozzer, D.; Invernizzi, R.W.; Blaauw, B.; Cantoni, O.; Zito, E. Ascorbic Acid Route to the Endoplasmic Reticulum: Function and Role in Disease. Antioxid Redox Signal. 2020. [Google Scholar] [CrossRef] [PubMed]
- Webb, E.A.; Balasubramanian, M.; Fratzl-Zelman, N.; Cabral, W.A.; Titheradge, H.; Alsaedi, A.; Saraff, V.; Vogt, J.; Cole, T.; Stewart, S.; et al. Phenotypic Spectrum in Osteogenesis Imperfecta Due to Mutations in TMEM38B: Unraveling a Complex Cellular Defect. J. Clin. Endocrinol. Metab. 2017, 102, 2019–2028. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Ito, S.; Nagata, K.; Sakai, L.Y.; Bachinger, H.P. Intracellular mechanisms of molecular recognition and sorting for transport of large extracellular matrix molecules. Proc. Natl. Acad. Sci. USA 2016, 113, E6036–E6044. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J.; Sieron, A.L.; Li, S.W. Procollagen N-proteinase and procollagen C-proteinase. Two unusual metalloproteinases that are essential for procollagen processing probably have important roles in development and cell signaling. Matrix Biol. 1998, 16, 399–408. [Google Scholar] [CrossRef]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef]
- Hohenester, E.; Sasaki, T.; Giudici, C.; Farndale, R.W.; Bachinger, H.P. Structural basis of sequence-specific collagen recognition by SPARC. Proc. Natl. Acad. Sci. USA 2008, 105, 18273–18277. [Google Scholar] [CrossRef]
- Dwan, K.; Phillipi, C.A.; Steiner, R.D.; Basel, D. Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database Syst. Rev. 2016, 10, CD005088. [Google Scholar] [CrossRef]
- Ralston, S.H.; Gaston, M.S. Management of Osteogenesis Imperfecta. Front. Endocrinol. 2019, 10, 924. [Google Scholar] [CrossRef] [PubMed]
- Marini, J.C.; Dang Do, A.N. Osteogenesis Imperfecta. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., de Herder, W.W., Dungan, K., Grossman, A., Hershman, J.M., Hofland, H.J., Kaltsas, G., et al., Eds.; MDText.xom, Inc.: South Dartmouth, MA, USA, 2020. [Google Scholar]
- Gotherstrom, C.; Walther-Jallow, L. Stem Cell Therapy as a Treatment for Osteogenesis Imperfecta. Curr. Osteoporos Rep. 2020, 18, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.; Kneissel, M. WNT signaling in bone homeostasis and disease: From human mutations to treatments. Nat. Med. 2013, 19, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Laine, C.M.; Joeng, K.S.; Campeau, P.M.; Kiviranta, R.; Tarkkonen, K.; Grover, M.; Lu, J.T.; Pekkinen, M.; Wessman, M.; Heino, T.J.; et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N. Engl. J. Med. 2013, 368, 1809–1816. [Google Scholar] [CrossRef]
- Westendorf, J.J.; Kahler, R.A.; Schroeder, T.M. Wnt signaling in osteoblasts and bone diseases. Gene 2004, 341, 19–39. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Makitie, R.E.; Kampe, A.J.; Taylan, F.; Makitie, O. Recent Discoveries in Monogenic Disorders of Childhood Bone Fragility. Curr. Osteoporos Rep. 2017, 15, 303–310. [Google Scholar] [CrossRef]
- Joeng, K.S.; Lee, Y.C.; Lim, J.; Chen, Y.; Jiang, M.M.; Munivez, E.; Ambrose, C.; Lee, B.H. Osteocyte-specific WNT1 regulates osteoblast function during bone homeostasis. J. Clin. Investig. 2017, 127, 2678–2688. [Google Scholar] [CrossRef]
- Thomas, K.R.; Musci, T.S.; Neumann, P.E.; Capecchi, M.R. Swaying is a mutant allele of the proto-oncogene Wnt-1. Cell 1991, 67, 969–976. [Google Scholar] [CrossRef]
- Joeng, K.S.; Lee, Y.C.; Jiang, M.M.; Bertin, T.K.; Chen, Y.; Abraham, A.M.; Ding, H.; Bi, X.; Ambrose, C.G.; Lee, B.H. The swaying mouse as a model of osteogenesis imperfecta caused by WNT1 mutations. Hum. Mol. Genet. 2014, 23, 4035–4042. [Google Scholar] [CrossRef] [PubMed]
- Day, T.F.; Guo, X.; Garrett-Beal, L.; Yang, Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 2005, 8, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Kramer, I.; Halleux, C.; Keller, H.; Pegurri, M.; Gooi, J.H.; Weber, P.B.; Feng, J.Q.; Bonewald, L.F.; Kneissel, M. Osteocyte Wnt/beta-catenin signaling is required for normal bone homeostasis. Mol. Cell Biol. 2010, 30, 3071–3085. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Slee, R.B.; Fukai, N.; Rawadi, G.; Roman-Roman, S.; Reginato, A.M.; Wang, H.; Cundy, T.; Glorieux, F.H.; Lev, D.; et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001, 107, 513–523. [Google Scholar] [CrossRef]
- Korvala, J.; Juppner, H.; Makitie, O.; Sochett, E.; Schnabel, D.; Mora, S.; Bartels, C.F.; Warman, M.L.; Deraska, D.; Cole, W.G.; et al. Mutations in LRP5 cause primary osteoporosis without features of OI by reducing Wnt signaling activity. BMC Med. Genet. 2012, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Boyden, L.M.; Mao, J.; Belsky, J.; Mitzner, L.; Farhi, A.; Mitnick, M.A.; Wu, D.; Insogna, K.; Lifton, R.P. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002, 346, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Kaveh, S.; Hosseinifard, H.; Ghadimi, N.; Vojdanian, M.; Aryankhesal, A. Efficacy and safety of Romosozumab in treatment for low bone mineral density: A systematic review and meta-analysis. Clin. Rheumatol. 2020, 39, 3261–3276. [Google Scholar] [CrossRef]
- Van Bezooijen, R.L.; ten Dijke, P.; Papapoulos, S.E.; Lowik, C.W. SOST/sclerostin, an osteocyte-derived negative regulator of bone formation. Cytokine Growth Factor Rev. 2005, 16, 319–327. [Google Scholar] [CrossRef]
- Balemans, W.; Ebeling, M.; Patel, N.; Van Hul, E.; Olson, P.; Dioszegi, M.; Lacza, C.; Wuyts, W.; Van Den Ende, J.; Willems, P.; et al. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum. Mol. Genet. 2001, 10, 537–543. [Google Scholar] [CrossRef]
- Delgado-Calle, J.; Sato, A.Y.; Bellido, T. Role and mechanism of action of sclerostin in bone. Bone 2017, 96, 29–37. [Google Scholar] [CrossRef]
- Markham, A. Romosozumab: First Global Approval. Drugs 2019, 79, 471–476. [Google Scholar] [CrossRef] [PubMed]
- McClung, M.R. Sclerostin antibodies in osteoporosis: Latest evidence and therapeutic potential. Ther. Adv. Musculoskelet Dis. 2017, 9, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Tauer, J.T.; Robinson, M.E.; Rauch, F. Osteogenesis Imperfecta: New Perspectives From Clinical and Translational Research. JBMR Plus 2019, 3, e10174. [Google Scholar] [CrossRef] [PubMed]
- Gaudio, A.; Fiore, V.; Rapisarda, R.; Sidoti, M.H.; Xourafa, A.; Catalano, A.; Tringali, G.; Zanoli, L.; Signorelli, S.S.; Fiore, C.E. Sclerostin is a possible candidate marker of arterial stiffness: Results from a cohort study in Catania. Mol. Med. Rep. 2017, 15, 3420–3424. [Google Scholar] [CrossRef] [PubMed]
- Van Hul, W.; Boudin, E.; Vanhoenacker, F.M.; Mortier, G. Camurati-Engelmann Disease. Calcif. Tissue Int. 2019, 104, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, A.; Alaerts, M.; Van Laer, L.; Loeys, B. Marfan Syndrome and Related Disorders: 25 Years of Gene Discovery. Hum. Mutat. 2016, 37, 524–531. [Google Scholar] [CrossRef]
- Tan, E.W.; Offoha, R.U.; Oswald, G.L.; Skolasky, R.L.; Dewan, A.K.; Zhen, G.; Shapiro, J.R.; Dietz, H.C.; Cao, X.; Sponseller, P.D. Increased fracture risk and low bone mineral density in patients with loeys-dietz syndrome. Am. J. Med. Genet. A 2013, 161A, 1910–1914. [Google Scholar] [CrossRef] [PubMed]
- Grafe, I.; Yang, T.; Alexander, S.; Homan, E.P.; Lietman, C.; Jiang, M.M.; Bertin, T.; Munivez, E.; Chen, Y.; Dawson, B.; et al. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nat. Med. 2014, 20, 670–675. [Google Scholar] [CrossRef]
- Tauer, J.T.; Abdullah, S.; Rauch, F. Effect of Anti-TGF-beta Treatment in a Mouse Model of Severe Osteogenesis Imperfecta. J. Bone Miner. Res. 2019, 34, 207–214. [Google Scholar] [CrossRef]
- Mo, C.; Ke, J.; Zhao, D.; Zhang, B. Role of the renin-angiotensin-aldosterone system in bone metabolism. J. Bone Miner. Metab. 2020, 38, 772–779. [Google Scholar] [CrossRef]
- Ayyavoo, A.; Derraik, J.G.; Cutfield, W.S.; Hofman, P.L. Elimination of pain and improvement of exercise capacity in Camurati-Engelmann disease with losartan. J. Clin. Endocrinol. Metab. 2014, 99, 3978–3982. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.; Cronin, S.J.F.; Penninger, J.M. Targeting the RANKL/RANK/OPG Axis for Cancer Therapy. Front. Oncol. 2020, 10, 1283. [Google Scholar] [CrossRef] [PubMed]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Whyte, M.P.; Obrecht, S.E.; Finnegan, P.M.; Jones, J.L.; Podgornik, M.N.; McAlister, W.H.; Mumm, S. Osteoprotegerin deficiency and juvenile Paget’s disease. N. Engl. J. Med. 2002, 347, 175–184. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Cundy, T.; Mantzoros, C.S. Juvenile Paget disease. Metabolism 2018, 80, 15–26. [Google Scholar] [CrossRef]
- Grasemann, C.; Unger, N.; Hovel, M.; Arweiler-Harbeck, D.; Herrmann, R.; Schundeln, M.M.; Muller, O.; Schweiger, B.; Lausch, E.; Meissner, T.; et al. Loss of Functional Osteoprotegerin: More Than a Skeletal Problem. J. Clin. Endocrinol. Metab. 2017, 102, 210–219. [Google Scholar] [CrossRef]
- Caffey, J. Familial hyperphosphatasemia with ateliosis and hypermetabolism of growing membranous bone; review of the clinical, radiographic and chemical features. Bull. Hosp. Joint Dis. 1972, 33, 81–110. [Google Scholar]
- Golob, D.S.; McAlister, W.H.; Mills, B.G.; Fedde, K.N.; Reinus, W.R.; Teitelbaum, S.L.; Beeki, S.; Whyte, M.P. Juvenile Paget disease: Life-long features of a mildly affected young woman. J. Bone Miner. Res. 1996, 11, 132–142. [Google Scholar] [CrossRef]
- Hughes, A.E.; Ralston, S.H.; Marken, J.; Bell, C.; MacPherson, H.; Wallace, R.G.; van Hul, W.; Whyte, M.P.; Nakatsuka, K.; Hovy, L.; et al. Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat. Genet. 2000, 24, 45–48. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Singhellakis, P.N.; Naot, D.; Adamidou, F.; Malandrinou, F.C.; Anastasilakis, A.D.; Polymerou, V.; Kita, M. Denosumab treatment for juvenile Paget's disease: Results from two adult patients with osteoprotegerin deficiency (“Balkan” mutation in the TNFRSF11B gene). J. Clin. Endocrinol. Metab. 2014, 99, 703–707. [Google Scholar] [CrossRef][Green Version]
- Grasemann, C.; Schundeln, M.M.; Hovel, M.; Schweiger, B.; Bergmann, C.; Herrmann, R.; Wieczorek, D.; Zabel, B.; Wieland, R.; Hauffa, B.P. Effects of RANK-ligand antibody (denosumab) treatment on bone turnover markers in a girl with juvenile Paget’s disease. J. Clin. Endocrinol. Metab. 2013, 98, 3121–3126. [Google Scholar] [CrossRef] [PubMed]
- Yayan, J. Denosumab for Effective Tumor Size Reduction in Patients With Giant Cell Tumors of the Bone: A Systematic Review and Meta-Analysis. Cancer Control. 2020, 27, 1073274820934822. [Google Scholar] [CrossRef] [PubMed]
- Uday, S.; Gaston, C.L.; Rogers, L.; Parry, M.; Joffe, J.; Pearson, J.; Sutton, D.; Grimer, R.; Hogler, W. Osteonecrosis of the Jaw and Rebound Hypercalcemia in Young People Treated With Denosumab for Giant Cell Tumor of Bone. J. Clin. Endocrinol. Metab. 2018, 103, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Anastasilakis, A.D.; Toulis, K.A.; Polyzos, S.A.; Terpos, E. RANKL inhibition for the management of patients with benign metabolic bone disorders. Expert. Opin. Investig. Drugs 2009, 18, 1085–1102. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, S.A.; Makras, P.; Tournis, S.; Anastasilakis, A.D. Off-label uses of denosumab in metabolic bone diseases. Bone 2019, 129, 115048. [Google Scholar] [CrossRef] [PubMed]
- Hilton, M.J.; Tu, X.; Wu, X.; Bai, S.; Zhao, H.; Kobayashi, T.; Kronenberg, H.M.; Teitelbaum, S.L.; Ross, F.P.; Kopan, R.; et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008, 14, 306–314. [Google Scholar] [CrossRef]
- Fukushima, H.; Nakao, A.; Okamoto, F.; Shin, M.; Kajiya, H.; Sakano, S.; Bigas, A.; Jimi, E.; Okabe, K. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol. Cell Biol. 2008, 28, 6402–6412. [Google Scholar] [CrossRef]
- Simpson, M.A.; Irving, M.D.; Asilmaz, E.; Gray, M.J.; Dafou, D.; Elmslie, F.V.; Mansour, S.; Holder, S.E.; Brain, C.E.; Burton, B.K.; et al. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 2011, 43, 303–305. [Google Scholar] [CrossRef]
- Hamada, Y.; Kadokawa, Y.; Okabe, M.; Ikawa, M.; Coleman, J.R.; Tsujimoto, Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 1999, 126, 3415–3424. [Google Scholar]
- Canalis, E. Clinical and experimental aspects of notch receptor signaling: Hajdu-Cheney syndrome and related disorders. Metabolism 2018, 80, 48–56. [Google Scholar] [CrossRef]
- Narumi, Y.; Min, B.J.; Shimizu, K.; Kazukawa, I.; Sameshima, K.; Nakamura, K.; Kosho, T.; Rhee, Y.; Chung, Y.S.; Kim, O.H.; et al. Clinical consequences in truncating mutations in exon 34 of NOTCH2: Report of six patients with Hajdu-Cheney syndrome and a patient with serpentine fibula polycystic kidney syndrome. Am. J. Med. Genet. A 2013, 161A, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Descartes, M.; Rojnueangnit, K.; Cole, L.; Sutton, A.; Morgan, S.L.; Patry, L.; Samuels, M.E. Hajdu-Cheney syndrome: Phenotypical progression with de-novo NOTCH2 mutation. Clin. Dysmorphol. 2014, 23, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Isidor, B.; Lindenbaum, P.; Pichon, O.; Bezieau, S.; Dina, C.; Jacquemont, S.; Martin-Coignard, D.; Thauvin-Robinet, C.; Le Merrer, M.; Mandel, J.L.; et al. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 2011, 43, 306–308. [Google Scholar] [CrossRef]
- Sakka, S.; Gafni, R.I.; Davies, J.H.; Clarke, B.; Tebben, P.; Samuels, M.; Saraff, V.; Klaushofer, K.; Fratzl-Zelman, N.; Roschger, P.; et al. Bone Structural Characteristics and Response to Bisphosphonate Treatment in Children With Hajdu-Cheney Syndrome. J. Clin. Endocrinol. Metab. 2017, 102, 4163–4172. [Google Scholar] [CrossRef] [PubMed]
- Binkley, T.; Johnson, J.; Vogel, L.; Kecskemethy, H.; Henderson, R.; Specker, B. Bone measurements by peripheral quantitative computed tomography (pQCT) in children with cerebral palsy. J. Pediatr. 2005, 147, 791–796. [Google Scholar] [CrossRef]
- Verbunt, J.A.; Seelen, H.A.; Vlaeyen, J.W.; van de Heijden, G.J.; Heuts, P.H.; Pons, K.; Knottnerus, J.A. Disuse and deconditioning in chronic low back pain: Concepts and hypotheses on contributing mechanisms. Eur. J. Pain. 2003, 7, 9–21. [Google Scholar] [CrossRef]
- Buenzli, P.R.; Sims, N.A. Quantifying the osteocyte network in the human skeleton. Bone 2015, 75, 144–150. [Google Scholar] [CrossRef]
- Bonewald, L.F. Osteocytes. In Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism, 9th ed.; American Society for Bone and Mineral Research: Washington, DC, USA, 2018; pp. 38–45. [Google Scholar] [CrossRef]
- Bacabac, R.G.; Smit, T.H.; Van Loon, J.J.; Doulabi, B.Z.; Helder, M.; Klein-Nulend, J. Bone cell responses to high-frequency vibration stress: Does the nucleus oscillate within the cytoplasm? FASEB J. 2006, 20, 858–864. [Google Scholar] [CrossRef]
- You, L.D.; Weinbaum, S.; Cowin, S.C.; Schaffler, M.B. Ultrastructure of the osteocyte process and its pericellular matrix. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004, 278, 505–513. [Google Scholar] [CrossRef]
- Plotkin, L.I.; Mathov, I.; Aguirre, J.I.; Parfitt, A.M.; Manolagas, S.C.; Bellido, T. Mechanical stimulation prevents osteocyte apoptosis: Requirement of integrins, Src kinases, and ERKs. Am. J. Physiol. Cell. Physiol. 2005, 289, C633–C643. [Google Scholar] [CrossRef]
- Aguirre, J.I.; Plotkin, L.I.; Stewart, S.A.; Weinstein, R.S.; Parfitt, A.M.; Manolagas, S.C.; Bellido, T. Osteocyte apoptosis is induced by weightlessness in mice and precedes osteoclast recruitment and bone loss. J. Bone Miner. Res. 2006, 21, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Bakker, A.; Klein-Nulend, J.; Burger, E. Shear stress inhibits while disuse promotes osteocyte apoptosis. Biochem. Biophys. Res. Commun. 2004, 320, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Dallas, S.L.; Prideaux, M.; Bonewald, L.F. The osteocyte: An endocrine cell … and more. Endocr. Rev. 2013, 34, 658–690. [Google Scholar] [CrossRef]
- Burger, E.H.; Klein-Nulend, J. Microgravity and bone cell mechanosensitivity. Bone 1998, 22 (Suppl. 5), 127S–130S. [Google Scholar] [CrossRef]
- Gaudio, A.; Pennisi, P.; Bratengeier, C.; Torrisi, V.; Lindner, B.; Mangiafico, R.A.; Pulvirenti, I.; Hawa, G.; Tringali, G.; Fiore, C.E. Increased sclerostin serum levels associated with bone formation and resorption markers in patients with immobilization-induced bone loss. J. Clin. Endocrinol. Metab. 2010, 95, 2248–2253. [Google Scholar] [CrossRef]
- Zhou, M.; Li, S.; Pathak, J.L. Pro-inflammatory Cytokines and Osteocytes. Curr. Osteoporos Rep. 2019, 17, 97–104. [Google Scholar] [CrossRef]
- Marin-Cascales, E.; Alcaraz, P.E.; Ramos-Campo, D.J.; Martinez-Rodriguez, A.; Chung, L.H.; Rubio-Arias, J.A. Whole-body vibration training and bone health in postmenopausal women: A systematic review and meta-analysis. Medicine 2018, 97, e11918. [Google Scholar] [CrossRef]
- Pathak, J.L.; Bravenboer, N.; Klein-Nulend, J. The Osteocyte as the New Discovery of Therapeutic Options in Rare Bone Diseases. Front. Endocrinol. 2020, 11, 405. [Google Scholar] [CrossRef]
- Van Dyne, S.; Holers, V.M.; Lublin, D.M.; Atkinson, J.P. The polymorphism of the C3b/C4b receptor in the normal population and in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 1987, 68, 570–579. [Google Scholar]
- Klein-Nulend, J.; van Oers, R.F.; Bakker, A.D.; Bacabac, R.G. Nitric oxide signaling in mechanical adaptation of bone. Osteoporos Int. 2014, 25, 1427–1437. [Google Scholar] [CrossRef]
- Ajubi, N.E.; Klein-Nulend, J.; Alblas, M.J.; Burger, E.H.; Nijweide, P.J. Signal transduction pathways involved in fluid flow-induced PGE2 production by cultured osteocytes. Am. J. Physiol. 1999, 276, E171–E178. [Google Scholar] [CrossRef] [PubMed]
- Lean, J.M.; Jagger, C.J.; Chambers, T.J.; Chow, J.W. Increased insulin-like growth factor I mRNA expression in rat osteocytes in response to mechanical stimulation. Am. J. Physiol. 1995, 268 Pt 2, E318–E327. [Google Scholar] [CrossRef] [PubMed]
- Kamel, M.A.; Picconi, J.L.; Lara-Castillo, N.; Johnson, M.L. Activation of beta-catenin signaling in MLO-Y4 osteocytic cells versus 2T3 osteoblastic cells by fluid flow shear stress and PGE2: Implications for the study of mechanosensation in bone. Bone 2010, 47, 872–881. [Google Scholar] [CrossRef]
- Ajubi, N.E.; Klein-Nulend, J.; Nijweide, P.J.; Vrijheid-Lammers, T.; Alblas, M.J.; Burger, E.H. Pulsating fluid flow increases prostaglandin production by cultured chicken osteocytes—A cytoskeleton-dependent process. Biochem. Biophys. Res. Commun. 1996, 225, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.C.; el-Haj, A.J.; Minter, S.L.; Tavares, I.A.; Bennett, A.; Lanyon, L.E. Loading-related increases in prostaglandin production in cores of adult canine cancellous bone in vitro: A role for prostacyclin in adaptive bone remodeling? J. Bone Miner. Res. 1991, 6, 1345–1351. [Google Scholar] [CrossRef]
- Kitase, Y.; Barragan, L.; Qing, H.; Kondoh, S.; Jiang, J.X.; Johnson, M.L.; Bonewald, L.F. Mechanical induction of PGE2 in osteocytes blocks glucocorticoid-induced apoptosis through both the beta-catenin and PKA pathways. J. Bone Miner. Res. 2010, 25, 2657–2668. [Google Scholar] [CrossRef]
- Gyori, D.S.; Mocsai, A. Osteoclast Signal Transduction During Bone Metastasis Formation. Front. Cell Dev. Biol. 2020, 8, 507. [Google Scholar] [CrossRef]
- Lacey, D.L.; Timms, E.; Tan, H.L.; Kelley, M.J.; Dunstan, C.R.; Burgess, T.; Elliott, R.; Colombero, A.; Elliott, G.; Scully, S.; et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 1998, 93, 165–176. [Google Scholar] [CrossRef]
- Kamalakar, A.; Washam, C.L.; Akel, N.S.; Allen, B.J.; Williams, D.K.; Swain, F.L.; Leitzel, K.; Lipton, A.; Gaddy, D.; Suva, L.J. PTHrP(12–48) Modulates the Bone Marrow Microenvironment and Suppresses Human Osteoclast Differentiation and Lifespan. J. Bone Miner. Res. 2017, 32, 1421–1431. [Google Scholar] [CrossRef]
- Kanazawa, K.; Kudo, A. Self-assembled RANK induces osteoclastogenesis ligand-independently. J. Bone Miner. Res. 2005, 20, 2053–2060. [Google Scholar] [CrossRef]
- Yasuda, H.; Shima, N.; Nakagawa, N.; Mochizuki, S.I.; Yano, K.; Fujise, N.; Sato, Y.; Goto, M.; Yamaguchi, K.; Kuriyama, M.; et al. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): A mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology 1998, 139, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Riggs, B.L.; Khosla, S.; Melton, L.J., 3rd. A unitary model for involutional osteoporosis: Estrogen deficiency causes both type I and type II osteoporosis in postmenopausal women and contributes to bone loss in aging men. J. Bone Miner. Res. 1998, 13, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Yang, Z.; Ma, Y.; Yue, Z.; Lin, H.; Qu, G.; Huang, J.; Dai, W.; Li, C.; Zheng, C.; et al. LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat. Med. 2016, 22, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Suva, L.J.; Winslow, G.A.; Wettenhall, R.E.; Hammonds, R.G.; Moseley, J.M.; Diefenbach-Jagger, H.; Rodda, C.P.; Kemp, B.E.; Rodriguez, H.; Chen, E.Y.; et al. A parathyroid hormone-related protein implicated in malignant hypercalcemia: Cloning and expression. Science 1987, 237, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Mudano, A.; Allison, J.; Hill, J.; Rothermel, T.; Saag, K. Variations in glucocorticoid induced osteoporosis prevention in a managed care cohort. J. Rheumatol. 2001, 28, 1298–1305. [Google Scholar]
- Overman, R.A.; Yeh, J.Y.; Deal, C.L. Prevalence of oral glucocorticoid usage in the United States: A general population perspective. Arthritis Care Res. 2013, 65, 294–298. [Google Scholar] [CrossRef]
- Coskun Benlidayi, I. Denosumab in the treatment of glucocorticoid-induced osteoporosis. Rheumatol. Int. 2018, 38, 1975–1984. [Google Scholar] [CrossRef]
- Kim, D.; Cho, S.K.; Park, B.; Jang, E.J.; Bae, S.C.; Sung, Y.K. Glucocorticoids Are Associated with an Increased Risk for Vertebral Fracture in Patients with Rheumatoid Arthritis. J. Rheumatol. 2018, 45, 612–620. [Google Scholar] [CrossRef]
- Adami, G.; Saag, K.G. Glucocorticoid-induced osteoporosis update. Curr. Opin. Rheumatol. 2019, 31, 388–393. [Google Scholar] [CrossRef]
- Adami, G.; Rahn, E.J.; Saag, K.G. Glucocorticoid-induced osteoporosis: From clinical trials to clinical practice. Ther. Adv. Musculoskelet Dis. 2019, 11, 1759720X19876468. [Google Scholar] [CrossRef]
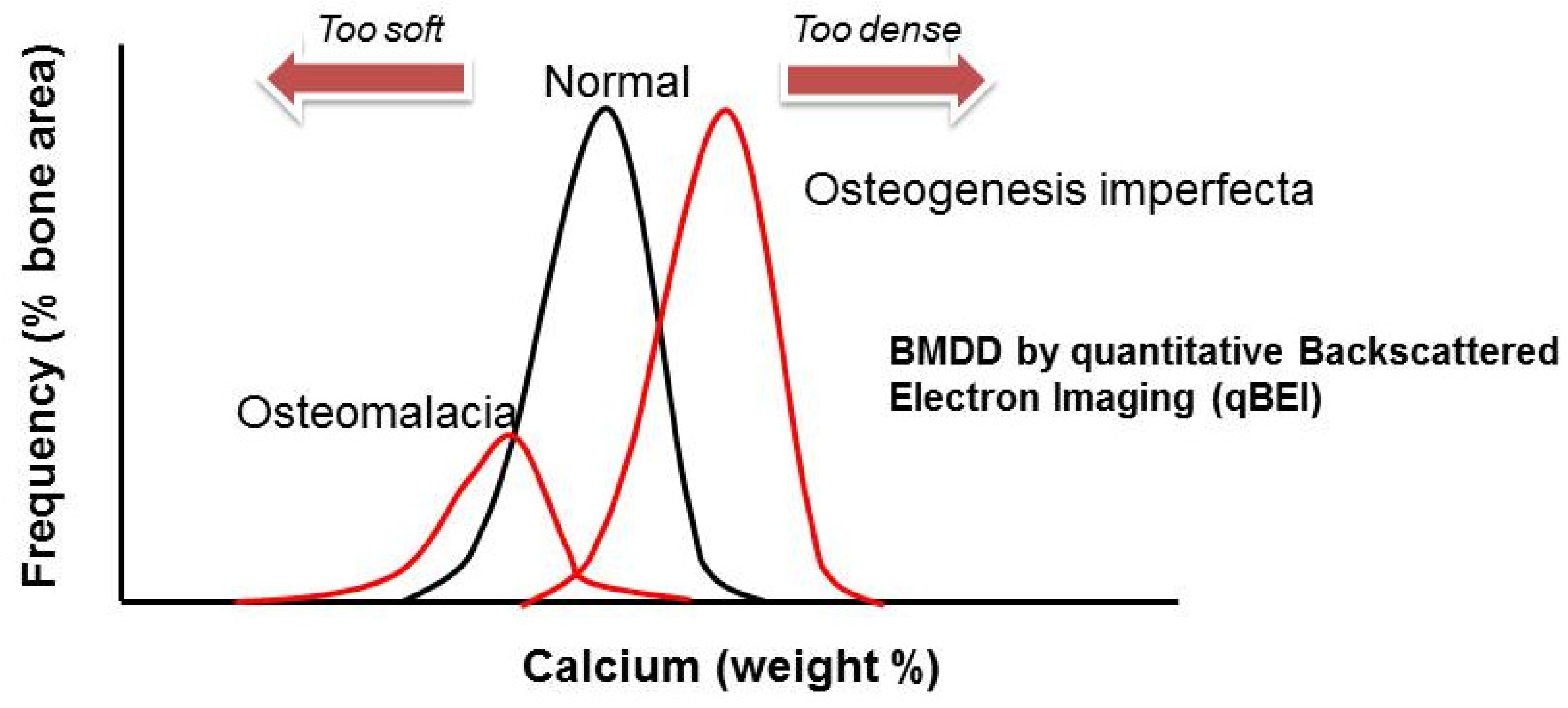
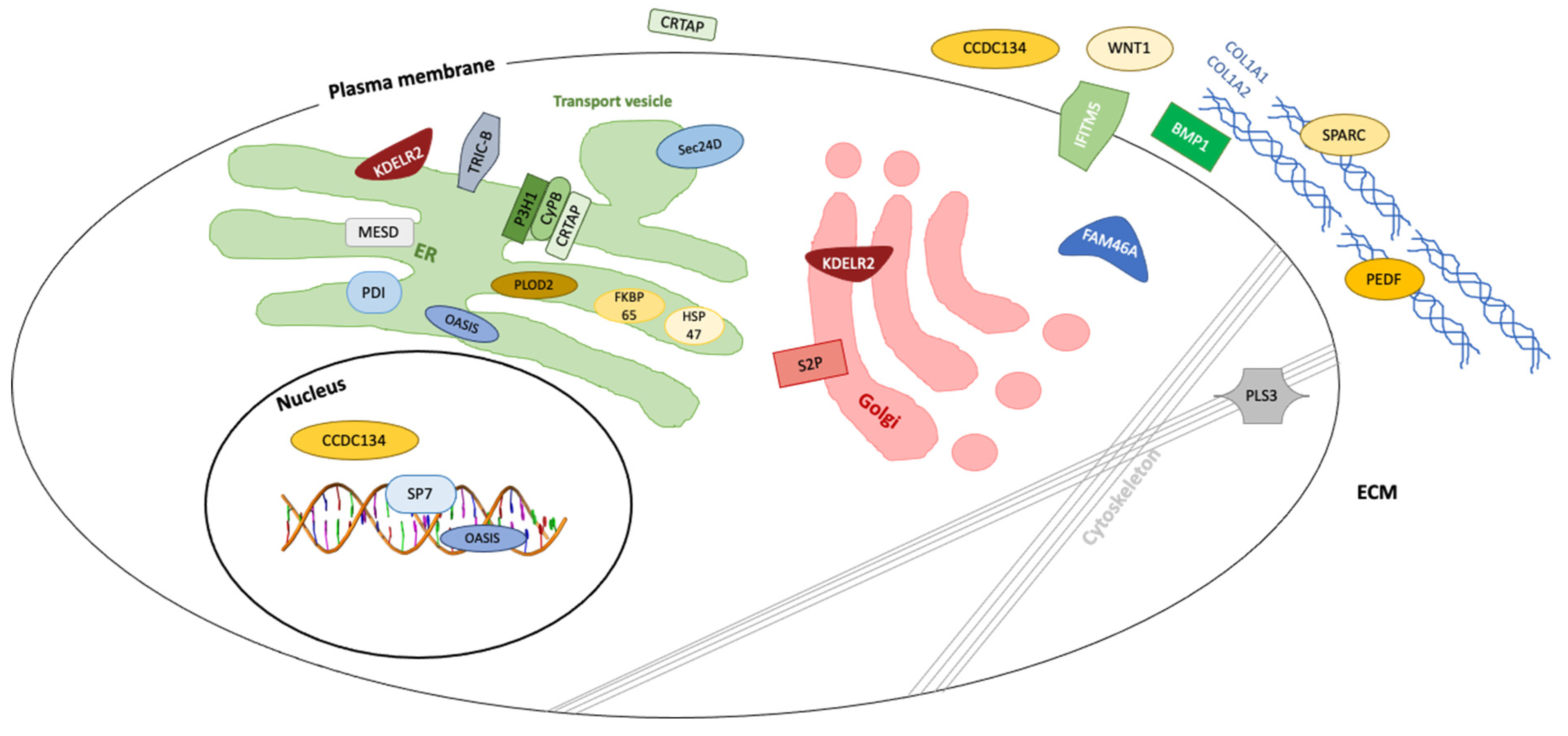
| Condition | OMIM | Inheritance | Gene | Mutation | Protein | Bone Pathway | Symptoms |
|---|---|---|---|---|---|---|---|
| Osteogenesis imperfecta and primary osteoporosis | 166200 | AD | COL1A1 COL1A2 | Loss of function | Collagen α1(I) chain Collagen α2(I) chain | Collagen synthesis | OI 1 (clinical type I, mild) |
| 166210 | OI 2 (clinical type II, perinatal lethal) | ||||||
| 259420 | OI 3 (clinical type III, severe) | ||||||
| 166220 | OI 4 (clinical type IV, moderate) | ||||||
| 610967 | AD | IFITM5 | Gain of function | Interferon-induced Transmembrane protein 5 (BRIL) | Mineralization | OI 5 (clinical types V; and III in atypical OI 6) | |
| 613982 | AR | SERPINF 1 | Loss of function | Pigment epithelium-derived factor (PEDF) | Mineralization | OI 6 (clinical type III) | |
| 610854 | AR | CRTAP | Loss of function | Cartilage-associated protein (CRTAP) | Collagen modification | OI 7 (clinical types II, III, IV) | |
| 610915 | AR | LEPRE1 (P3H1) | Loss of function | Leucine proline enriched proteoglycan1/Prolyl 3-hydroxylase 1 (P3H1) | Collagen modification | OI 8 (clinical types II, III) | |
| 259440 | AR | PPIB | Loss of function | Cyclophilin B (CyPB) | Collagen modification | OI 9 (clinical types II, III) | |
| 613848 | AR | SERPINH1 | Loss of function | Serpin peptidase inhibitor, clade H, member 1/heat shock protein 47 | Collagen folding and cross-linking | OI 10 (clinical type III) | |
| 610968 | AR | FKBP10 | Loss of function | Peptidyl-prolyl cis-transisomerase FKBP10 | Collagen folding and cross-linking | OI 11 (clinical types III, IV) | |
| 259450 | AR | Bruck Syndrome Type 1 (BS1) | |||||
| 613849 | AR | SP7 | Loss of function | Zinc-finger transcription factor, Osterix | Osteoblast differentiation and maturation | OI 12 (clinical type IV) | |
| 112264 | AR | BMP1 | Loss of function | Bone morphogenic protein1/procollagen C proteinase | Collagen processing | OI 13 (clinical Type III) | |
| 615066 | AR | TMEM38B | Loss of function | Trimeric intracellular cation channel B (TRIC-B) | ER calcium flux | OI 14 (clinical type I, III, IV) | |
| 615220 | AR | WNT1 | Loss of function | Wingless-type MMTV integration site family, member 1 | WNT signaling | OI 15 (clinical type III, IV) | |
| AD | Primary osteoporosis | ||||||
| 616229 | AR | CREB3L1 | Loss of function | Old astrocyte specifically induced substance (OASIS) | ER UPR response, ER-Golgi trafficking | OI 16 (clinical type III) | |
| AD | OI 16 (clinical type I) | ||||||
| 616507 | AR | SPARC | Loss of function | Secreted protein, acidic, cysteine-rich (SPARC, or osteonectin) | Procollagen processing and extracellular assembly | OI 17 (clinical type III, IV) | |
| 617952 | AR | TENT5A (FAM46A) | Loss of function | Terminal nucleotidyltransferase 46, Member A (FAM46A) | BMP signaling | OI 18 (clinical type III), overlap with Stuve-Wiedemann syndrome | |
| 601559 | |||||||
| 301014 | XR | MBTPS2 | Loss of function | Site 2 protease (S2P) | Golgi Regulated intramembrane proteolysis | OI 19 (clinical type III, IV) | |
| 607782 | AR | MESD | Loss of function | Mesoderm development LRP chaperon | WNT signaling | OI 20 (clinical type III) | |
| 607186 | AR | SEC24D | Loss of function | SEC24D | ER COPII Transport of procollagen | OI (clinical type III), overlap with | |
| Cole-Carpenter Syndrome 2 | |||||||
| 618788 | AR | CCDC134 | Loss of function | Coiled-coil domain containing 134 | MAPK pathway | OI (clinical type III) | |
| 609024 | AR | KDELR2 | Loss of function | KDEL endoplasmic reticulum protein retention receptor 2 | Regulate the trafficking of proteins between the Golgi apparatus and the ER | OI (clinical type IIB/III) | |
| Other Primary Osteoporosis | 259770 | AR | LRP5 | Loss of function | Low density lipoprotein receptor 5 (LRP5) | WNT signaling | Osteoporosis pseudoglioma syndrome |
| 166710 | AD | Primary osteoporosis | |||||
| 300910 | XL | PLS3 | Loss of function | Plastin 3 | Formation of F-actin bundles | Primary osteoporosis | |
| 609220 | AR | PLOD2 | Loss of function | Telopeptide lysyl hydroxylase | Collagen crosslinking | Bruck Syndrome 2 (BS2) | |
| 126550 | AD | SGMS2 | Loss of function | Phosphatidylcholine:ceramide cholinephosphotransferase 2 | Mineralization | Calvarial doughnut lesions with bone fragility without (CDL) or with spondylometaphyseal dysplasia (CDLSMD) | |
| 112240 | AD | P4HB | Loss of function | Protein disulfide-isomerase | Catalyzes rearrangement of disulfid bonds | Cole-Carpenter syndrome 1 | |
| 605822 | AR | XYLT2 | Loss of function | Xylosyltransferase 2 | Proteoglycan biosynthesis | Spondylo-ocular dysplasia | |
| 166260 | AD | ANO5 | Loss of function | Anoctamin-5 | Unclear (chloride channel) | Gnathodiaphyseal dysplasia | |
| 231070 | AR | GORAB | Loss of function | RAB6-interacting golgin | Unclear | Geroderma osteodysplasticum | |
| 612940 | AR | PYCR1 | Loss of function | Pyrroline-5-carboxylate reductase 1, mitochondrial | Unclear (Prolin biosynthesis) | Cutis laxa (ARCL2B) | |
| 182250 | AD | IFIH1 | Gain of function | Interferon-induced helicase C domain-containing protein 1 | Unclear (Antiviral innate immunity) | Singleton-Mertin dysplasia Type 1 | |
| 616298 | AD | DDX58 | Gain of function | Antiviral innate immune response receptor RIG-I | Unclear (antiviral innate immunity) | Singleton-Mertin dysplasia Type 2 | |
| 616866 | AR | TRIP4 | Loss of function | Activating signal cointegrator 1 | Unclear (transcription coactivator) | Spinal muscular atrophy with congenital bone fractures-1 (SMABF1) | |
| 616867 | AR | ASCC1 | Loss of function | Activating signal cointegrator 1 complex subunit 1 | Unclear (DNA damage repair) | Spinal muscular atrophy with congenital bone fractures-2 (SMABF2) | |
| 603109 | AD | SMAD3 | Loss of function | Smad family member 3 | TGF-ß pathway | Loeys-Dietz syndrome | |
| Osteolysis Group | 174810 602080 | AD | TNFRSF11A | Gain of function | Tumor necrosis factor receptor superfamily member 11A | RANK overactivation | Familial expansile osteolysis (FEO) Juvenile Paget’s Disease (PDB2) |
| 239000 | AR | TNFRSF11B | Loss of function | Tumor necrosis factor receptor superfamily member 11B | OPG deficiency with Increased RANKL-mediated osteoclastogenesis | Juvenile Paget’s Disease (PDB5) | |
| 259600 | AR | MMP2 | Loss of function | Matrix metalloproteinase 2 | Unclear (collagenolysis) | Multicentric osteolysis, nodulosis and arthropathy (MANO) | |
| 277950 | MMP14 | Matrix metalloproteinase 14 | |||||
| 102500 | AD | NOTCH2 | Gain of function | Neurogenic locus notch homolog protein 2 | Regulate cell fate; osteoblast and osteoclast function | Hajdu-Cheney Syndrome |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Gazzar, A.; Högler, W. Mechanisms of Bone Fragility: From Osteogenesis Imperfecta to Secondary Osteoporosis. Int. J. Mol. Sci. 2021, 22, 625. https://doi.org/10.3390/ijms22020625
El-Gazzar A, Högler W. Mechanisms of Bone Fragility: From Osteogenesis Imperfecta to Secondary Osteoporosis. International Journal of Molecular Sciences. 2021; 22(2):625. https://doi.org/10.3390/ijms22020625
Chicago/Turabian StyleEl-Gazzar, Ahmed, and Wolfgang Högler. 2021. "Mechanisms of Bone Fragility: From Osteogenesis Imperfecta to Secondary Osteoporosis" International Journal of Molecular Sciences 22, no. 2: 625. https://doi.org/10.3390/ijms22020625
APA StyleEl-Gazzar, A., & Högler, W. (2021). Mechanisms of Bone Fragility: From Osteogenesis Imperfecta to Secondary Osteoporosis. International Journal of Molecular Sciences, 22(2), 625. https://doi.org/10.3390/ijms22020625






