A New Look at the Enterobacterial Common Antigen Forms Obtained during Rough Lipopolysaccharides Purification
Abstract
:1. Introduction
2. Results
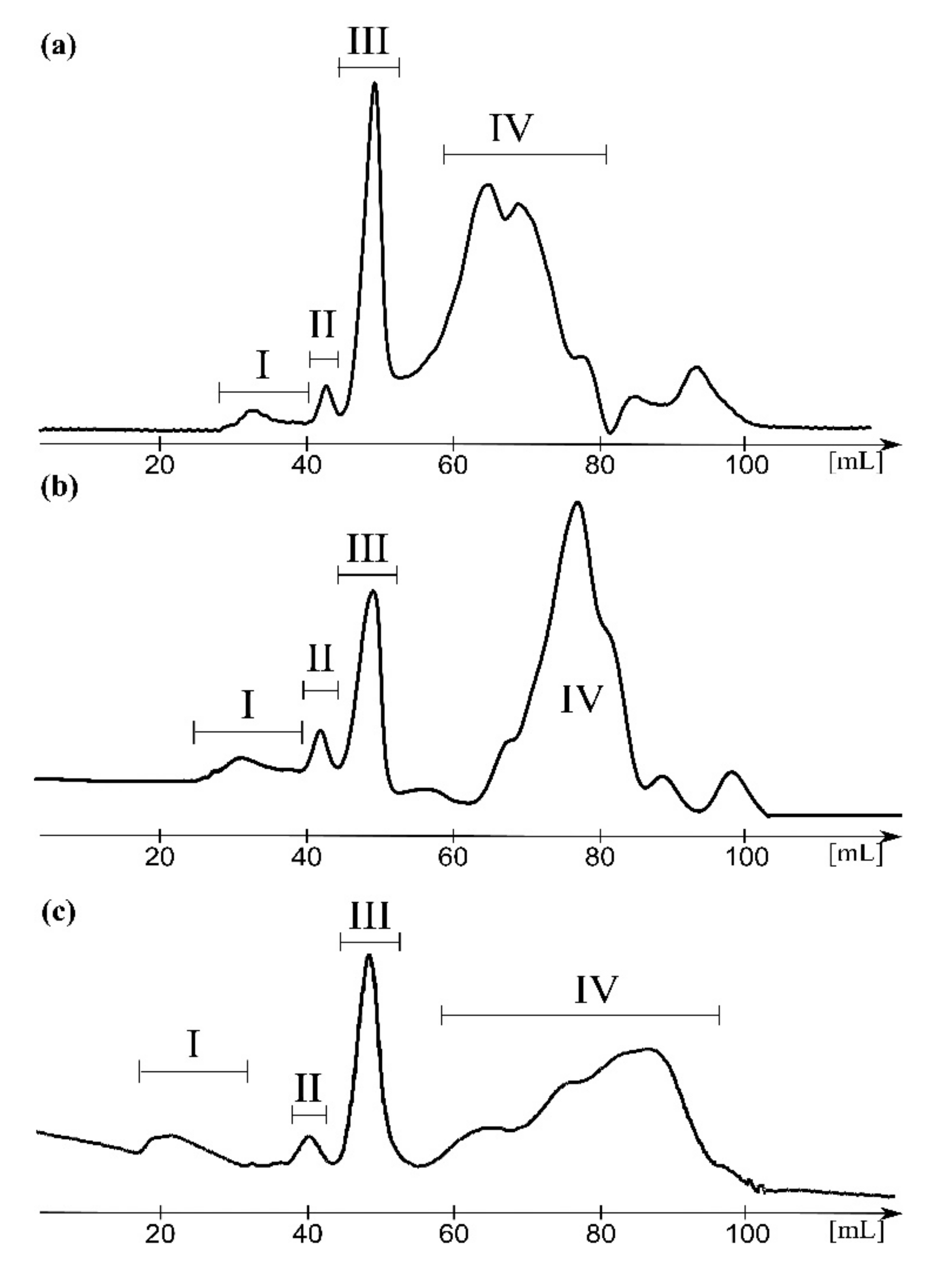
2.1. Supernatants Isolated upon LPS Ultracentrifugation Are Reach in Linear and Cyclic ECA
2.2. ECA Structure Verification by Mass Spectrometry
2.3. ZIC®HILIC Fractionation of ECA Forms
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. LPS Preparation
4.3. Isolation of ECA Forms
4.4. Mass Spectrometry
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ECA | Enterobacterial common antigen |
| c[ECA]n | Cyclic ECA composed of n repeating units |
| [ECA]n | Linear ECA composed of n repeating units |
| ECAPG | Phosphoglyceride-linked ECA |
| ECACYC | Cyclic ECA |
| ECALPS | Lipopolysaccharide-associated ECA |
| ESI-IT MS | Electrospray ionization-ion trap mass spectrometry |
| LPS | Lipopolysaccharide |
| MALDI-TOF MS | Matrix-assisted laser-desorption/ionization-time of flight mass spectrometry |
| MS | Mass spectrometry |
| NAc | N-acetyl group |
| NMR | nuclear magnetic resonance (spectroscopy) |
| OAc | O-acetyl group |
| PCP | phenol-chloroform-petroleum (extraction) |
| ZIC®HILIC | zwitterionic-hydrophilic interaction liquid chromatography |
References
- Kunin, C.M.; Beard, M.V.; Halmagyi, N.E. Evidence for a common hapten associated with endotoxin fractions of E. coli and other Enterobacteriaceae. Exp. Biol. Med. 1962, 111, 160–166. [Google Scholar] [CrossRef]
- Bottger, E.C.; Jurs, M.; Barrett, T.; Wachsmuth, K.; Metzger, S.; Bitter-Suermann, D. Qualitative and quantitative determination of enterobacterial common antigen (ECA) with monoclonal antibodies: Expression of ECA by two Actinobacillus species. J. Clin. Microbiol. 1987, 25, 377–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, A.K.; Mitchell, A.M. Enterobacterial common antigen: Aynthesis and function of an enigmatic molecule. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Lugowski, C.; Romanowska, E.; Kenne, L.; Lindberg, B. Identification of a trisaccharide repeating-unit in the enterobacterial common-antigen. Carbohydr. Res. 1983, 118, 173–181. [Google Scholar] [CrossRef]
- Vinogradov, E.V.; Knirel, Y.A.; Thomas-Oates, J.E.; Shashkov, A.S.; L’Vov, V.L. The structure of the cyclic enterobacterial common antigen (ECA) from Yersinia pestis. Carbohydr. Res. 1994, 258, 223–232. [Google Scholar] [CrossRef]
- Kajimura, J.; Rahman, A.; Hsu, J.; Evans, M.R.; Gardner, K.H.; Rick, P.D. O acetylation of the enterobacterial common antigen polysaccharide is catalyzed by the product of the yiaH gene of Escherichia coli K-12. J. Bacteriol. 2006, 188, 7542–7550. [Google Scholar] [CrossRef] [Green Version]
- Kiss, P.; Rinno, J.; Schmidt, G.; Mayer, H. Structural studies on the immunogenic form of the enterobacterial common antigen. Eur. J. Biochem. 1978, 88, 211–218. [Google Scholar] [CrossRef]
- Dell, A.; Oates, J.; Lugowski, C.; Romanowska, E.; Kenne, L.; Lindberg, B. The enterobacterial common-antigen, a cyclic polysaccharide. Carbohydr. Res. 1984, 133, 95–104. [Google Scholar] [CrossRef]
- Rick, P.D.; Hubbard, G.L.; Kitaoka, M.; Nagaki, H.; Kinoshita, T.; Dowd, S.; Simplaceanu, V.; Ho, C. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology 1998, 8, 557–567. [Google Scholar] [CrossRef]
- De Vlugt, J.E.; Xiao, P.; Munro, R.; Charchoglyan, A.; Brewer, D.; Al-Abdul-Wahid, M.S.; Brown, L.S.; Ladizhansky, V. Identifying lipids tightly bound to an integral membrane protein. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183345. [Google Scholar] [CrossRef]
- Gozdziewicz, T.K.; Lugowski, C.; Lukasiewicz, J. First evidence for a covalent linkage between enterobacterial common antigen and lipopolysaccharide in Shigella sonnei phase II ECALPS. J. Biol. Chem. 2014, 289, 2745–2754. [Google Scholar] [CrossRef] [Green Version]
- Maciejewska, A.; Kaszowska, M.; Jachymek, W.; Lugowski, C.; Lukasiewicz, J. Lipopolysaccharide-linked enterobacterial common antigen (ECA(LPS)) occurs in rough strains of Escherichia coli R1, R2, and R4. Int. J. Mol. Sci. 2020, 21, 6038. [Google Scholar] [CrossRef]
- Kunin, C.M. Separation, characterization, and biological significance of a common antigen in Enterobacteriaceae. J. Exp. Med. 1963, 118, 565–586. [Google Scholar] [CrossRef] [Green Version]
- Kunin, C.M.; Beard, M.V. Serological studies of O antigens of Escherichia coli by means of the hemagglutination test. J. Bacteriol. 1963, 85, 541–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.M.; Srikumar, T.; Silhavy, T.J. Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muszynski, A.; Rabsztyn, K.; Knapska, K.; Duda, K.A.; Duda-Grychtol, K.; Kasperkiewicz, K.; Radziejewska-Lebrecht, J.; Holst, O.; Skurnik, M. Enterobacterial common antigen and O-specific polysaccharide coexist in the lipopolysaccharide of Yersinia enterocolitica serotype O:3. Microbiology 2013, 159, 1782–1793. [Google Scholar] [CrossRef] [PubMed]
- Paunova-Krasteva, T.S.; Pavlova, V.A.; De Castro, C.; Ivanova, R.M.; Molinaro, A.; Nikolova, E.B.; Stoitsova, S.R. Cyclic enterobacterial common antigens from Escherichia coli O157 as microbe-associated molecular patterns. Can. J. Microbiol. 2014, 60, 173–176. [Google Scholar] [CrossRef]
- Liu, L.; Zha, J.; DiGiandomenico, A.; McAllister, D.; Stover, C.K.; Wang, Q.; Boons, G.J. Synthetic enterobacterial common antigen (ECA) for the development of a universal immunotherapy for drug-resistant Enterobacteriaceae. Angew. Chem. Int. Ed. Engl. 2015, 54, 10953–10957. [Google Scholar] [CrossRef] [Green Version]
- Mannel, D.; Mayer, H. Isolation and chemical characterization of the enterobacterial common antigen. Eur. J. Biochem. 1978, 86, 361–370. [Google Scholar] [CrossRef]
- Lugowski, C.; Romanowska, E. Enterobacterial common antigen: Isolation from Shigella sonnei, purification and immunochemical characterization. Eur. J. Biochem. 1978, 91, 89–97. [Google Scholar] [CrossRef]
- Peters, H.; Jurs, M.; Jann, B.; Jann, K.; Timmis, K.N.; Bitter-Suermann, D. Monoclonal antibodies to enterobacterial common antigen and to Escherichia coli lipopolysaccharide outer core: Demonstration of an antigenic determinant shared by enterobacterial common antigen and E. coli K5 capsular polysaccharide. Infect. Immun. 1985, 50, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, H.M.; Basu, S.; Mayer, H. Comparison of enterobacterial common antigen from different species by serological techniques. Eur. J. Biochem. 1987, 162, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, H.M.; Neter, E.; Mayer, H. Modification of the lipid moiety of the enterobacterial common antigen by the “Pseudomonas factor”. Infect. Immun. 1983, 40, 696–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farnback, M.; Eriksson, L.; Senchenkova, S.; Zych, K.; Knirel, Y.A.; Sidorczyk, Z.; Widmalm, G. Crystal structure of a cyclic enterobacterial common antigen. Angew. Chem. Int. Ed. Engl. 2003, 42, 2543–2546. [Google Scholar] [CrossRef]
- Fregolino, E.; Ivanova, R.; Lanzetta, R.; Molinaro, A.; Parrilli, M.; Paunova-Krasteva, T.; Stoitsova, S.R.; De Castro, C. Occurrence and structure of cyclic enterobacterial common antigen in Escherichia coli O157:H(-). Carbohydr. Res. 2012, 363, 29–32. [Google Scholar] [CrossRef]
- Westphal, O. Bacterial lipopolysaccharides extraction with phenol-water and further applications of the procedure. Met. Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Duda, K.A.; Duda, K.T.; Beczala, A.; Kasperkiewicz, K.; Radziejewska-Lebrecht, J.; Skurnik, M. ECA-immunogenicity of Proteus mirabilis strains. Arch. Immunol. Ther. Exp. 2009, 57, 147–151. [Google Scholar] [CrossRef] [Green Version]
- Weber, G.; von Wirén, N.; Hayen, H. Hydrophilic interaction chromatography of small metal species in plants using sulfobetaine- and phosphorylcholine-type zwitterionic stationary phases. J. Sep. Sci. 2008, 31, 1615–1622. [Google Scholar] [CrossRef]
- Man-Kupisinska, A.; Bobko, E.; Gozdziewicz, T.K.; Maciejewska, A.; Jachymek, W.; Lugowski, C.; Lukasiewicz, J. Fractionation and analysis of lipopolysaccharide-derived oligosaccharides by zwitterionic-type hydrophilic interaction liquid chromatography coupled with electrospray ionisation mass spectrometry. Carbohydr. Res. 2016, 427, 29–37. [Google Scholar] [CrossRef]
- Ceroni, A.; Maass, K.; Geyer, H.; Geyer, R.; Dell, A.; Haslam, S.M. GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 2008, 7, 1650–1659. [Google Scholar] [CrossRef] [Green Version]





| Fraction | Polysaccharide Composition | Observed Ion (m/z) | Calculated Ion (m/z) | Theoretical Monoisotopic Mass (Da) | Interpretation of the Ion |
|---|---|---|---|---|---|
| I | [ECA]7 + 4OAc | 1108.15 | 1108.15 | 4436.61 | [M-4H]4− |
| 1477.86 | 1477.86 | [M-3H]3− | |||
| [ECA]8 + 4OAc | 1259.95 | 1259.95 | 5043.83 | [M-4H]4− | |
| [ECA]8 + 5OAc | 1694.27 | 1694.27 | 5085.84 | [M-3H]3− | |
| [ECA]9 + 5OAc | 1422.27 | 1422.26 | 5693.07 | [M-4H]4− | |
| 1896.69 | 1896.68 | [M-3H]3− | |||
| [ECA]10 + 6OAc | 1584.57 | 1584.57 | 6342.30 | [M-4H]4− | |
| 2113.09 | 2113.09 | [M-3H]3− | |||
| [ECA]11 + 7OAc | 1746.88 | 1746.88 | 6991.53 | [M-4H]4− | |
| II | c[ECA]5 + 3OAc | 1053.05 | 1053.04 | 3162.14 | [M-3H]3− |
| 1580.07 | 1580.06 | [M-2H]2− | |||
| c[ECA]6 + 4OAc | 1269.46 | 1269.45 | 3811.37 | [M-3H]3− | |
| III | c[ECA]4 | 808.63 | 808.62 | 2428.89 | [M-3H]3− |
| 1213.44 | 1213.44 | [M-2H]2− | |||
| c[ECA]4 − NAc | 794.62 | 794.62 | 2386.87 | [M-3H]3− | |
| 1192.44 | 1192.43 | [M-2H]2− |
| Fraction | Polysaccharide Composition | Observed Ion (m/z) | Calculated Ion (m/z) | Theoretical Monoisotopic Mass (Da) | Interpretationof the Ion |
|---|---|---|---|---|---|
| I | [ECA]7 | 1076.64 | 1076.64 | 4310.58 | [M-4H]4− |
| 1435.85 | 1435.85 | [M-3H]3− | |||
| [ECA]8 + 2OAc | 990.96 | 990.96 | 4959.81 | [M-5H]5− | |
| 1238.95 | 1238.95 | [M-4H]4− | |||
| 1652.27 | 1652.26 | [M-3H]3− | |||
| [ECA]9 + 2OAc | 1112.41 | 1112.40 | 5567.03 | [M-5H]5− | |
| 1390.76 | 1390.75 | [M-4H]4− | |||
| 1854.69 | 1854.67 | [M-3H]3− | |||
| [ECA]10 + 2OAc | 1542.57 | 1542.56 | 6174.26 | [M-4H]4− | |
| 2057.10 | 2057.08 | [M-3H]3− | |||
| II | c[ECA]5 + 2OAc | 779.03 | 779.02 | 3120.13 | [M-4H]4− |
| 1039.04 | 1039.04 | [M-3H]3− | |||
| 1559.06 | 1559.06 | [M-2H]2− | |||
| c[ECA]6 + 2OAc | 1241.45 | 1241.44 | 3727.35 | [M-3H]3− | |
| III | c[ECA]4 | 808.63 | 808.62 | 2428.89 | [M-3H]3− |
| 1213.44 | 1213.44 | [M-2H]2− |
| Fraction | Polysaccharide Composition | Observed Ion (m/z) | Calculated Ion (m/z) | Theoretical Monoisotopic Mass (Da) | Interpretation of the Ion |
|---|---|---|---|---|---|
| II | c[ECA]5 + OAc | 3079.46 | 3079.13 | 3078.12 | [M + H]+ |
| c[ECA]5 + 2OAc | 3121.40 | 3121.14 | 3120.13 | [M + H]+ | |
| c[ECA]5 + 3OAc | 3163.36 | 3163.15 | 3162.14 | [M + H]+ | |
| c[ECA]5 + 4OAc | 3205.29 | 3205.16 | 3204.15 | [M + H]+ | |
| c[ECA]5 + 5OAc | 3247.95 | 3247.17 | 3246.16 | [M + H]+ | |
| c[ECA]6 + 2OAc | 3728.27 | 3728.35 | 3727.35 | [M + H]+ | |
| c[ECA]6 + 3OAc | 3770.18 | 3770.37 | 3769.36 | [M + H]+ | |
| c[ECA]6 + 4OAc | 3812.16 | 3812.38 | 3811.37 | [M + H]+ | |
| c[ECA]6 + 5OAc | 3854.00 | 3854.39 | 3853.38 | [M + H]+ | |
| III | c[ECA]4 − NAc * | 2409.98 | 2409.86 | 2386.87 | [M + H, Na]+ |
| c[ECA]4 | 2451.98 | 2451.87 | 2428.89 | [M + H, Na]+ | |
| c[ECA]4 + OAc | 2493.99 | 2493.88 | 2470.90 | [M + H, Na]+ | |
| c[ECA]4 + 2OAc | 2535.98 | 2535.90 | 2512.91 | [M + H, Na]+ | |
| c[ECA]5 + OAc | 3101.48 | 3101.11 | 3078.12 | [M + H, Na]+ | |
| c[ECA]5 + 2OAc | 3143.65 | 3143.12 | 3120.13 | [M + H, Na]+ | |
| c[ECA]5 + 3OAc | 3185.45 | 3185.13 | 3162.14 | [M + H, Na]+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gozdziewicz, T.K.; Maciejewska, A.; Tsybulska, A.; Lugowski, C.; Lukasiewicz, J. A New Look at the Enterobacterial Common Antigen Forms Obtained during Rough Lipopolysaccharides Purification. Int. J. Mol. Sci. 2021, 22, 701. https://doi.org/10.3390/ijms22020701
Gozdziewicz TK, Maciejewska A, Tsybulska A, Lugowski C, Lukasiewicz J. A New Look at the Enterobacterial Common Antigen Forms Obtained during Rough Lipopolysaccharides Purification. International Journal of Molecular Sciences. 2021; 22(2):701. https://doi.org/10.3390/ijms22020701
Chicago/Turabian StyleGozdziewicz, Tomasz K, Anna Maciejewska, Alona Tsybulska, Czeslaw Lugowski, and Jolanta Lukasiewicz. 2021. "A New Look at the Enterobacterial Common Antigen Forms Obtained during Rough Lipopolysaccharides Purification" International Journal of Molecular Sciences 22, no. 2: 701. https://doi.org/10.3390/ijms22020701
APA StyleGozdziewicz, T. K., Maciejewska, A., Tsybulska, A., Lugowski, C., & Lukasiewicz, J. (2021). A New Look at the Enterobacterial Common Antigen Forms Obtained during Rough Lipopolysaccharides Purification. International Journal of Molecular Sciences, 22(2), 701. https://doi.org/10.3390/ijms22020701






