Mining Fiskeby III and Mandarin (Ottawa) Expression Profiles to Understand Iron Stress Tolerant Responses in Soybean
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Analyses
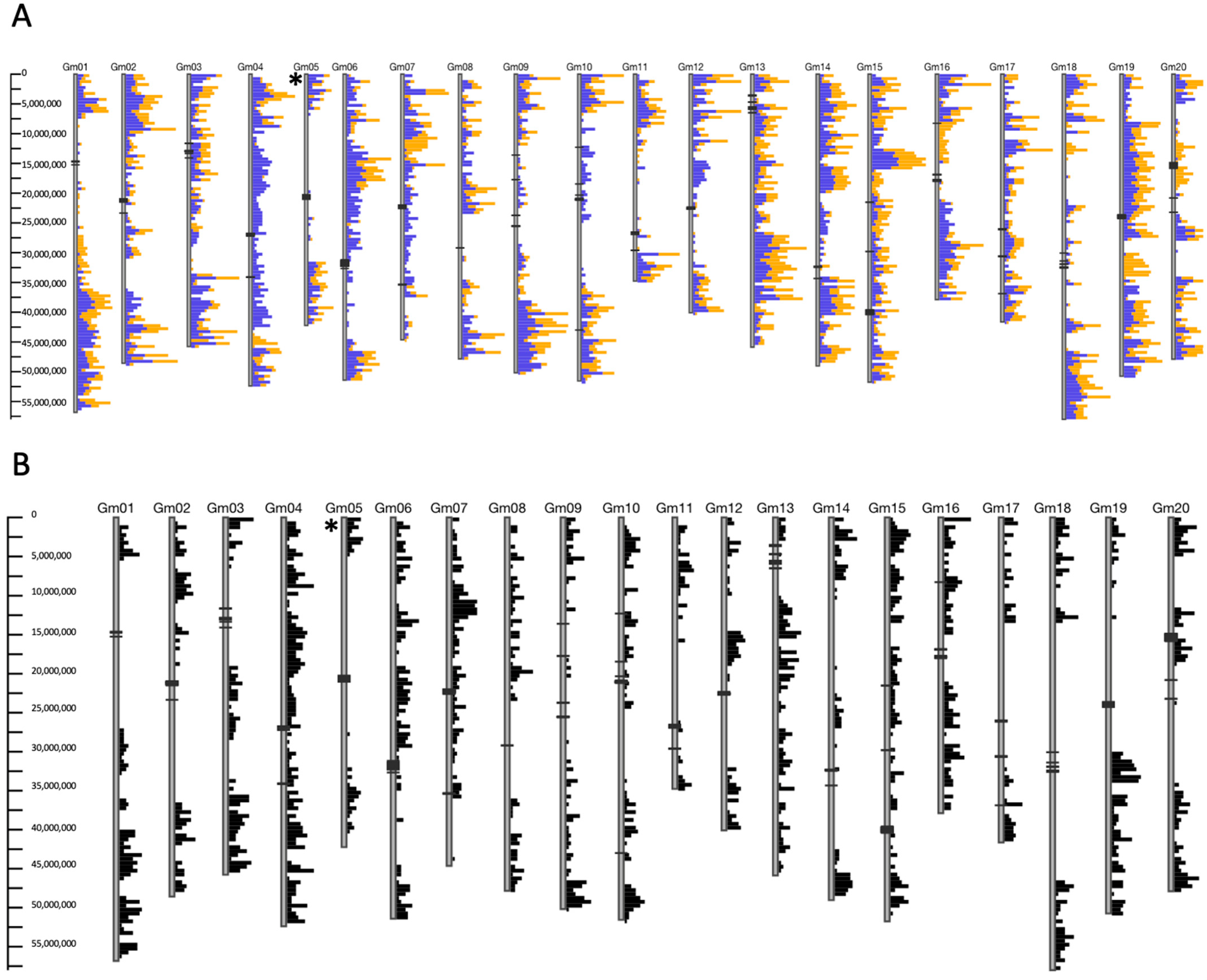
2.2. SNP Analysis of Genotypes of Interest
2.3. RNA-Seq Analysis
2.3.1. Mandarin RNA-Seq
2.3.2. Fiskeby RNA-Seq
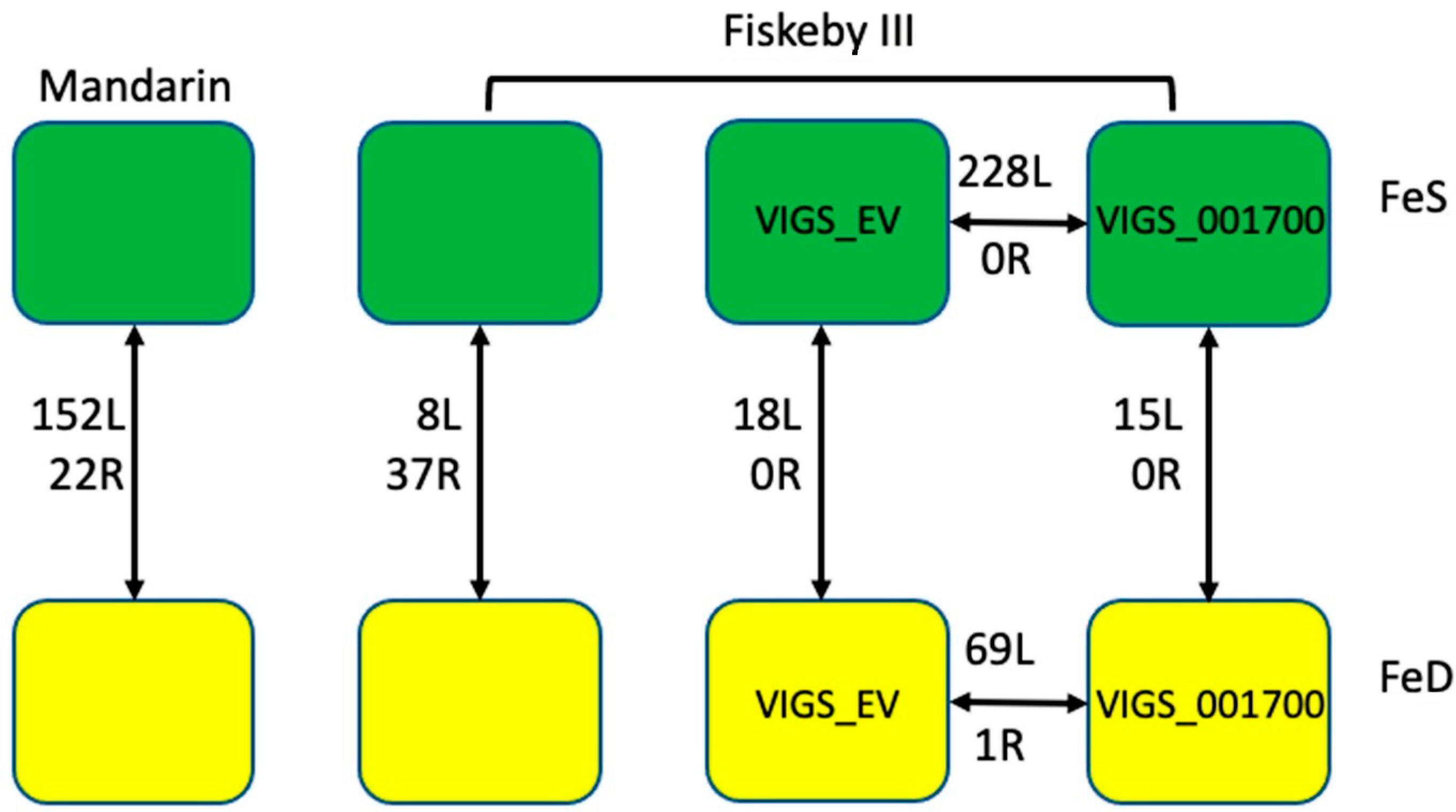
2.4. VIGS Plants
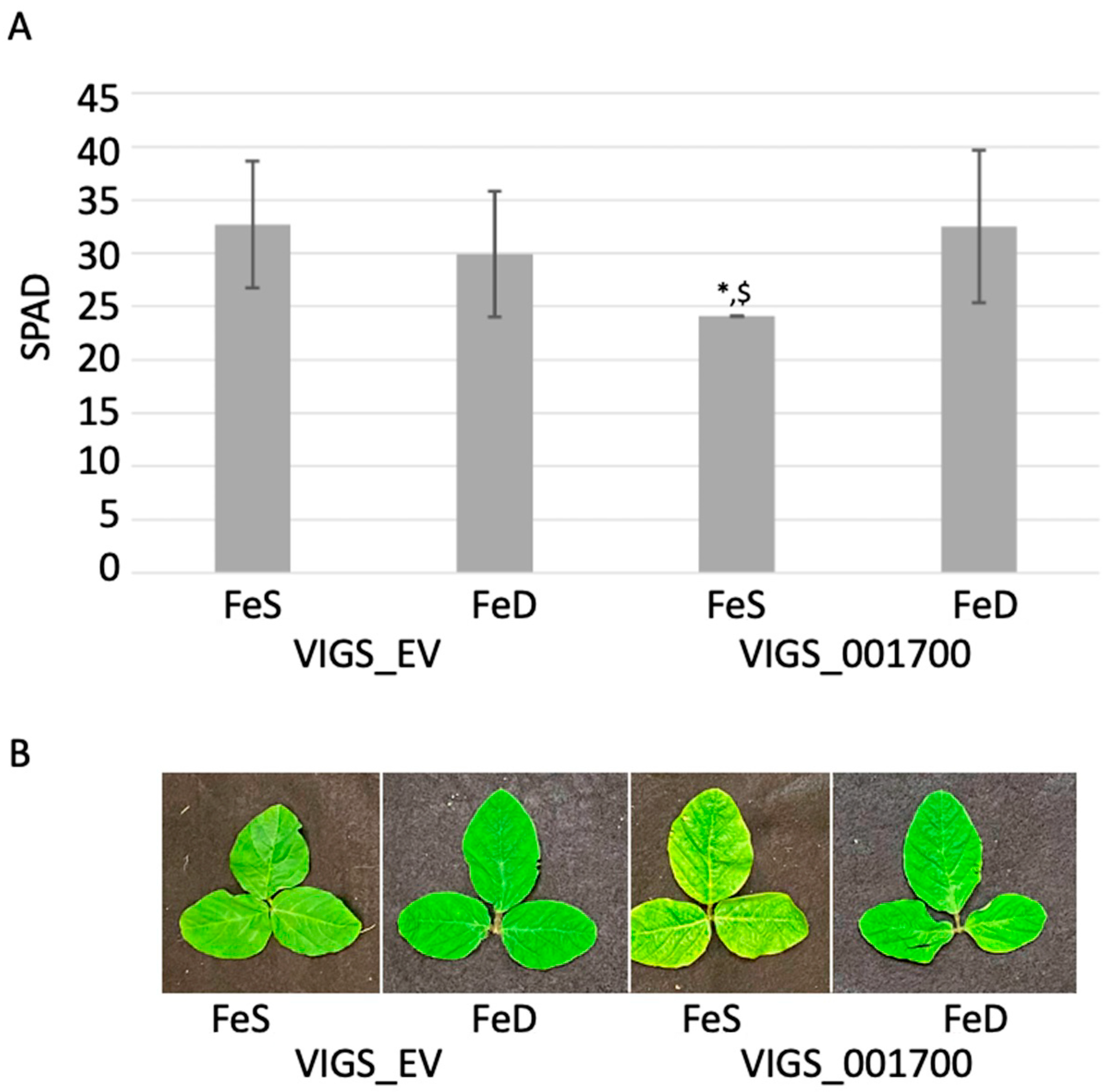
2.4.1. Phenotypic Analysis of VIGS Plants
2.4.2. Identifying DEGs between VIGS_EV and VIGS_Glyma.05G001700
2.4.3. DEGs VIGS_EV Response to Iron Treatment
2.4.4. VIGS_Glyma.05G001700 Response to Iron Treatment
3. Discussion
3.1. Comparing Mandarin (Ottawa) and Fiskeby III Gene Expression
3.2. Gene Expression in Mandarin (Ottawa) Leaves and Roots
3.3. Gene Expression in Fiskeby III Leaves and Roots
3.4. Candidate Gene Underlying Gm05 IDC QTL
3.5. Comparing Gene Expression in EV and Glyma.05001700 Silenced Plants
3.6. Effect of Iron Treatment on Transcriptome of VIGS Infected Plants
3.7. Conclusions
4. Materials and Methods
4.1. Virus-Induced Gene Silencing (VIGS) Constructs
4.2. Phenotypic Analyses
4.3. Hydroponic Growth Conditions
4.4. RNA Extraction and Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Romheld, V.; Marschner, H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986, 80, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Santi, S.; Schmidt, W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009, 183, 1072–1084. [Google Scholar] [CrossRef]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.H.; Schmidt, W. Mobilization of iron by plant-borne coumarins. Trends Plant Sci. 2017, 22, 538–548. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brumbarova, T.; Bauer, P.; Ivanov, R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015, 20, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Guerinot, M.L. All together now: Regulation of the iron deficiency response. J. Exp. Bot. 2021, 72, 2045–2055. [Google Scholar] [CrossRef]
- Tsai, H.H.; Schmidt, W. The enigma of environmental pH sensing in plants. Nat. Plants 2021, 7, 106–115. [Google Scholar] [CrossRef]
- Froechlich, D.; Fehr, W. Agronomic performance of soybeans with differing levels of iron deficiency chlorosis on calcareous soil. Crop Sci. 1981, 21, 438–441. [Google Scholar] [CrossRef]
- Hansen, N.C.; Jolley, V.D.; Naeve, S.L.; Goos, R.J. Iron deficiency of soybean in the north central U.S. and associated soil properties. Soil Sci. Plant Nutr. 2004, 50, 983–987. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Cianzio, S.; Shoemaker, R. Mapping genetic loci for iron deficiency chlorosis in soybean. Mol. Breed. 1997, 3, 219–229. [Google Scholar] [CrossRef]
- Assefa, T.; Zhang, J.; Chowda-Reddy, R.V.; Lauter, A.N.M.; Singh, A.; O’Rourke, J.A.; Graham, M.A.; Singh, A.K. Deconstrcting the genetic architecture of iron deficiency chlorosis in soybean using genome-wide approaches. BMC Plant Biol. 2020, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Merry, R.; Butenhoff, K.; Campbell, B.W.; Michno, J.M.; Wang, D.; Orf, J.H.; Lorenz, A.J.; Stupar, R.M. Identification and fine-mapping of a soybean quantitative trait locus on chromosome 5 confering tolerance to iron deficiency chlorosis. Plant Genome 2019, 12, 190007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Whitham, S.A.; Hill, J.H. Virus-induced gene silencing in soybean and common bean. Methods Mol. Biol. 2013, 975, 149–156. [Google Scholar] [CrossRef]
- Morales, A.M.A.P.; O’Rourke, J.A.; van de Mortel, M.; Scheider, K.T.; Bancroft, T.J.; Borem, A.; Nelson, R.T.; Nettleton, D.; Baum, T.J.; Shoemaker, R.C.; et al. Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway. Funct. Plant Biol. 2013, 40, 1029–1047. [Google Scholar] [CrossRef] [Green Version]
- Meyer, J.D.F.; Silva, D.C.G.; Yang, C.; Pedley, K.F.; Zhang, C.; van de Mortel, M.; Hill, J.H.; Shoemaker, R.C.; Abdelnoor, R.V.; Whitham, S.A.; et al. Identification and analyses of candidate genes for Rpp4-mediated resistance to Asian soybean rust in soybean. Plant Physiol 2009, 150, 295–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atwood, S.E.; O’Rourke, J.A.; Peiffer, G.A.; Yin, T.; Majumder, M.; Zhang, C.; Cianzio, S.R.; Hill, J.H.; Cook, D.; Whitham, S.A.; et al. Replication protein A subunit 3 and the iron efficiency response in soybean. Plant Cell Environ. 2014, 37, 213–234. [Google Scholar] [CrossRef] [Green Version]
- Ogata, T.; Nagatoshi, Y.; Yamagishi, N.; Yoshikawa, N.; Fujita, Y. Virus-induced down-regulation of GmERA1A and GmERA1B genes enhances the stomatal response to abscisic acid and drought resistance in soybea. PLoS ONE 2017, 12, e0175650. [Google Scholar] [CrossRef] [Green Version]
- Kandoth, P.K.; Heinz, R.; Yeckel, G.; Gross, N.W.; Juvale, P.S.; Hill, J.; Whitham, S.A.; Baum, T.J.; Mitchum, M.G. A virus-induced gene silencing method to study soybean cyst nematode parasitism in Glycine max. BMC Res. Notes 2013, 6, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.Y.; Zhang, C.; Li, Z.C.; Wang, Z.R.; Jiang, X.X.; Shi, Y.F.; Tian, S.N.; Braun, E.; Mei, Y.; Qiu, W.L.; et al. The MAPK kinase kinase GmMEKK1 regulates cell death and defense responses. Plant Physiol. 2018, 178, 907–922. [Google Scholar] [CrossRef] [Green Version]
- Pedley, K.F.; Pandey, A.K.; Ruck, A.L.; Lincoln, L.M.; Whitham, S.A.; Graham, M.A. Rpp1 encodes a ULP1-NBS-LRR protein that controls immunity to Phakospora pachyrhizi in soybean. Mol. Plant-Microbe Interact. 2018, 32, 120–133. [Google Scholar] [CrossRef] [Green Version]
- Burton, A.L.; Burkey, K.O.; Carter, T.E., Jr.; Orf, J.; Cregan, P.B. Phenotypic variation and identification of quantitative trait loci for ozone tolerance in a Fiskeby III x Mandarin (Ottawa) soybean population. Theor. Appl. Genet. 2016, 129, 1113–1125. [Google Scholar] [CrossRef]
- Do, T.D.; Vuong, T.D.; Dunn, D.; Smothers, S.; Patil, G.; Yungbluth, D.C.; Chen, P.; Scaboo, A.; Xu, D.; Carter, T.E.; et al. Mapping and confirmation of loci for salt tolerance in a novel soybean germplasm, Fiskeby III. Theor. Appl. Genet. 2018, 131, 513–524. [Google Scholar] [CrossRef]
- Whaley, A.; Sheridan, J.; Safari, S.; Burton, A.; Burkey, K.; Schlueter, J. RNA-seq analysis reveals genetic response and tolerance mechanisms to ozone exposure in soybean. BMC Genom. 2015, 16, 426. [Google Scholar] [CrossRef] [Green Version]
- Rod, K.S.; Walker, D.R.; Bradley, C.A. Evaluation of major ancestors of North American soybean cultivars for resistance to three Pythium species that cause seedling blight. Plant Dis. 2018, 102, 2241–2252. [Google Scholar] [CrossRef] [Green Version]
- Bailey, A.; Burkey, K.; Taggart, M.; Rufty, T. Leaf traits that contribute to differential ozone response in ozone-tolerant and sensitive soybean genotypes. Plants 2019, 8, 235. [Google Scholar] [CrossRef] [Green Version]
- Severin, A.; Woody, J.L.; Bolon, Y.T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Hyten, D.; Song, Q.; Zhu, Y.; Choi, I.Y.; Nelson, R.L.; Costa, J.M.; Specht, J.E.; Shoemaker, R.C.; Cregan, P.B. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 2006, 103, 16666–16671. [Google Scholar] [CrossRef] [Green Version]
- GRIN-Global. Germplasm Resources Information Network-Global, US National Plant Germplasm System. Available online: https://npgsweb.ars-grin.gov/gringlobal/search (accessed on 19 May 2021).
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhang, J.; Jin, H.; Feng, D.; Wang, J.; Wang, H.B.; Liu, B. The iron deficiency response regulators IAA-LEUCINE RESISTANT3 and bHLH104 possess different targets and have distinct effects on photosynthesis in Arabidopsis. J. Plant Biol. 2019, 62, 109–119. [Google Scholar] [CrossRef]
- Tissot, N.; Robe, K.; Gaqo, F.; Grant-Grant, S.; Boucherez, J.; Bellegarde, F.; Maghiaoui, A.; Marcelin, R.; Izquierdo, E.; Benhamed, M.; et al. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 2019, 223, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Martinoia, E.; Maeshima, M.; Nehaus, H.E. Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 2007, 58, 83–102. [Google Scholar] [CrossRef] [PubMed]
- Gollhofer, J.; Timofeev, R.; Lan, P.; Schmidt, W.; Buckhout, T.J. Vacuolar-iron-transporter1-like proteins mediate iron homeostasis in Arabidopsis. PLoS ONE 2014, 9, e110468. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Wu, Q.; Meng, Y.T.; Tao, Y.; Shen, R.F. AtHAP5A regulates iron translocation in iron-deficient Arabidopsis thaliana. J. Integr. Plant Biol. 2020, 62, 1910–1925. [Google Scholar] [CrossRef] [PubMed]
- Nozoye, T.; Kim, S.; Kakei, Y.; Takahashi, M.; Nakanishi, H.; Nishizawa, N.K. Enhanced levels of nicotianamine promote iron accumulation, and tolerance to calcareous soil in soybean. Biosci. Biotechnol. Biochem. 2014, 78, 1677–1684. [Google Scholar] [CrossRef] [PubMed]
- Nozoye, T.; Otani, M.; Senoura, T.; Nakanishi, H.; Nishizawa, N.K. Overexpression of barley nicotianamine synthase 1 confers tolerance in the sweet potato to iron deficiency in calcareous soil. Plant Soil 2017, 418, 75–88. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuler, M.; Bauer, P. Heavy metals need assistance: The contribution of nicotianamine to metal circulation throughout the plant and the Arabidopsis NAS gene family. Front. Plant Sci. 2011, 2, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remy, E.; Cabrito, T.R.; Baster, P.; Batista, R.A.; Teixeira, M.C.; Firimi, J.; Sà-Correia, I.; Duque, P. A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 2013, 25, 901–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haydon, M.J.; Cobbett, C.S. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 2017, 143, 1705–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niño-González, M.; Novo-Uzal, E.; Richardson, D.N.; Barros, P.M.; Duque, P. More transporters, more substrates: The Arabidopsis major facilitator superfamily revisited. Mol. Plant 2019, 12, 1182–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remy, E.; Niño-González, M.; Godinho, C.P.; Cabrito, T.R.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. Heterologous expression of the yeast Tpo1p or Pdr5p membrane transporters in Arabidopsis confers plant xenobiotic tolerance. Sci. Rep. 2017, 7, 4529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Cao, L.; Mwimba, M.; Zhao, Y.; Li, L.; Zhou, M.; Schnable, P.S.; O’Rourke, J.A.; Dong, X.; Wang, W. Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 23840–23849. [Google Scholar] [CrossRef] [Green Version]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Durrett, T.P.; Gassmann, W.; Rogers, E.E. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007, 144, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Murgia, I.; Vigani, G.; Silvestre, D.D.; Mauri, P.; Rossi, R. Formate dehydrogenase takes part in molybdenum and iron homeostasis and affects dark-induced senescence in plants. J. Plant Interact. 2020, 15, 386–397. [Google Scholar] [CrossRef]
- Tsai, H.H.; Rodríguez-Celma, J.; Lan, P.; Wu, Y.C.; Vélez-Bermúdez, I.C.; Schmidt, W. Scopoletin 8-hydroxylase-mediated fraxetin production is crucial for iron mobilization. Plant Physiol. 2018, 177, 194–207. [Google Scholar] [CrossRef] [Green Version]
- Galstyan, A.; Cifuentes-Esquivel, N.; Bou-Torrent, J.; Martinez-Garcia, J.F. The shade avoidance syndrome in Arabidopsis: A fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 2011, 66, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Liang, H.-M.; Yang, S.-Y.; Boch, A.; Clemens, S.; Chen, C.-C.; Wu, J.-F.; Huang, J.-L.; Yeh, K.-C. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009, 182, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Kurt, F.; Filiz, E. Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100, and bHLH101 genes in Arabidopsis, tomato, rice, soybean and maize: Insights into iron (Fe) homeostasis. BioMetals 2018, 31, 489–504. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.-Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef]
- Moran Lauter, A.N.; Rutter, L.; Cook, D.; O’Rourke, J.A.; Graham, M.A. Examining short-term responses to a long-term problem: RNA-seq analyses of iron deficiency chlorosis tolerant soybean. Int. J. Mol. Sci. 2020, 21, 3591. [Google Scholar] [CrossRef]
- Zientara, K.; Wawrzyńska, A.; Łukomska, J.; López-Moya, J.R.; Liszewska, F.; Assunção, A.G.; Aarts, M.G.; Sirko, A. Activity of the AtMRP3 promoter in transgenic Arabidopsis thaliana and Nicotiana tabacum plants is increased by cadmium, nickel, arsenic, cobalt and lead but not by zinc and iron. J. Biotechnol. 2009, 139, 258–263. [Google Scholar] [CrossRef]
- Peiffer, G.A.; King, K.E.; Severin, A.J.; May, G.D.; Cianzio, S.R.; Lin, S.F.; Lauter, N.C.; Shoemaker, R.C. Identification of candidate genes underlying an iron efficiency quantitative trait locus in soybean. Plant Physiol. 2012, 158, 1745–1754. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rather, B.A.; Masood, A.; Sehar, Z.; Majid, A.; Anjum, N.A.; Khan, N.A. Mechanisms and role of nitric oxide in phytotoxicity mitigation of copper. Front. Plant Sci. 2020, 11, 675. [Google Scholar] [CrossRef] [PubMed]
- Kathare, P.K.; Dharmasiri, S.; Vincill, E.D.; Routray, P.; Ahmad, I.; Roberts, D.M.; Dharmasiri, N. Arabidopisis PIC30 encodes a major facility superfamily transporter responsible for the uptake of picolinate herbicides. Plant J. 2019, 102, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The roles of Arabidopsis C1-2i subclass of C2H2 zinc-finger transcription factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Tax, F.E. Receptor-like kinases: Key regulators of plant development and defense. J. Integr. Plant Biol. 2013, 55, 1184–1187. [Google Scholar] [CrossRef]
- Califar, B.; Sng, N.J.; Zupanska, A.; Paul, A.L.; Ferl, R.J. Root skewing-associated genes impact the spaceflight response of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.C. Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 1–10. [Google Scholar] [CrossRef]
- Atencio, L.; Salazar, J.; Lauter, A.N.M.; Gonzales, M.D.; O’Rourke, J.A.; Graham, M.A. Characterizing short and long term iron stress responses in iron deficiency tolerant and susceptible soybean (Glycine max L. Merr). Plant Stress 2021, 2, 100012. [Google Scholar] [CrossRef]
- Moran Lauter, A.N.; Peiffer, G.A.; Yin, T.; Whitham, S.A.; Cook, D.; Shoemaker, R.C.; Graham, M.A. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Takatsuka, H.; Takahashi, N.; Kurata, R.; Fukao, Y.; Kobayashi, K.; Ito, M.; Umeda, M. Arabidopsis R1R2R3-Myb proteins are essential for inhibiting cell division in response to DNA damage. Nat. Commun. 2017, 8, 635. [Google Scholar] [CrossRef] [Green Version]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Gao, Z.; Liu, X.; Sun, D.; Tang, W. Transcriptional profiling reveals a time-of-day specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis. Plant Cell 2019, 31, 2353–2369. [Google Scholar] [CrossRef] [PubMed]
- Murmu, J.; Wilton, M.; Allard, G.; Pandeya, R.; Desveaux, D.; Singh, J.; Subramaniam, R. Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol. Plant Pathol. 2013, 15, 174–184. [Google Scholar] [CrossRef]
- Nagatoshi, Y.; Mitsuda, N.; Hayashi, M.; Inoue, S.I.; Okuma, E.; Kubo, A.; Murata, Y.; Seo, M.; Saji, H.; Kinoshita, T.; et al. GOLDEN2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc. Natl. Acad. Sci. USA 2016, 113, 4218–4223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Hoai, N.N.; Jeong, C.Y.; Trinh, N.N.; Hong, S.W.; Lee, H. Loss of the R2R3 MYB, AtMYB73, causes hyper induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- Pang, X.; Xue, M.; Ren, M.; Nan, D.; Wu, Y.; Guo, H. Ammopiptanthus mongolicus stress-responsive NAC gene enhances the tolerance of transgenic Arabidopsis thaliana to drought and cold stress. Genet. Mol. Biol. 2019, 42, 624–634. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, H.-L.; Li, Z.; Guo, H. Genetic network between leaf senescence and plant immunity: Crucial regulatory nodes and new insights. Plants 2020, 9, 495. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Kmiecik, P.; Leonardelli, M.; Teige, M. Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 2016, 67, 3793–3807. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Wang, B.; Li, Z.; Peng, Z.; Zhang, J. TsNAC1 is a key transcription factor in abiotic stress resistance and growth. Plant Physiol. 2017, 176, 742–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Sun, Q.; Zhao, L.; Li, Z.; Peng, Z.; Zhang, J. Heterologous expression of the transcription factor EsNAC1 in Arabidopsis enhances abiotic stress resistance and retards growth by regulating the expression of different target genes. Front. Plant Sci. 2018, 9, 1495. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, J.A.; McCabe, C.E.; Graham, M.A. Dynamic gene expression changes in response to micronutrient, macronutrient, and multiple stress exposure in soybean. Funct. Integr. Genom. 2020, 20, 321–341. [Google Scholar] [CrossRef] [Green Version]
- Yeon Moon, J.; Belloeil, C.; Ianna, M.L.; Shin, R. Arabidopsis CNGC family members contribute to heavy metal ion uptake in plants. Int. J. Mol. Sci. 2019, 20, 413. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, M.A.; Rajewski, A.; He, J.; Castaneda, O.G.; Litt, A.; Kaloshian, I. Classification and phylogenetic analyses of the Arabidopsis and tomato G-type lectin receptor kinases. BMC Genom. 2018, 19, 239. [Google Scholar] [CrossRef] [Green Version]
- Rogers, E.E.; Guerinot, M.L. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 2002, 14, 1787–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grillet, L.; Schmidt, W. Iron acquisition strategies in land plants: Not so different after all. New Phytol. 2019, 224, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Robe, K.; Conejero, G.; Gao, F.; Lefebvre-Legendre, L.; Sylvestre-Gonon, E.; Rofial, V.; Hem, S.; Rouhier, N.; Barberon, M.; Hecker, A.; et al. Coumarin accumulation and trafficking in Arabidopsis thaliana: A complex and dynamic process. New Phytol. 2020, 229, 2062–2079. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 3, 950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 223. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, N.; Kar, D.; Mahajan, B.D.; Nanda, S.; Rahiman, R.; Panchakshari, N.; Bhagavatula, L.; Datta, S. The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 2019, 70, 4643–4656. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.E.; Wu, X.; Stacey, G.; Nguyen, H.T. Two MATE proteins play a role in iron efficiency in soybean. J. Plant Physiol. 2009, 166, 1453–1459. [Google Scholar] [CrossRef]
- Charlson, D.V.; Bailey, T.B.; Cianzio, S.R.; Shoemaker, R.C. Molecular marker Satt481 is associated with iron deficiency chlorosis resistance in a soybean breeding population. Crop Sci. 2005, 46, 2394–2399. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.F.; Baumer, J.; Ivers, D.; Cianzio, S.R.; Shoemaker, R.C. Nutrient solution screening of Fe chlorosis resistance in soybean evaluated by molecular characterization. J. Plant Nutr. 2000, 23, 1915–1928. [Google Scholar] [CrossRef]
- Lu, P.; Magwanga, R.O.; Kirungu, J.N.; Hu, Y.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Zhang, Z.; Hou, Y.; et al. Overexpression of cotton a DTX/MATE gene enhances drought, salt, and cold stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 299. [Google Scholar] [CrossRef] [Green Version]
- Simmons, A.R.; Davies, K.A.; Wang, W.; Liu, Z.; Bergmann, D.C. SOL1 and SOL2 regulate fate transition and cell divisions in the Arabidopsis stomatal lineage. Development 2019, 146, dev171066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, F.; Zhang, F. Cell cycle regulation in the plant response to stress. Front. Plant Sci. 2019, 10, 1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Chen, H.; Wang, C.; Hu, Z.; Yan, S. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc. Natl. Acad. Sci. USA 2018, 115, E3837–E3845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, M.; van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graves, J.D.; Lee, Y.J.; Liu, K.; Li, G.; Lin, F.T.; Lin, W.C. E2F1 sumoylation as a protective cellular mechanism in oxidative stress response. Proc. Natl. Acad. Sci. USA 2020, 117, 14958–14969. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-K.; Torii, K.U. Linking cell cycle to stomatal differentiation. Curr. Opin. Plant Biol. 2019, 51, 66–73. [Google Scholar] [CrossRef]
- Cho, H.; Zhu, Y.; Ma, Y.; Berkowitz, G.A. The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca2+ as a secondary cystosolic messenger. Plant J. 2016, 85, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Sarowar, S.; Kim, Y.J.; Kim, E.N.; Kim, K.D.; Hwang, B.K.; Islam, R.; Shin, J.S. Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stress. Plant Cell Rep. 2005, 24, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Hindt, M.N.; Akmakjian, G.Z.; Pivarski, K.L.; Punshon, T.; Baxter, I.; Salt, D.E.; Guerinot, M.L. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode imortant negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 2017, 9, 876–890. [Google Scholar] [CrossRef]
- Zheng, L.; Huang, F.; Narsai, R.; Wu, J.; Giraud, E.; He, F.; Cheng, L.; Wang, F.; Wu, P.; Whelan, J.; et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 2009, 151, 262–274. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Lan, P. Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopdsis roots. BMC Res. Notes 2015, 8, 555. [Google Scholar] [CrossRef] [Green Version]
- Rai, V.; Sanagala, R.; Sinilal, B.; Yadav, S.; Sarkar, A.K.; Dantu, P.K.; Jain, A. Iron availability affects phosphate deficiency-mediated responses, and evidence of cross-talk with auxin and zinc in Arabidopsis. Plant Cell Physiol. 2015, 56, 1107–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chutia, R.; Scharfenberg, S.; Neumann, S.; Abel, S.; Ziegler, J. Modulation of phosphate deficiency-induced metabolic changes by iron availability in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 7609. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.; Marin, E.; Floriani, M.; Chiarenza, S.; Richaud, P.; Nussaume, L.; Thibaud, M. Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 2006, 88, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Bournier, M.; Tissot, N.; Mari, S.; Boucherez, J.; Lacombe, E.; Briat, J.-F.; Gaymard, F. Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 2013, 288, 22670–22680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiavi, N.B. Characterization of the Arabidopsis thaliana RING-Type Ubiquitin Ligase XBAT31.1 and Its Role in Response to Iron Deficiency Stress; Dalhousie University: Halifax, NS, Canada, 2017. [Google Scholar]
- Bastow, E.L.; de la Torre, V.S.G.; Maclean, A.E.; Green, R.T.; Merlot, S.; Thomine, S.; Balk, J. Vacuolar iron stores gated by NRAMP3 and NRAMP4 are the primary source of iron in germinating seeds. Plant Physiol. 2018, 177, 1267–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitham, S.A.; Lincoln, L.M.; Chowda-Reddy, R.; Dittman, J.D.; O’Rourke, J.A.; Graham, M.A. Virus-Induced Gene Silencing and Transient Gene Expression in Soybean (Glycine max) Using Bean Pod Mottle Virus Infectious Clones. Curr. Protoc. Plant Biol. 2016, 1, 263–283. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Bradshaw, J.D.; Whitham, S.A.; Hill, J.H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010, 153, 52–65. [Google Scholar] [CrossRef] [Green Version]
- Chaney, R.L.; Coulombe, B.A.; Bell, P.F.; Angle, J.S. Detailed method to screen dicot cultivars for resistance to Fe-chlorosis using FeDTPA and bicarbonate in nutrient solutions. J. Plant Nutr. 1992, 15, 2063–2083. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Graham, M.A.; Vodkin, L.; Gonzalez, D.O.; Cianzio, S.R.; Shoemaker, R.C. Recovering from iron deficiency chlorosis in near-isogenic soybeans: A microarray study. Plant Physiol. Biochem. 2007, 45, 287–292. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinform. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 January 2020).
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- RStudio Team. RStudio: Integrated Development for R; RStudio Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Lawrence, M.; Gentleman, R.; Carey, V. rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 2009, 25, 1841–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Nature: Houston, TX, USA, 2016. [Google Scholar]
- Fisher, R. The Design of Experiments, 8th ed.; London Oliver and Boyd: Edinburgh, UK, 1966. [Google Scholar]
- Bonferroni, C. III Calcolo delle assicurazioni su gruppi di teste. Studi Onore Del Profr. Salvatore Ortu Carboni 1935, 13–60. [Google Scholar]
- Wang, Z.; Libault, M.; Joshi, T.; Valliyodan, B.; Nguyen, H.T.; Xu, D.; Stacey, G.; Cheng, J. SoyDB: A knowledge database of soybean transcription factors. BMC Plant Biol. 2010, 10, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environmnet for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Gribskov, M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; Van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]


| GO ID | # of DEGs | Corrected p-Value | Description |
|---|---|---|---|
| GO:0016036 | 16 | 3.02 × 10−6 | Cellular response to phosphate starvation |
| GO:0030643 | 3 | 0.005 | Cellular phosphate ion homeostasis |
| GO:0019375 | 10 | 0.005 | Galactolipid biosynthetic process |
| GO:0055072 | 6 | 0.007 | Iron ion homeostasis |
| GO:0006879 | 4 | 0.011 | Cellular iron ion homeostasis |
| GO:0006091 | 7 | 0.012 | Generation of precursor metabolites and energy |
| GO:0006817 | 5 | 0.018 | Phosphate ion transport |
| GO:0015979 | 13 | 0.020 | Photosynthesis |
| GO:0010043 | 7 | 0.035 | Response to zinc ion |
| GO ID | # of DEGs | Corrected p-Value | Description |
|---|---|---|---|
| GO:0019375 | 6 | 0.0001 | Galactolipid biosynthetic process |
| GO:0016036 | 6 | 0.001 | Cellular response to -Pi stress |
| GO:0030643 | 2 | 0.002 | Cellular phosphate ion homeostasis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Rourke, J.A.; Morrisey, M.J.; Merry, R.; Espina, M.J.; Lorenz, A.J.; Stupar, R.M.; Graham, M.A. Mining Fiskeby III and Mandarin (Ottawa) Expression Profiles to Understand Iron Stress Tolerant Responses in Soybean. Int. J. Mol. Sci. 2021, 22, 11032. https://doi.org/10.3390/ijms222011032
O’Rourke JA, Morrisey MJ, Merry R, Espina MJ, Lorenz AJ, Stupar RM, Graham MA. Mining Fiskeby III and Mandarin (Ottawa) Expression Profiles to Understand Iron Stress Tolerant Responses in Soybean. International Journal of Molecular Sciences. 2021; 22(20):11032. https://doi.org/10.3390/ijms222011032
Chicago/Turabian StyleO’Rourke, Jamie A., Michael J. Morrisey, Ryan Merry, Mary Jane Espina, Aaron J. Lorenz, Robert M. Stupar, and Michelle A. Graham. 2021. "Mining Fiskeby III and Mandarin (Ottawa) Expression Profiles to Understand Iron Stress Tolerant Responses in Soybean" International Journal of Molecular Sciences 22, no. 20: 11032. https://doi.org/10.3390/ijms222011032
APA StyleO’Rourke, J. A., Morrisey, M. J., Merry, R., Espina, M. J., Lorenz, A. J., Stupar, R. M., & Graham, M. A. (2021). Mining Fiskeby III and Mandarin (Ottawa) Expression Profiles to Understand Iron Stress Tolerant Responses in Soybean. International Journal of Molecular Sciences, 22(20), 11032. https://doi.org/10.3390/ijms222011032






