β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis
Abstract
:1. Introduction
2. Aggregation-Prone β-Barrel Proteins: Structure, Diversity, and Biological Roles
2.1. DNA-Binding Proteins of Viruses
2.2. Outer Membrane Proteins of Gram-Negative Bacteria
2.3. Proteins Containing the Cold-Shock Domain
2.4. Cupins
2.5. GFP-like Proteins
2.6. Other Amyloidogenic Eukaryotic β-Barrel Proteins
3. Folding of β-Barrel Proteins and Their Aggregation
3.1. Particularities of the β-Barrels’ Folding
3.2. Formation of the Quaternary Structure of β-Barrels as a Result of Their Oligomerization
4. The Transition of β-Barrel Proteins to Amyloids
4.1. Misfolding of Proteins: Pathological and Functional Amyloids
4.2. Diversity and Possible Biological Roles of Amyloids Formed by β-Barrel Proteins
4.2.1. Amyloid Formation from β-Barrel Proteins of Viruses
4.2.2. Amyloid Formation by β-Barrel Proteins of Prokaryotes
4.2.3. Amyloid Formation by β-Barrel Proteins of Eukaryotes
5. Interrelation between Amyloid Pathogenesis and β-Barrel Formation
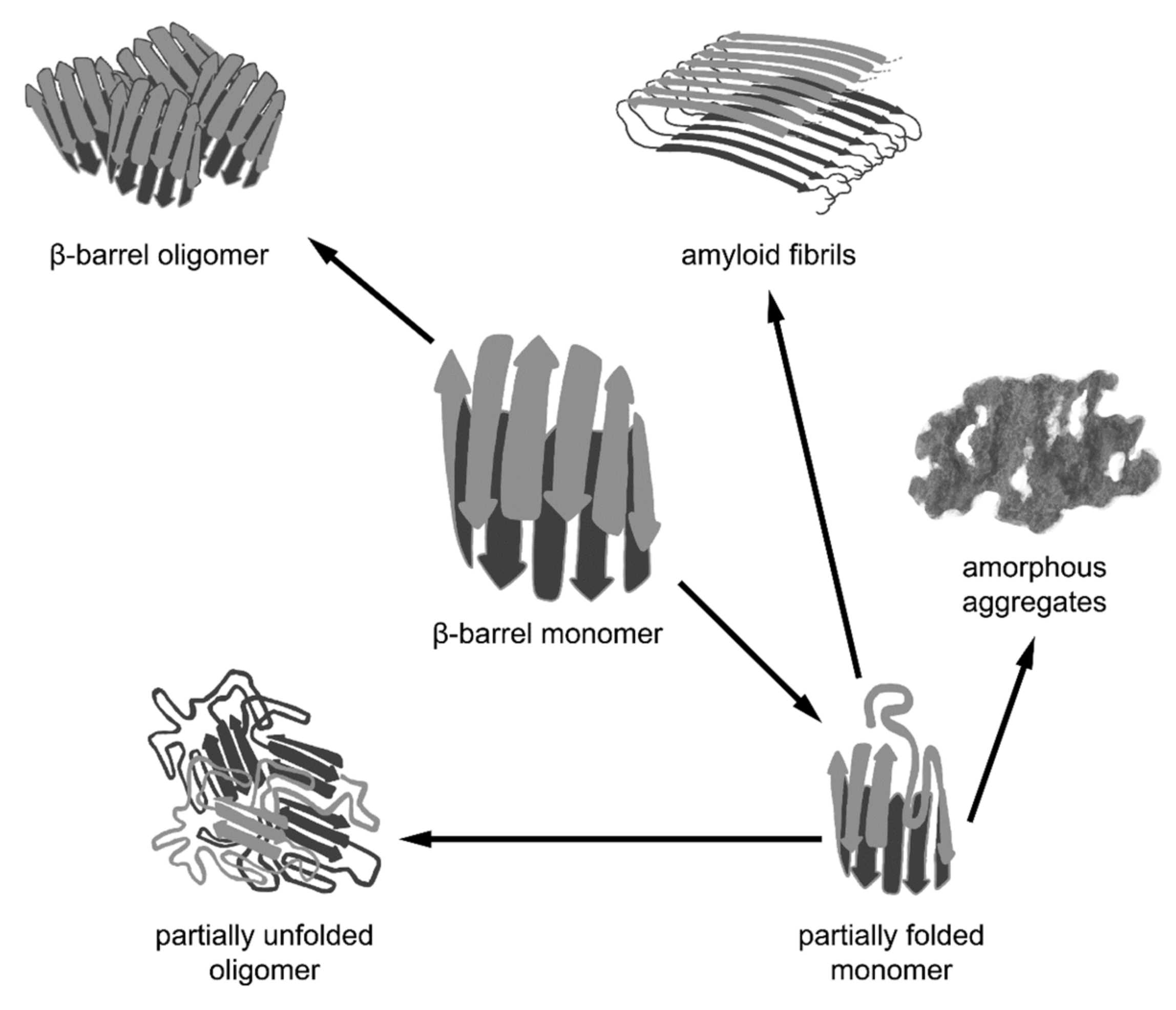
5.1. β-Barrel Formation as the ”Off-Pathway” of Fibrillogenesis
5.2. β-Barrels as a Universal Intermediate State in Fibrillogenesis
6. Conclusions
| Amyloid Properties | Methods * | Protein |
|---|---|---|
| Formation of fibrillar structures | TEM or AFM | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], HPV16 E2 [47], RopA [64], RopB [64], CspA [185], SOD1 [208], YB-1 [209] |
| High turbidity and Rayleigh light scattering compared to monomeric proteins | Absorption and fluorescence spectroscopy | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], HPV16 E2 [47], EBNA-1 [48], RopA [64], RopB [64] |
| High content of β-sheets and β-turns | CD, FTIR | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], RopA [64], RopB [64], YB-1 [209], HPV16 E2 [47], EBNA-1 [48], nFGF-1 [186] |
| Resistance to treatment with ionic detergents and proteases | Treatment with denaturants/proteases | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], EBNA-1 [48], RopA [64], RopB [64] |
| Interaction with amyloid-specific fluorescent probes (ThT, CR);significant increase in the fluorescence quantum yield and fluorescence lifetime, as well as a shift in the absorption and fluorescence excitation spectra of the dyes upon incorporation into amyloid fibrils;ability to visualize aggregates in the presence of fluorescent probes using confocal microscopy | Tinctorial methods (including using spectroscopic approaches and confocal fluorescence microscopy) | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], HPV16 E2 [47], EBNA-1 [48], RopA [64], RopB [64], SOD1 [208] |
| Apple-green birefringence in polarized light when stained with CR | Polarized light microscopy | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], RopA [64], RopB [64], YB-1 [209] |
| The presence of two scattering diffraction signals indicative of the cross-β structure | XRD | Vicilin [18], cupin-1.1 [18], cupin-1.2 [18], YB-1 [209] |
| Protein or Peptide | Structure of Monomer | Polymorphism | Secondary Structure Changes * | Method(s) by Which Changes in the Secondary Structure Were Detected * | Ref. |
|---|---|---|---|---|---|
| RopA | Predicted β-barrel structure | Mostly unstructured aggregates with the admixture of more ordered fibril-like structures | Before aggregation, more than 40% of β-structures; after aggregation, 42% of β-structures | CD | [64] |
| Amyloid fibrils | Before aggregation, more than 40% of β-structures; after aggregation, 48% of β-structures | CD | [64] | ||
| RopB | Predicted β-barrel structure | Mostly unstructured aggregates with the admixture of more ordered fibril-like structures | Before aggregation, more than 30% of β-structures; after aggregation, 38% of β-structures | CD | [64] |
| Amyloid fibrils | Before aggregation, more than 30% of β-structures; after aggregation, 44% of β-structures | CD | [64] | ||
| β-Lactoglobulin | 162 amino acid residues that fold up into an 8-stranded, antiparallel β-barrel with a 3-turn α-helix on the outer surface and a ninth β-strand flanking the first strand | Spherical particles | Increase in β-sheet content in the particulate form relative to the native form of the protein, as well as a shift in the β-sheet band from ∼1629 cm−1 to ∼1625 cm−1 | FTIR | [213,248] |
| Fibrillar structure | Intramolecular β-sheets (Delta 1632 cm−1) decreased and intermolecular β-sheets (Delta 1622 cm−1) increased | FTIR | [123,245,249,250,251,252] | ||
| OmpA | Disordered structure in the absence of lipid bilayers | Unfolded monomer | No regular structure | CD | [61] |
| Oligomeric amyloid-like state | Oligomeric form of the protein exhibited a spectrum indicative of β-sheet structure, but with a different shape and intensity than the native β-barrel spectrum | CD | [61] | ||
| YB-1 | β-structure and random coil | Fibrillar structure | Increase in the content of β-structures in comparison with monomeric protein | CD | [209] |
| Globular particles | CD spectra typical for unfolded proteins | CD | [209] | ||
| Vicilin (full-length) | α-helix, β-structure, and random coil | Fibrillar structure containing a fraction of less structured aggregates | Before aggregation, ~39% of β-structures; after aggregation, 41% of β-structures | CD | [18] |
| Cupin-1.1 (the domain of vicilin) | α -helix, β-structure, and random coil | Fibrillar structure | Before aggregation, ~4% of β-structures; after aggregation, 40% of β-structures | CD | [18] |
| Cupin-1.2 (the domain of vicilin) | α-helix, β-structure, and random coil | Fibrillar structure | Before aggregation, ~12% of β-structures; after aggregation, 42% of β-structures | CD | [18] |
| Papillomavirus HPV16 E2 | α-helix, β-structure, and random coil | Monomer | Two negative bands at 212 and 225 nm indicative of β-barrel structure | CD | [47] |
| Granular structures and small annuli with diameters of ∼5 nm and 10 nm, respectively | No data | No data | [47] | ||
| Amyloid-like fibrils | Increase in the content of β-structures in comparison with monomer | CD | [47] | ||
| Epstein–Barr virus EBNA1 | α-helix, β-structure, and random coil | Dimer or monomer | Characteristic bands for α-helix at 208 and 222 nm | CD | [48] |
| Spherical oligomers | A decrease in α-helical content | CD | [48] | ||
| nFGF-1 | All β-sheet structure | Monomer | CD data: two bands at 228 nm and 205 nm indicative of β-barrel structure; FTIR data: 1618 and 1639 cm−1 amide I bands indicating the β-barrel structure | CD, FTIR | [186] |
| Fibrillar structure | CD data: the β-barrel conformation is disorganized (the 228 nm ellipticity band disappears), resulting in the formation of extended β-sheet conformation (formation of the negative band at 218 nm); FTIR data: 1618 and 1639 cm−1 amide I bands disappear; new band at1625 cm−1 (indicating the formation of extended β-sheets) is formed | CD, FTIR | [186] |
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sipe, J.D.; Benson, M.D.; Buxbaum, J.N.; Ikeda, S.; Merlini, G.; Saraiva, M.J.M.; Westermark, P. Amyloid fibril proteins and amyloidosis: Chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid 2016, 23, 209–213. [Google Scholar] [CrossRef]
- Nelson, R.; Eisenberg, D. Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 2006, 16, 260–265. [Google Scholar] [CrossRef]
- Ivanova, M.I.; Thompson, M.J.; Eisenberg, D. A systematic screen of beta(2)-microglobulin and insulin for amyloid-like segments. Proc. Natl. Acad. Sci. USA 2006, 103, 4079–4082. [Google Scholar] [CrossRef] [Green Version]
- Sunde, M.; Serpell, L.C.; Bartlam, M.; Fraser, P.E.; Pepys, M.B.; Blake, C.C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [Google Scholar] [CrossRef] [Green Version]
- Sipe, J.D.; Cohen, A.S. Review: History of the amyloid fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Eanes, E.D.; Glenner, G.G. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968, 16, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.; McKinley, M.; Prusiner, S. Identification of a protein that purifies with the scrapie prion. Science 1982, 218, 1309–1311. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.; Ihara, Y.; Salazar, F. Alzheimer’s disease: Insolubility of partially purified paired helical filaments in sodium dodecyl sulfate and urea. Science 1982, 215, 1243–1245. [Google Scholar] [CrossRef] [PubMed]
- Bennhold, H. Specific staining of amyloid by Congo red (in German). Muenchen. Med. Wochenschr. 1922, 69, 1537–1538. Available online: https://archive.org/details/munchenermedizin6921unse/page/1536/mode/2up (accessed on 8 September 2021). (in German).
- Divry, P.; Florkin, M. Sur les propriétés optiques de l’amyloïde. C. R. Soc. Biol. 1927, 97, 1808–1810. [Google Scholar]
- Naiki, H.; Higuchi, K.; Hosokawa, M.; Takeda, T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavine T. Anal. Biochem. 1989, 177, 244–249. [Google Scholar] [CrossRef]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2020: Update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [Green Version]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid—From bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef]
- Nizhnikov, A.A.; Antonets, K.S.; Inge-Vechtomov, S.G. Amyloids: From pathogenesis to function. Biochem. Mosc. 2015, 80, 1127–1144. [Google Scholar] [CrossRef]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef]
- Kosolapova, A.O.; Antonets, K.S.; Belousov, M.V.; Nizhnikov, A.A. Biological Functions of Prokaryotic Amyloids in Interspecies Interactions: Facts and Assumptions. Int. J. Mol. Sci. 2020, 21, 7240. [Google Scholar] [CrossRef] [PubMed]
- Antonets, K.S.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Kosolapova, A.O.; Sulatsky, M.I.; Andreeva, E.A.; Zykin, P.A.; Malovichko, Y.V.; Shtark, O.Y.; et al. Accumulation of storage proteins in plant seeds is mediated by amyloid formation. PLoS Biol. 2020, 18, e3000564. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.B.; Kajava, A.V. Breaking the amyloidogenicity code: Methods to predict amyloids from amino acid sequence. FEBS Lett. 2013, 587, 1089–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelitsch, M.D.; Weissman, J.S. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 2000, 97, 11910–11915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurer-Stroh, S.; Debulpaep, M.; Kuemmerer, N.; Lopez de la Paz, M.; Martins, I.C.; Reumers, J.; Morris, K.L.; Copland, A.; Serpell, L.; Serrano, L.; et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 2010, 7, 237–242. [Google Scholar] [CrossRef]
- Olzscha, H. Posttranslational modifications and proteinopathies: How guardians of the proteome are defeated. Biol. Chem. 2019, 400, 895–915. [Google Scholar] [CrossRef]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Prions, chaperones, and proteostasis in yeast. Cold Spring Harb. Perspect. Biol. 2017, 9, a023663. [Google Scholar] [CrossRef] [PubMed]
- Shafrir, Y.; Durell, S.R.; Anishkin, A.; Guy, H.R. Beta-barrel models of soluble amyloid beta oligomers and annular protofibrils. Proteins Struct. Funct. Bioinf. 2010, 78, 3458–3472. [Google Scholar] [CrossRef] [Green Version]
- Fairman, J.W.; Noinaj, N.; Buchanan, S.K. The structural biology of β-barrel membrane proteins: A summary of recent reports. Curr. Opin. Struct. Biol. 2011, 21, 523–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, R.M.; Mizuguchi, K.; Dhanaraj, V.; Albert, A.; Blundell, T.L.; Murzin, A.G. A six-stranded double-psi β barrel is shared by several protein superfamilies. Structure 1999, 7, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Thangappan, J.; Wu, S.; Lee, S.G. Comparative Analysis of TM and Cytoplasmic β-barrel Conformations Using Joint Descriptor. Sci. Rep. 2018, 8, 14185. [Google Scholar] [CrossRef] [PubMed]
- Dhar, R.; Feehan, R.; Slusky, J.S.G. Membrane Barrels Are Taller, Fatter, Inside-Out Soluble Barrels. J. Phys. Chem. B 2021, 125, 3622–3628. [Google Scholar] [CrossRef]
- Delcour, A.H. Structure and function of pore-forming beta-barrels from bacteria. J. Mol. Microbiol. Biotechnol. 2002, 4, 1–10. [Google Scholar]
- Bishop, R.E. Structural biology of membrane-intrinsic β-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim. Biophys. Acta Biomembr. 2008, 1778, 1881–1896. [Google Scholar] [CrossRef] [Green Version]
- Noinaj, N.; Kuszak, A.J.; Gumbart, J.C.; Lukacik, P.; Chang, H.; Easley, N.C.; Lithgow, T.; Buchanan, S.K. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 2013, 501, 385–390. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of Staphylococcal alpha -Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1865. [Google Scholar] [CrossRef]
- Kikuchi, N.; Fujiwara, K.; Ikeguchi, M. β-Strand twisting/bending in soluble and transmembrane β-barrel structures. Proteins Struct. Funct. Bioinf. 2018, 86, 1231–1241. [Google Scholar] [CrossRef]
- Tamm, L.K.; Arora, A.; Kleinschmidt, J.H. Structure and assembly of beta-barrel membrane proteins. J. Biol. Chem. 2001, 276, 32399–32402. [Google Scholar] [CrossRef] [Green Version]
- Remmert, M.; Biegert, A.; Linke, D.; Lupas, A.N.; Söding, J. Evolution of Outer Membrane β-Barrels from an Ancestral ββ Hairpin. Mol. Biol. Evol. 2010, 27, 1348–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorobieva, A.A.; White, P.; Liang, B.; Horne, J.E.; Bera, A.K.; Chow, C.M.; Gerben, S.; Marx, S.; Kang, A.; Stiving, A.Q.; et al. De novo design of transmembrane β barrels. Science 2021, 371, eabc8182. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D.E.; Andersen, K.K. Folding of outer membrane proteins. Arch. Biochem. Biophys. 2013, 531, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Roske, Y. Cold-Shock Domains-Abundance, Structure, Properties, and Nucleic-Acid Binding. Cancers 2021, 13, 190. [Google Scholar] [CrossRef]
- Agarwal, G.; Rajavel, M.; Gopal, B.; Srinivasan, N. Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold. PLoS ONE 2009, 4, e5736. [Google Scholar] [CrossRef]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef]
- Brito, N.F.; Oliveira, D.S.; Santos, T.C.; Moreira, M.F.; Melo, A.C.A. Current and potential biotechnological applications of odorant-binding proteins. Appl. Microbiol. Biotechnol. 2020, 104, 8631–8648. [Google Scholar] [CrossRef] [PubMed]
- Forest, K.T.; Langford, P.R.; Kroll, J.S.; Getzoff, E.D. Cu, Zn superoxide dismutase structure from a microbial pathogen establishes a class with a conserved dimer interface. J. Mol. Biol. 2000, 296, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, M.; Marcinkowska, E.; Wiedlocha, A. FGF-1: From biology through engineering to potential medical applications. Crit. Rev. Clin. Lab. Sci. 2008, 45, 91–135. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Barwell, J.A.; Pfuetzner, R.A.; Furey, W., Jr.; Edwards, A.M.; Frappier, L. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 1995, 83, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Hegde, R.S.; Grossman, S.R.; Laimins, L.A.; Sigler, P.B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 1992, 359, 505–512. [Google Scholar] [CrossRef]
- Dheekollu, J.; Wiedmer, A.; Ayyanathan, K.; Deakyne, J.S.; Messick, T.E.; Lieberman, P.M. Cell-cycle-dependent EBNA1-DNA crosslinking promotes replication termination at oriP and viral episome maintenance. Cell 2021, 184, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Wetzler, D.E.; Castano, E.M.; de Prat-Gay, G. A quasi-spontaneous amyloid route in a DNA binding gene regulatory domain: The papillomavirus HPV16 E2 protein. Protein Sci. 2007, 16, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Freire, E.; Oddo, C.; Frappier, L.; de Prat-Gay, G. Kinetically driven refolding of the hyperstable EBNA1 origin DNA-binding dimeric beta-barrel domain into amyloid-like spherical oligomers. Proteins 2008, 70, 450–461. [Google Scholar] [CrossRef]
- Rollauer, S.E.; Sooreshjani, M.A.; Noinaj, N.; Buchanan, S.K. Outer membrane protein biogenesis in Gram-negative bacteria. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150023. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagès, J.M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef] [Green Version]
- Dekker, N. Outer-membrane phospholipase A: Known structure, unknown biological function. Mol. Microbiol. 2000, 35, 711–717. [Google Scholar] [CrossRef] [Green Version]
- Choi, U.; Lee, C.R. Distinct Roles of Outer Membrane Porins in Antibiotic Resistance and Membrane Integrity in Escherichia coli. Front. Microbiol. 2019, 10, 953. [Google Scholar] [CrossRef] [PubMed]
- Krewulak, K.D.; Vogel, H.J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta Biomembr. 2008, 1778, 1781–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moeck, G.S.; Coulton, J.W. TonB-dependent iron acquisition: Mechanisms of siderophore-mediated active transport. Mol. Microbiol. 1998, 28, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Mach, T.; Neves, P.; Spiga, E.; Weingart, H.; Winterhalter, M.; Ruggerone, P.; Ceccarelli, M.; Gameiro, P. Facilitated permeation of antibiotics across membrane channels—Interaction of the quinolone moxifloxacin with the OmpF channel. J. Am. Chem. Soc. 2008, 130, 13301–13309. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, S.; Prasadarao, N.V. Outer membrane protein A and OprF: Versatile roles in Gram-negative bacterial infections. FEBS J. 2012, 279, 919–931. [Google Scholar] [CrossRef]
- Kim, K.; Kim, K.P.; Choi, J.; Lim, J.A.; Lee, J.; Hwang, S.; Ryu, S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of cronobacter sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kim, K.S. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Pediatr. Res. 2002, 51, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Danoff, E.J.; Fleming, K.G. Aqueous, Unfolded OmpA Forms Amyloid-Like Fibrils upon Self-Association. PLoS ONE 2015, 10, e0132301. [Google Scholar] [CrossRef] [Green Version]
- Joseph Sahaya Rajan, J.; Chinnappan Santiago, T.; Singaravel, R.; Ignacimuthu, S. Outer membrane protein C (OmpC) of Escherichia coli induces neurodegeneration in mice by acting as an amyloid. Biotechnol. Lett. 2016, 38, 689–700. [Google Scholar] [CrossRef]
- Montes García, J.F.; Vaca, S.; Delgado, N.L.; Uribe-García, A.; Vázquez, C.; Sánchez Alonso, P.; Xicohtencatl Cortes, J.; Cruz Cordoba, A.; Negrete Abascal, E. Mannheimia haemolytica OmpP2-like is an amyloid-like protein, forms filaments, takes part in cell adhesion and is part of biofilms. Antonie Van Leeuwenhoek 2018, 111, 2311–2321. [Google Scholar] [CrossRef] [PubMed]
- Kosolapova, A.O.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Sulatsky, M.I.; Antonets, K.S.; Volkov, K.V.; Lykholay, A.N.; Shtark, O.Y.; Vasileva, E.N.; et al. Two novel amyloid proteins, RopA and RopB, from the root nodule bacterium Rhizobium leguminosarum. Biomolecules 2019, 9, 694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Pautsch, A.; Schulz, G.E. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 1998, 5, 1013–1017. [Google Scholar] [CrossRef]
- Pautsch, A.; Schulz, G.E. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 2000, 298, 273–282. [Google Scholar] [CrossRef] [Green Version]
- De Mot, R.; Vanderleyden, J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 1994, 12, 333–334. [Google Scholar] [CrossRef]
- Stathopoulos, C. An alternative topological model for Escherichia coli OmpA. Protein Sci. 1996, 5, 170–173. [Google Scholar] [CrossRef] [Green Version]
- Rao Movva, N.; Nakamura, K.; Inouye, M. Amino acid sequence of the signal peptide of ompA protein, a major outer membrane protein of Escherichia coli. J. Biol. Chem. 1980, 255, 27–29. [Google Scholar] [CrossRef]
- Smith, S.G.J.; Mahon, V.; Lambert, M.A.; Fagan, R.P. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2007, 273, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Snijder, H.J.; Ubarretxena-Belandia, I.; Blaauw, M.; Kalk, K.H.; Verheij, H.M.; Egmond, M.R.; Dekker, N.; Dijkstra, B.W. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 1999, 401, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Dekker, N.; Tommassen, J.; Lustig, A.; Rosenbusch, J.P.; Verheij, H.M. Dimerization regulates the enzymatic activity of Escherichia coli outer membrane phospholipase A. J. Biol. Chem. 1997, 272, 3179–3184. [Google Scholar] [CrossRef] [Green Version]
- Wu, E.L.; Fleming, P.J.; Yeom, M.S.; Widmalm, G.; Klauda, J.B.; Fleming, K.G.; Im, W. E. coli outer membrane and interactions with OmpLA. Biophys. J. 2014, 106, 2493–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graumann, P.L.; Marahiel, M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998, 23, 286–290. [Google Scholar] [CrossRef]
- Chaikam, V.; Karlson, D.T. Comparison of structure, function and regulation of plant cold shock domain proteins to bacterial and animal cold shock domain proteins. BMB Rep. 2010, 43, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.H.; Bloch, D.B. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA 2007, 13, 704–712. [Google Scholar] [CrossRef]
- Kozak, S.L.; Marin, M.; Rose, K.M.; Bystrom, C.; Kabat, D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006, 281, 29105–29119. [Google Scholar] [CrossRef] [Green Version]
- Gonda, K.; Wudel, J.; Nelson, D.; Katoku-Kikyo, N.; Reed, P.; Tamada, H.; Kikyo, N. Requirement of the protein B23 for nucleolar disassembly induced by the FRGY2a family proteins. J. Biol. Chem. 2006, 281, 8153–8160. [Google Scholar] [CrossRef] [Green Version]
- Janz, M.; Jurchott, K.; Karawajew, L.; Royer, H. Y-box factor YB-1 is associated with the centrosome during mitosis. Gene Funct. Dis. 2000, 1, 57–59. [Google Scholar] [CrossRef]
- Chernov, K.G.; Mechulam, A.; Popova, N.V.; Pastre, D.; Nadezhdina, E.S.; Skabkina, O.V.; Shanina, N.A.; Vasiliev, V.D.; Tarrade, A.; Melki, J.; et al. YB-1 promotes microtubule assembly in vitro through interaction with tubulin and microtubules. BMC Biochem. 2008, 9, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruzanov, P.V.; Evdokimova, V.M.; Korneeva, N.L.; Hershey, J.W.; Ovchinnikov, L.P. Interaction of the universal mRNA-binding protein, p50, with actin: A possible link between mRNA and microfilaments. J. Cell Sci. 1999, 112, 3487–3496. [Google Scholar] [CrossRef]
- Eliseeva, I.A.; Kim, E.R.; Guryanov, S.G.; Ovchinnikov, L.P.; Lyabin, D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry 2011, 76, 1402–1433. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M. Cupins: A New Superfamily of Functionally Diverse Proteins that Include Germins and Plant Storage Proteins. Biotechnol. Genet. Eng. Rev. 1998, 15, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Radauer, C.; Breiteneder, H. Evolutionary biology of plant food allergens. J. Allergy Clin. Immunol. 2007, 120, 518–525. [Google Scholar] [CrossRef]
- Shutov, A.D.; Kakhovskaya, I.A. Evolution of seed storage globulins and cupin superfamily. Mol. Biol. 2011, 45, 529–535. [Google Scholar] [CrossRef]
- Hussain, Z.; Wiedner, R.; Steiner, K.; Hajek, T.; Avi, M.; Hecher, B.; Sessitsch, A.; Schwab, H. Characterization of two bacterial hydroxynitrile lyases with high similarity to cupin superfamily proteins. Appl. Environ. Microbiol. 2012, 78, 2053–2055. [Google Scholar] [CrossRef] [Green Version]
- Galperin, M.Y.; Koonin, E.V. Divergence and convergence in enzyme evolution. J. Biol. Chem. 2012, 287, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Lane, B.G.; Dunwell, J.M.; Ray, J.A.; Schmitt, M.R.; Cuming, A.C. Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 1993, 268, 12239–12242. [Google Scholar] [CrossRef]
- Ferreira, R.B.; Monteiro, S.; Freitas, R.; Santos, C.N.; Chen, Z.; Batista, L.M.; Duarte, J.; Borges, A.; Teixeira, A.R. The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 2007, 8, 677–700. [Google Scholar] [CrossRef]
- Woo, E.J.; Dunwell, J.M.; Goodenough, P.W.; Marvier, A.C.; Pickersgill, R.W. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 2000, 7, 1036–1040. [Google Scholar] [CrossRef] [PubMed]
- Bäumlein, H.; Braun, H.; Kakhovskaya, I.A.; Shutov, A.D. Seed storage proteins of spermatophytes share a common ancestor with desiccation proteins of fungi. J. Mol. Evol. 1995, 41, 1070–1075. [Google Scholar] [CrossRef]
- Shutov, A.D.; Kakhovskaya, I.A.; Braun, H.; Bäumlein, H.; Müntz, K. Legumin-like and vicilin-like seed storage proteins: Evidence for a common single-domain ancestral gene. J. Mol. Evol. 1995, 41, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Monge, R.; Lopez-Torrejón, G.; Pascual, C.Y.; Varela, J.; Martin-Esteban, M.; Salcedo, G. Vicilin and convicilin are potential major allergens from pea. Clin. Exp. Allergy 2004, 34, 1747–1753. [Google Scholar] [CrossRef]
- Bar-El Dadon, S.; Pascual, C.Y.; Eshel, D.; Teper-Bamnolker, P.; Paloma Ibáñez, M.D.; Reifen, R. Vicilin and the basic subunit of legumin are putative chickpea allergens. Food Chem. 2013, 138, 13–18. [Google Scholar] [CrossRef]
- Sales, M.P.; Gerhardt, I.R.; Grossi-de-Sa, M.F.; Xavier-Filho, J. Do legume storage proteins play a role in defending seeds against bruchids? Plant Physiol. 2000, 124, 515–522. [Google Scholar] [CrossRef] [Green Version]
- Gomes, V.M.; Mosqueda, M.I.; Blanco-Labra, A.; Sales, M.P.; Fernandes, K.V.S.; Cordeiro, R.A.; Xavier-Filho, J. Vicilin Storage Proteins from Vigna unguiculata (Legume) Seeds Inhibit Fungal Growth. J. Agric. Food Chem. 1997, 45, 4110–4115. [Google Scholar] [CrossRef]
- Craggs, T.D. Green fluorescent protein: Structure, folding and chromophore maturation. Chem. Soc. Rev. 2009, 38, 2865–2875. [Google Scholar] [CrossRef]
- Pakhomov, A.A.; Martynov, V.I. GFP family: Structural insights into spectral tuning. Chem. Biol. 2008, 15, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Remington, S.J. Fluorescent proteins: Maturation, photochemistry and photophysics. Curr. Opin. Struct. Biol. 2006, 16, 714–721. [Google Scholar] [CrossRef]
- Van Thor, J.J. Photoreactions and dynamics of the green fluorescent protein. Chem. Soc. Rev. 2009, 38, 2935–2950. [Google Scholar] [CrossRef]
- Day, R.N.; Davidson, M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009, 38, 2887–2921. [Google Scholar] [CrossRef] [Green Version]
- Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Verkhusha, V.V.; Turoverov, K.K. Beta-barrel scaffold of fluorescent proteins: Folding, stability and role in chromophore formation. Int. Rev. Cell Mol. Biol. 2013, 302, 221–278. [Google Scholar] [CrossRef] [Green Version]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef]
- Macel, M.L.; Ristoratore, F.; Locascio, A.; Spagnuolo, A.; Sordino, P.; D’Aniello, S. Sea as a color palette: The ecology and evolution of fluorescence. Zool. Lett. 2020, 6, 9. [Google Scholar] [CrossRef]
- Tsutsui, K.; Shimada, E.; Ogawa, T.; Tsuruwaka, Y. A novel fluorescent protein from the deep-sea anemone Cribrinopsis japonica (Anthozoa: Actiniaria). Sci. Rep. 2016, 6, 23493. [Google Scholar] [CrossRef] [Green Version]
- Hunt, M.E.; Scherrer, M.P.; Ferrari, F.D.; Matz, M.V. Very bright green fluorescent proteins from the Pontellid copepod Pontella mimocerami. PLoS ONE 2010, 5, e11517. [Google Scholar] [CrossRef] [Green Version]
- Shagin, D.A.; Barsova, E.V.; Yanushevich, Y.G.; Fradkov, A.F.; Lukyanov, K.A.; Labas, Y.A.; Semenova, T.N.; Ugalde, J.A.; Meyers, A.; Nunez, J.M.; et al. GFP-like proteins as ubiquitous metazoan superfamily: Evolution of functional features and structural complexity. Mol. Biol. Evol. 2004, 21, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Marshall, N.J.; Vorobyev, M. Are corals colorful? Photochem. Photobiol. 2006, 82, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Scucchia, F.; Nativ, H.; Neder, M.; Goodbody-Gringley, G.; Mass, T. Physiological characteristics of Stylophora pistillata larvae across a depth gradient. Front. Mar. Sci. 2020, 7, 13. [Google Scholar] [CrossRef]
- Salih, A.; Larkum, A.; Cox, G.; Kuhl, M.; Hoegh-Guldberg, O. Fluorescent pigments in corals are photoprotective. Nature 2000, 408, 850–853. [Google Scholar] [CrossRef]
- Lyndby, N.H.; Kuhl, M.; Wangpraseurt, D. Heat generation and light scattering of green fluorescent protein-like pigments in coral tissue. Sci. Rep. 2016, 6, 26599. [Google Scholar] [CrossRef] [Green Version]
- Bou-Abdallah, F.; Chasteen, N.D.; Lesser, M.P. Quenching of superoxide radicals by green fluorescent protein. Biochim. Biophys. Acta Gen. Subj. 2006, 1760, 1690–1695. [Google Scholar] [CrossRef] [Green Version]
- Bogdanov, A.M.; Mishin, A.S.; Yampolsky, I.V.; Belousov, V.V.; Chudakov, D.M.; Subach, F.V.; Verkhusha, V.V.; Lukyanov, S.; Lukyanov, K.A. Green fluorescent proteins are light-induced electron donors. Nat. Chem. Biol. 2009, 5, 459–461. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Stepanenko, O.V.; Shcherbakova, D.M.; Kuznetsova, I.M.; Turoverov, K.K.; Verkhusha, V. V Modern fluorescent proteins: From chromophore formation to novel intracellular applications. Biotechniques 2011, 51, 313–327. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Verkhusha, V.V.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Fluorescent proteins as biomarkers and biosensors: Throwing color lights on molecular and cellular processes. Curr. Protein. Pept. Sci. 2008, 9, 338–369. [Google Scholar] [CrossRef] [Green Version]
- Zimmer, M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem. Rev. 2002, 102, 759–781. [Google Scholar] [CrossRef]
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, A.E.; Cousens, L.S.; Weaver, L.H.; Matthews, B.W. Three-dimensional structure of human basic fibroblast growth factor. Proc. Natl. Acad. Sci. USA 1991, 88, 3441–3445. [Google Scholar] [CrossRef] [Green Version]
- Gimenez-Gallego, G.; Rodkey, J.; Bennett, C.; Rios-Candelore, M.; DiSalvo, J.; Thomas, K. Brain-derived acidic fibroblast growth factor: Complete amino acid sequence and homologies. Science 1985, 230, 1385–1388. [Google Scholar] [CrossRef]
- Xiang, L.-W.; Melton, L.D.; Leung, I.K.H. Interactions of β-Lactoglobulin with Small Molecules. In Encyclopedia of Food Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 560–565. [Google Scholar] [CrossRef]
- Heyn, T.R.; Mayer, J.; Neumann, H.R.; Selhuber-Unkel, C.; Kwade, A.; Schwarz, K.; Keppler, J.K. The threshold of amyloid aggregation of beta-lactoglobulin: Relevant factor combinations. J. Food Eng. 2020, 283, 110005. [Google Scholar] [CrossRef]
- Franklin, M.W.; Nepomnyachiy, S.; Feehan, R.; Ben-Tal, N.; Kolodny, R.; Slusky, J.S.G. Efflux Pumps Represent Possible Evolutionary Convergence onto the beta-Barrel Fold. Structure 2018, 26, 1266–1274. [Google Scholar] [CrossRef] [Green Version]
- Pande, V.S.; Grosberg, A.; Tanaka, T.; Rokhsar, D.S. Pathways for protein folding: Is a new view needed? Curr. Opin. Struct. Biol. 1998, 8, 68–79. [Google Scholar] [CrossRef]
- Dinner, A.R.; Sali, A.; Smith, L.J.; Dobson, C.M.; Karplus, M. Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem. Sci. 2000, 25, 331–339. [Google Scholar] [CrossRef]
- Jahn, T.R.; Radford, S.E. The Yin and Yang of protein folding. FEBS J. 2005, 272, 5962–5970. [Google Scholar] [CrossRef]
- Tamm, L.K.; Hong, H.; Liang, B. Folding and assembly of beta-barrel membrane proteins. Biochim. Biophys. Acta Biomembr. 2004, 1666, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, D.; Mahalakshmi, R. Transmembrane beta-barrels: Evolution, folding and energetics. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2467–2482. [Google Scholar] [CrossRef]
- Kleinschmidt, J.H. Folding of beta-barrel membrane proteins in lipid bilayers—Unassisted and assisted folding and insertion. Biochim. Biophys. Acta Biomembr. 2015, 1848, 1927–1943. [Google Scholar] [CrossRef] [Green Version]
- Doyle, M.T.; Bernstein, H.D. Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel. Nat. Commun. 2019, 10, 3358. [Google Scholar] [CrossRef] [Green Version]
- Ugrinov, K.G.; Clark, P.L. Cotranslational folding increases GFP folding yield. Biophys. J. 2010, 98, 1312–1320. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.S.A. Molecular and pharmacological chaperones for SOD1. Biochem. Soc. Trans. 2020, 48, 1795–1806. [Google Scholar] [CrossRef]
- Watanabe, M.; Dykes-Hoberg, M.; Cizewski Culotta, V.; Price, D.L.; Wong, P.C.; Rothstein, J.D. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 2001, 8, 933–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisberg, S.J.; Lyakhovetsky, R.; Werdiger, A.C.; Gitler, A.D.; Soen, Y.; Kaganovich, D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc. Natl. Acad. Sci. USA 2012, 109, 15811–15816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Close, D.W.; Paul, C.D.; Langan, P.S.; Wilce, M.C.J.; Traore, D.A.K.; Halfmann, R.; Rocha, R.C.; Waldo, G.S.; Payne, R.J.; Rucker, J.B.; et al. Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering. Proteins Struct. Funct. Bioinf. 2015, 83, 1225–1237. [Google Scholar] [CrossRef] [Green Version]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Yanushevich, Y.G.; Staroverov, D.B.; Savitsky, A.P.; Fradkov, A.F.; Gurskaya, N.G.; Bulina, M.E.; Lukyanov, K.A.; Lukyanov, S.A. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 2002, 511, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Roldan-Salgado, A.; Sanchez-Barreto, C.; Gaytan, P. LanFP10-A, first functional fluorescent protein whose chromophore contains the elusive mutation G67A. Gene 2016, 592, 281–290. [Google Scholar] [CrossRef]
- Aglyamova, G.V.; Hunt, M.E.; Modi, C.K.; Matz, M.V. Multi-colored homologs of the green fluorescent protein from hydromedusa Obelia sp. Photochem. Photobiol. Sci. 2011, 10, 1303–1309. [Google Scholar] [CrossRef]
- Hunt, M.E.; Modi, C.K.; Aglyamova, G.V.; Ravikant, D.V.; Meyer, E.; Matz, M.V. Multi-domain GFP-like proteins from two species of marine hydrozoans. Photochem. Photobiol. Sci. 2012, 11, 637–644. [Google Scholar] [CrossRef]
- Cranfill, P.J.; Sell, B.R.; Baird, M.A.; Allen, J.R.; Lavagnino, Z.; de Gruiter, H.M.; Kremers, G.J.; Davidson, M.W.; Ustione, A.; Piston, D.W. Quantitative assessment of fluorescent proteins. Nat. Methods 2016, 13, 557–562. [Google Scholar] [CrossRef] [Green Version]
- Tsutsui, H.; Jinno, Y.; Shoda, K.; Tomita, A.; Matsuda, M.; Yamashita, E.; Katayama, H.; Nakagawa, A.; Miyawaki, A. A diffraction-quality protein crystal processed as an autophagic cargo. Mol. Cell 2015, 58, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Povarova, N.V.; Petri, N.D.; Blokhina, A.E.; Bogdanov, A.M.; Gurskaya, N.G.; Lukyanov, K.A. Functioning of Fluorescent Proteins in Aggregates in Anthozoa Species and in Recombinant Artificial Models. Int. J. Mol. Sci. 2017, 18, 1503. [Google Scholar] [CrossRef] [Green Version]
- Deakyne, J.S.; Malecka, K.A.; Messick, T.E.; Lieberman, P.M. Structural and Functional Basis for an EBNA1 Hexameric Ring in Epstein-Barr Virus Episome Maintenance. J. Virol. 2017, 91, e01046-17. [Google Scholar] [CrossRef] [Green Version]
- Malecka, K.A.; Dheekollu, J.; Deakyne, J.S.; Wiedmer, A.; Ramirez, U.D.; Lieberman, P.M.; Messick, T.E. Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication. J. Virol. 2019, 93, e00487-19. [Google Scholar] [CrossRef]
- Leroy, L.; Barbosa, J.; de Prat-Gay, G.; Polikarpov, I.; Pinheiro, C.B. The structure of the extended E2 DNA-binding domain of the bovine papillomavirus-1. Proteins Struct. Funct. Bioinf. 2020, 88, 106–112. [Google Scholar] [CrossRef]
- Hegde, R.S.; Wang, A.F.; Kim, S.S.; Schapira, M. Subunit rearrangement accompanies sequence-specific DNA binding by the bovine papillomavirus-1 E2 protein. J. Mol. Biol. 1998, 276, 797–808. [Google Scholar] [CrossRef]
- Sim, J.; Ozgur, S.; Lin, B.Y.; Yu, J.H.; Broker, T.R.; Chow, L.T.; Griffith, J. Remodeling of the human papillomavirus type 11 replication origin into discrete nucleoprotein particles and looped structures by the E2 protein. J. Mol. Biol. 2008, 375, 1165–1177. [Google Scholar] [CrossRef] [Green Version]
- Makhatadze, G.I.; Marahiel, M.A. Effect of pH and phosphate ions on self-association properties of the major cold-shock protein from Bacillus subtilis. Protein Sci. 1994, 3, 2144–2147. [Google Scholar] [CrossRef] [Green Version]
- Morgan, H.P.; Wear, M.A.; McNae, I.; Gallagher, M.P.; Walkinshaw, M.D. Crystallization and X-ray structure of cold-shock protein E from Salmonella typhimurium. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 1240–1245. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, H.; Marahiel, M.A.; Heinemann, U. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 1993, 364, 164–168. [Google Scholar] [CrossRef]
- Max, K.E.; Zeeb, M.; Bienert, R.; Balbach, J.; Heinemann, U. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J. 2007, 274, 1265–1279. [Google Scholar] [CrossRef]
- Ren, J.; Nettleship, J.E.; Sainsbury, S.; Saunders, N.J.; Owens, R.J. Structure of the cold-shock domain protein from Neisseria meningitidis reveals a strand-exchanged dimer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 247–251. [Google Scholar] [CrossRef] [Green Version]
- Evdokimova, V.M.; Wei, C.L.; Sitikov, A.S.; Simonenko, P.N.; Lazarev, O.A.; Vasilenko, K.S.; Ustinov, V.A.; Hershey, J.W.; Ovchinnikov, L.P. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem. 1995, 270, 3186–3192. [Google Scholar] [CrossRef] [Green Version]
- Gaudreault, I.; Guay, D.; Lebel, M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004, 32, 316–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Oury, T.D.; Crapo, J.D.; Valnickova, Z.; Enghild, J.J. Human extracellular superoxide dismutase is a tetramer composed of two disulphide-linked dimers: A simplified, high-yield purification of extracellular superoxide dismutase. Biochem. J. 1996, 317, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Due, A.V.; Petersen, S.V.; Valnickova, Z.; Ostergaard, L.; Oury, T.D.; Crapo, J.D.; Enghild, J.J. Extracellular superoxide dismutase exists as an octamer. FEBS Lett. 2006, 580, 1485–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stromqvist, M. Characterization of recombinant human extracellular superoxide dismutase. J. Chromatogr. B Biomed. Sci. Appl. 1993, 621, 139–148. [Google Scholar] [CrossRef]
- Dobson, C.M. Unfolded proteins, compact states and molten globules. Curr. Opin. Struct. Biol. 1992, 2, 6–12. [Google Scholar] [CrossRef]
- Uversky, V.N. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry 1993, 32, 13288–13298. [Google Scholar] [CrossRef]
- Uversky, V.N.; Winter, S.; Lober, G. Use of fluorescence decay times of 8-ANS-protein complexes to study the conformational transitions in proteins which unfold through the molten globule state. Biophys. Chem. 1996, 60, 79–88. [Google Scholar] [CrossRef]
- Goldschmidt, L.; Teng, P.K.; Riek, R.; Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2010, 107, 3487–3492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balbirnie, M.; Grothe, R.; Eisenberg, D.S. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc. Natl. Acad. Sci. USA 2001, 98, 2375–2380. [Google Scholar] [CrossRef] [Green Version]
- Jahn, T.R.; Radford, S.E. Folding versus aggregation: Polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008, 469, 100–117. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, D.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [Green Version]
- Westermark, P.; Benson, M.D.; Buxbaum, J.N.; Cohen, A.S.; Frangione, B.; Ikeda, S.; Masters, C.L.; Merlini, G.; Saraiva, M.J.; Sipe, J.D.; et al. Amyloid: Toward terminology clarification. Report from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid 2005, 12, 1–4. [Google Scholar] [CrossRef]
- Vassar, R.; Bennett, B.D.; Babu-Khan, S.; Kahn, S.; Mendiaz, E.A.; Denis, P.; Teplow, D.B.; Ross, S.; Amarante, P.; Loeloff, R.; et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999, 286, 735–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prusiner, S.B.; McKinley, M.P.; Bowman, K.A.; Bolton, D.C.; Bendheim, P.E.; Groth, D.F.; Glenner, G.G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 1983, 35, 349–358. [Google Scholar] [CrossRef]
- Chartier-Harlin, M.C.; Kachergus, J.; Roumier, C.; Mouroux, V.; Douay, X.; Lincoln, S.; Levecque, C.; Larvor, L.; Andrieux, J.; Hulihan, M.; et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004, 364, 1167–1169. [Google Scholar] [CrossRef]
- Ano Bom, A.P.; Rangel, L.P.; Costa, D.C.; de Oliveira, G.A.; Sanches, D.; Braga, C.A.; Gava, L.M.; Ramos, C.H.; Cepeda, A.O.; Stumbo, A.C.; et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J. Biol. Chem. 2012, 287, 28152–28162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navalkar, A.; Ghosh, S.; Pandey, S.; Paul, A.; Datta, D.; Maji, S.K. Prion-like p53 Amyloids in Cancer. Biochemistry 2020, 59, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef] [Green Version]
- Pham, C.L.; Kwan, A.H.; Sunde, M. Functional amyloid: Widespread in Nature, diverse in purpose. Essays Biochem. 2014, 56, 207–219. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collinson, S.K.; Emody, L.; Muller, K.H.; Trust, T.J.; Kay, W.W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 1991, 173, 4773–4781. [Google Scholar] [CrossRef] [Green Version]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Yin, M.; Chen, J.; Li, X. Assembly and substrate recognition of curli biogenesis system. Nat. Commun. 2020, 11, 241. [Google Scholar] [CrossRef] [Green Version]
- Dean, D.N.; Lee, J.C. Modulating functional amyloid formation via alternative splicing of the premelanosomal protein PMEL17. J. Biol. Chem. 2020, 295, 7544–7553. [Google Scholar] [CrossRef] [Green Version]
- Rouse, S.L.; Matthews, S.J.; Dueholm, M.S. Ecology and Biogenesis of Functional Amyloids in Pseudomonas. J. Mol. Biol. 2018, 430, 3685–3695. [Google Scholar] [CrossRef]
- Hejair, H.M.A.; Zhu, Y.; Ma, J.; Zhang, Y.; Pan, Z.; Zhang, W.; Yao, H. Functional role of ompF and ompC porins in pathogenesis of avian pathogenic Escherichia coli. Microb. Pathog. 2017, 107, 29–37. [Google Scholar] [CrossRef]
- Nizhnikov, A.A.; Alexandrov, A.I.; Ryzhova, T.A.; Mitkevich, O.V.; Dergalev, A.A.; Ter-Avanesyan, M.D.; Galkin, A.P. Proteomic screening for amyloid proteins. PLoS ONE 2014, 9, e116003. [Google Scholar] [CrossRef] [Green Version]
- Nizhnikov, A.A.; Ryzhova, T.A.; Volkov, K.V.; Zadorsky, S.P.; Sopova, J.V.; Inge-Vechtomov, S.G.; Galkin, A.P. Interaction of Prions Causes Heritable Traits in Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrescu, A.T.; Rathgeb-Szabo, K. An NMR investigation of solution aggregation reactions preceding the misassembly of acid-denatured cold shock protein A into fibrils. J. Mol. Biol. 1999, 291, 1191–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srisailam, S.; Kumar, T.K.; Rajalingam, D.; Kathir, K.M.; Sheu, H.S.; Jan, F.J.; Chao, P.C.; Yu, C. Amyloid-like fibril formation in an all beta-barrel protein. Partially structured intermediate state(s) is a precursor for fibril formation. J. Biol. Chem. 2003, 278, 17701–17709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanenko, O.V.; Sulatsky, M.I.; Mikhailova, E.V.; Kuznetsova, I.M.; Turoverov, K.K.; Stepanenko, O.V.; Sulatskaya, A.I. New findings on GFP-like protein application as fluorescent tags: Fibrillogenesis, oligomerization, and amorphous aggregation. Int. J. Biol. Macromol. 2021, 192, 1304–1310. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.T.; Schoenfish, A.R.; Roy, M.; Waldo, G.; Jennings, P.A. The rough energy landscape of superfolder GFP is linked to the chromophore. J. Mol. Biol. 2007, 373, 476–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enoki, S.; Maki, K.; Inobe, T.; Takahashi, K.; Kamagata, K.; Oroguchi, T.; Nakatani, H.; Tomoyori, K.; Kuwajima, K. The equilibrium unfolding intermediate observed at pH 4 and its relationship with the kinetic folding intermediates in green fluorescent protein. J. Mol. Biol. 2006, 361, 969–982. [Google Scholar] [CrossRef]
- Fukuda, H.; Arai, M.; Kuwajima, K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry 2000, 39, 12025–12032. [Google Scholar] [CrossRef]
- Hsu, S.T.; Blaser, G.; Jackson, S.E. The folding, stability and conformational dynamics of beta-barrel fluorescent proteins. Chem. Soc. Rev. 2009, 38, 2951–2965. [Google Scholar] [CrossRef]
- Huang, J.R.; Craggs, T.D.; Christodoulou, J.; Jackson, S.E. Stable intermediate states and high energy barriers in the unfolding of GFP. J. Mol. Biol. 2007, 370, 356–371. [Google Scholar] [CrossRef]
- Hsu, S.T.; Blaser, G.; Behrens, C.; Cabrita, L.D.; Dobson, C.M.; Jackson, S.E. Folding study of Venus reveals a strong ion dependence of its yellow fluorescence under mildly acidic conditions. J. Biol. Chem. 2010, 285, 4859–4869. [Google Scholar] [CrossRef] [Green Version]
- Melnik, T.N.; Povarnitsyna, T.V.; Glukhov, A.S.; Uversky, V.N.; Melnik, B.S. Sequential melting of two hydrophobic clusters within the green fluorescent protein GFP-cycle3. Biochemistry 2011, 50, 7735–7744. [Google Scholar] [CrossRef] [PubMed]
- Melnik, T.N.; Povarnitsyna, T.V.; Glukhov, A.S.; Melnik, B.S. Multi-state proteins: Approach allowing experimental determination of the formation order of structure elements in the green fluorescent protein. PLoS ONE 2012, 7, e48604. [Google Scholar] [CrossRef] [Green Version]
- Enoki, S.; Saeki, K.; Maki, K.; Kuwajima, K. Acid denaturation and refolding of green fluorescent protein. Biochemistry 2004, 43, 14238–14248. [Google Scholar] [CrossRef]
- Melnik, B.S.; Molochkov, N.V.; Prokhorov, D.A.; Uversky, V.N.; Kutyshenko, V.P. Molecular mechanisms of the anomalous thermal aggregation of green fluorescent protein. Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 1930–1939. [Google Scholar] [CrossRef]
- Xu, S.C.S.; LoRicco, J.G.; Bishop, A.C.; James, N.A.; Huynh, W.H.; McCallum, S.A.; Roan, N.R.; Makhatadze, G.I. Sequence-independent recognition of the amyloid structural motif by GFP protein family. Proc. Natl. Acad. Sci. USA 2020, 117, 22122–22127. [Google Scholar] [CrossRef]
- Pansarasa, O.; Bordoni, M.; Diamanti, L.; Sproviero, D.; Gagliardi, S.; Cereda, C. Sod1 in amyotrophic lateral sclerosis: “ambivalent” behavior connected to the disease. Int. J. Mol. Sci. 2018, 19, 1345. [Google Scholar] [CrossRef] [Green Version]
- Musteikyte, G.; Ziaunys, M.; Smirnovas, V. Methylene blue inhibits nucleation and elongation of SOD1 amyloid fibrils. PeerJ 2020, 8, e9719. [Google Scholar] [CrossRef]
- Forsberg, K.; Andersen, P.M.; Marklund, S.L.; Brännström, T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011, 121, 623–634. [Google Scholar] [CrossRef] [Green Version]
- Durazo, A.; Shaw, B.F.; Chattopadhyay, M.; Faull, K.F.; Nersissian, A.M.; Valentine, J.S.; Whitelegge, J.P. Metal-free superoxide dismutase-1 and three different amyotrophic lateral sclerosis variants share a similar partially unfolded beta-barrel at physiological temperature. J. Biol. Chem. 2009, 284, 34382–34389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiDonato, M.; Craig, L.; Huff, M.E.; Thayer, M.M.; Cardoso, R.M.; Kassmann, C.J.; Lo, T.P.; Bruns, C.K.; Powers, E.T.; Kelly, J.W.; et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003, 332, 601–615. [Google Scholar] [CrossRef]
- Banci, L.; Blazevits, O.; Cantini, F.; Danielsson, J.; Lang, L.; Luchinat, C.; Mao, J.; Oliveberg, M.; Ravera, E. Solid-state NMR studies of metal-free SOD1 fibrillar structures. J. Biol. Inorg. Chem. 2014, 19, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Estacio, S.G.; Leal, S.S.; Cristovao, J.S.; Faisca, P.F.; Gomes, C.M. Calcium binding to gatekeeper residues flanking aggregation-prone segments underlies non-fibrillar amyloid traits in superoxide dismutase 1 (SOD1). Biochim. Biophys. Acta Proteins Proteom. 2015, 1854, 118–126. [Google Scholar] [CrossRef]
- Strange, R.W.; Antonyuk, S.; Hough, M.A.; Doucette, P.A.; Rodriguez, J.A.; Hart, P.J.; Hayward, L.J.; Valentine, J.S.; Hasnain, S.S. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 2003, 328, 877–891. [Google Scholar] [CrossRef]
- Wang, J.; Slunt, H.; Gonzales, V.; Fromholt, D.; Coonfield, M.; Copeland, N.G.; Jenkins, N.A.; Borchelt, D.R. Copper-binding-site-null SOD1 causes ALS in transgenic mice: Aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003, 12, 2753–2764. [Google Scholar] [CrossRef] [Green Version]
- Basso, M.; Massignan, T.; Samengo, G.; Cheroni, C.; De Biasi, S.; Salmona, M.; Bendotti, C.; Bonetto, V. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J. Biol. Chem. 2006, 281, 33325–33335. [Google Scholar] [CrossRef] [Green Version]
- Guryanov, S.G.; Selivanova, O.M.; Nikulin, A.D.; Enin, G.A.; Melnik, B.S.; Kretov, D.A.; Serdyuk, I.N.; Ovchinnikov, L.P. Formation of amyloid-like fibrils by Y-box binding protein 1 (YB-1) is mediated by its cold shock domain and modulated by disordered terminal domains. PLoS ONE 2012, 7, e36969. [Google Scholar] [CrossRef] [Green Version]
- Stenina, O.I.; Shaneyfelt, K.M.; DiCorleto, P.E. Thrombin induces the release of the Y-box protein dbpB from mRNA: A mechanism of transcriptional activation. Proc. Natl. Acad. Sci. USA 2001, 98, 7277–7282. [Google Scholar] [CrossRef] [Green Version]
- Tacke, F.; Kanig, N.; En-Nia, A.; Kaehne, T.; Eberhardt, C.S.; Shpacovitch, V.; Trautwein, C.; Mertens, P.R. Y-box protein-1/p18 fragment identifies malignancies in patients with chronic liver disease. BMC Cancer 2011, 11, 185. [Google Scholar] [CrossRef] [Green Version]
- Cannon, D.; Eichhorn, S.J.; Donald, A.M. Structure of Spherulites in Insulin, β-Lactoglobulin, and Amyloid β. ACS Omega 2016, 1, 915–922. [Google Scholar] [CrossRef]
- Krebs, M.R.H.; Devlin, G.L.; Donald, A.M. Amyloid fibril-like structure underlies the aggregate structure across the pH range for β-lactoglobulin. Biophys. J. 2009, 96, 5013–5019. [Google Scholar] [CrossRef] [Green Version]
- Maity, S.; Sepay, N.; Pal, S.; Sardar, S.; Parvej, H.; Pal, S.; Chakraborty, J.; Pradhan, A.; Halder, U.C. Modulation of amyloid fibrillation of bovine β-lactoglobulin by selective methionine oxidation. RSC Adv. 2021, 11, 11192–11203. [Google Scholar] [CrossRef]
- Lambrecht, M.A.; Jansens, K.J.A.; Rombouts, I.; Brijs, K.; Rousseau, F.; Schymkowitz, J.; Delcour, J.A. Conditions Governing Food Protein Amyloid Fibril Formation. Part II: Milk and Legume Proteins. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1277–1291. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Peculiarities of the Super-Folder GFP Folding in a Crowded Milieu. Int. J. Mol. Sci. 2016, 17, 1805. [Google Scholar] [CrossRef] [Green Version]
- Munishkina, L.A.; Ahmad, A.; Fink, A.L.; Uversky, V.N. Guiding protein aggregation with macromolecular crowding. Biochemistry 2008, 47, 8993–9006. [Google Scholar] [CrossRef] [Green Version]
- Munishkina, L.A.; Cooper, E.M.; Uversky, V.N.; Fink, A.L. The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J. Mol. Recognit. 2004, 17, 456–464. [Google Scholar] [CrossRef]
- Ma, Q.; Fan, J.B.; Zhou, Z.; Zhou, B.R.; Meng, S.R.; Hu, J.Y.; Chen, J.; Liang, Y. The contrasting effect of macromolecular crowding on amyloid fibril formation. PLoS ONE 2012, 7, e36288. [Google Scholar] [CrossRef] [Green Version]
- Stathopulos, P.B.; Rumfeldt, J.A.O.; Scholz, G.A.; Irani, R.A.; Frey, H.E.; Hallewell, R.A.; Lepock, J.R.; Meiering, E.M. Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc. Natl. Acad. Sci. USA 2003, 100, 7021–7026. [Google Scholar] [CrossRef] [Green Version]
- Antonets, K.S.; Volkov, K.V.; Maltseva, A.L.; Arshakian, L.M.; Galkin, A.P.; Nizhnikov, A.A. Proteomic analysis of Escherichia coli protein fractions resistant to solubilization by ionic detergents. Biochem. Mosc. 2016, 81, 34–46. [Google Scholar] [CrossRef]
- Antonets, K.S.; Nizhnikov, A.A. Predicting amyloidogenic proteins in the proteomes of plants. Int. J. Mol. Sci. 2017, 18, 2155. [Google Scholar] [CrossRef] [Green Version]
- Antonets, K.S.; Kliver, S.F.; Nizhnikov, A.A. Exploring Proteins Containing Amyloidogenic Regions in the Proteomes of Bacteria of the Order Rhizobiales. Evol. Bioinform. 2018, 14, 117693431876878. [Google Scholar] [CrossRef] [Green Version]
- Meehan, S.; Knowles, T.P.J.; Baldwin, A.J.; Smith, J.F.; Squires, A.M.; Clements, P.; Treweek, T.M.; Ecroyd, H.; Tartaglia, G.G.; Vendruscolo, M.; et al. Characterisation of Amyloid Fibril Formation by Small Heat-shock Chaperone Proteins Human αA-, αB- and R120G αB-Crystallins. J. Mol. Biol. 2007, 372, 470–484. [Google Scholar] [CrossRef]
- Laganowsky, A.; Liu, C.; Sawaya, M.R.; Whitelegge, J.P.; Park, J.; Zhao, M.; Pensalfini, A.; Soriaga, A.B.; Landau, M.; Teng, P.K.; et al. Atomic view of a toxic amyloid small oligomer. Science 2012, 335, 1228–1231. [Google Scholar] [CrossRef] [Green Version]
- Soriaga, A.B.; Sangwan, S.; MacDonald, R.; Sawaya, M.R.; Eisenberg, D. Crystal structures of IAPP amyloidogenic segments reveal a novel packing motif of out-of-register beta sheets. J. Phys. Chem. B 2016, 120, 5810–5816. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhao, M.; Jiang, L.; Cheng, P.N.; Park, J.; Sawaya, M.R.; Pensalfini, A.; Gou, D.; Berk, A.J.; Glabe, C.G.; et al. Out-of-register β-sheets suggest a pathway to toxic amyloid aggregates. Proc. Natl. Acad. Sci. USA 2012, 109, 20913–20918. [Google Scholar] [CrossRef] [Green Version]
- Riek, R. The three-dimensional structures of amyloids. Cold Spring Harb. Perspect. Biol. 2017, 9, a023572. [Google Scholar] [CrossRef] [Green Version]
- Berhanu, W.M.; Hansmann, U.H. The stability of cylindrin beta-barrel amyloid oligomer models-a molecular dynamics study. Proteins Struct. Funct. Bioinf. 2013, 81, 1542–1555. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xi, W.; Hansmann, U.H.E.; Wei, Y. Fibril-Barrel Transitions in Cylindrin Amyloids. J. Chem. Theory Comput. 2017, 13, 3936–3944. [Google Scholar] [CrossRef]
- Shao, Q.; Wong, K.M.; Seroski, D.T.; Wang, Y.; Liu, R.; Paravastu, A.K.; Hudalla, G.A.; Hall, C.K. Anatomy of a selectively coassembled beta-sheet peptide nanofiber. Proc. Natl. Acad. Sci. USA 2020, 117, 4710–4717. [Google Scholar] [CrossRef]
- Sun, Y.; Ge, X.; Xing, Y.; Wang, B.; Ding, F. beta-barrel Oligomers as Common Intermediates of Peptides Self-Assembling into Cross-beta Aggregates. Sci. Rep. 2018, 8, 10353. [Google Scholar] [CrossRef] [Green Version]
- Sepehri, A.; Nepal, B.; Lazaridis, T. Distinct Modes of Action of IAPP Oligomers on Membranes. J. Chem. Inf. Model. 2021, 61, 4645–4655. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.; Duan, X.; Ding, F. Direct Observation of beta-Barrel Intermediates in the Self-Assembly of Toxic SOD128-38 and Absence in Nontoxic Glycine Mutants. J. Chem. Inf. Model. 2021, 61, 966–975. [Google Scholar] [CrossRef]
- Do, T.D.; LaPointe, N.E.; Nelson, R.; Krotee, P.; Hayden, E.Y.; Ulrich, B.; Quan, S.; Feinstein, S.C.; Teplow, D.B.; Eisenberg, D.; et al. Amyloid β-Protein C-Terminal Fragments: Formation of Cylindrins and β-Barrels. J. Am. Chem. Soc. 2016, 138, 549–557. [Google Scholar] [CrossRef] [Green Version]
- Kandel, N.; Matos, J.O.; Tatulian, S.A. Structure of amyloid β25–35 in lipid environment and cholesterol-dependent membrane pore formation. Sci. Rep. 2019, 9, 2689. [Google Scholar] [CrossRef] [Green Version]
- Chang, Z.; Luo, Y.; Zhang, Y.; Wei, G. Interactions of Aβ25−35 β-Barrel-like Oligomers with Anionic Lipid Bilayer and Resulting Membrane Leakage: An All-Atom Molecular Dynamics Study. J. Phys. Chem. B 2011, 115, 1165–1174. [Google Scholar] [CrossRef]
- Jang, H.; Connelly, L.; Arce, F.T.; Ramachandran, S.; Kagan, B.L.; Lal, R.; Nussinov, R. Mechanisms for the Insertion of Toxic, Fibril-like beta-Amyloid Oligomers into the Membrane. J. Chem. Theory Comput. 2013, 9, 822–833. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Han, J.; Borchers, C.H.; Konermann, L. Structure and Dynamics of Small Soluble Aβ(1–40) Oligomers Studied by Top-Down Hydrogen Exchange Mass Spectrometry. Biochemistry 2012, 51, 3694–3703. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Campanera, J.M.; Ngo, S.T.; Loquet, A.; Derreumaux, P. Tetrameric Aβ40 and Aβ42 β-Barrel Structures by Extensive Atomistic Simulations. II. In Aqueous Solution. J. Phys. Chem. B 2019, 123, 6750–6756. [Google Scholar] [CrossRef]
- Osterlund, N.; Moons, R.; Ilag, L.L.; Sobott, F.; Graslund, A. Native Ion Mobility-Mass Spectrometry Reveals the Formation of β-Barrel Shaped Amyloid-β Hexamers in a Membrane-Mimicking Environment. J. Am. Chem. Soc. 2019, 141, 10440–10450. [Google Scholar] [CrossRef] [Green Version]
- Ngo, S.T.; Nguyen, P.H.; Derreumaux, P. Impact of A2T and D23N Mutations on Tetrameric Aβ42 Barrel within a Dipalmitoylphosphatidylcholine Lipid Bilayer Membrane by Replica Exchange Molecular Dynamics. J. Phys. Chem. B 2020, 124, 1175–1182. [Google Scholar] [CrossRef]
- Serra-Batiste, M.; Ninot-Pedrosa, M.; Bayoumi, M.; Gairi, M.; Maglia, G.; Carulla, N. Aβ42 assembles into specific β-barrel pore-forming oligomers in membrane-mimicking environments. Proc. Natl. Acad. Sci. USA 2016, 113, 10866–10871. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Kakinen, A.; Wan, X.; Moriarty, N.; Hunt, C.P.J.; Li, Y.; Andrikopoulos, N.; Nandakumar, A.; Davis, T.P.; Parish, C.L.; et al. Spontaneous formation of β-sheet nano-barrels during the early aggregation of Alzheimer’s amyloid beta. Nano Today 2021, 38, 101125. [Google Scholar] [CrossRef]
- Gosal, W.S.; Clark, A.H.; Pudney, P.D.A.; Ross-Murphy, S.B. Novel Amyloid Fibrillar Networks Derived from a Globular Protein: β-Lactoglobulin. Langmuir 2002, 18, 7174–7181. [Google Scholar] [CrossRef]
- Orme, R.; Douglas, C.W.I.; Rimmer, S.; Webb, M. Proteomic analysis of Escherichia coli biofilms reveals the overexpression of the outer membrane protein OmpA. Proteomics 2006, 6, 4269–4277. [Google Scholar] [CrossRef]
- Tolin, S.; Arrigoni, G.; Moscatiello, R.; Masi, A.; Navazio, L.; Sablok, G.; Squartini, A. Quantitative analysis of the naringenin-inducible proteome in Rhizobium leguminosarum by isobaric tagging and mass spectrometry. Proteomics 2013, 13, 1961–1972. [Google Scholar] [CrossRef]
- Bromley, E.H.C.; Krebs, M.R.H.; Donald, A.M. Mechanisms of structure formation in particulate gels of beta-lactoglobulin formed near the isoelectric point. Eur. Phys. J. E Soft Matter 2006, 21, 145–152. [Google Scholar] [CrossRef]
- Loveday, S.M.; Anema, S.G.; Singh, H. β-Lactoglobulin nanofibrils: The long and the short of it. Int. Dairy J. 2017, 67, 35–45. [Google Scholar] [CrossRef]
- Loveday, S.M.; Wang, X.L.; Rao, M.A.; Anema, S.G.; Creamer, L.K.; Singh, H. Tuning the properties of β-lactoglobulin nanofibrils with pH, NaCl and CaCl2. Int. Dairy J. 2010, 20, 571–579. [Google Scholar] [CrossRef]
- Loveday, S.M.; Wang, X.L.; Rao, M.A.; Anema, S.G.; Singh, H. β-Lactoglobulin nanofibrils: Effect of temperature on fibril formation kinetics, fibril morphology and the rheological properties of fibril dispersions. Food Hydrocoll. 2012, 27, 242–249. [Google Scholar] [CrossRef]
- Hamada, D.; Dobson, C.M. A kinetic study of β-lactoglobulin amyloid fibril formation promoted by urea. Protein Sci. 2009, 11, 2417–2426. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulatskaya, A.I.; Kosolapova, A.O.; Bobylev, A.G.; Belousov, M.V.; Antonets, K.S.; Sulatsky, M.I.; Kuznetsova, I.M.; Turoverov, K.K.; Stepanenko, O.V.; Nizhnikov, A.A. β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis. Int. J. Mol. Sci. 2021, 22, 11316. https://doi.org/10.3390/ijms222111316
Sulatskaya AI, Kosolapova AO, Bobylev AG, Belousov MV, Antonets KS, Sulatsky MI, Kuznetsova IM, Turoverov KK, Stepanenko OV, Nizhnikov AA. β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis. International Journal of Molecular Sciences. 2021; 22(21):11316. https://doi.org/10.3390/ijms222111316
Chicago/Turabian StyleSulatskaya, Anna I., Anastasiia O. Kosolapova, Alexander G. Bobylev, Mikhail V. Belousov, Kirill S. Antonets, Maksim I. Sulatsky, Irina M. Kuznetsova, Konstantin K. Turoverov, Olesya V. Stepanenko, and Anton A. Nizhnikov. 2021. "β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis" International Journal of Molecular Sciences 22, no. 21: 11316. https://doi.org/10.3390/ijms222111316
APA StyleSulatskaya, A. I., Kosolapova, A. O., Bobylev, A. G., Belousov, M. V., Antonets, K. S., Sulatsky, M. I., Kuznetsova, I. M., Turoverov, K. K., Stepanenko, O. V., & Nizhnikov, A. A. (2021). β-Barrels and Amyloids: Structural Transitions, Biological Functions, and Pathogenesis. International Journal of Molecular Sciences, 22(21), 11316. https://doi.org/10.3390/ijms222111316











