Screening and Identification of Candidate GUN1-Interacting Proteins in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results and Discussion
2.1. Y2H Screening
2.2. Pair-Wise Y2H Assay
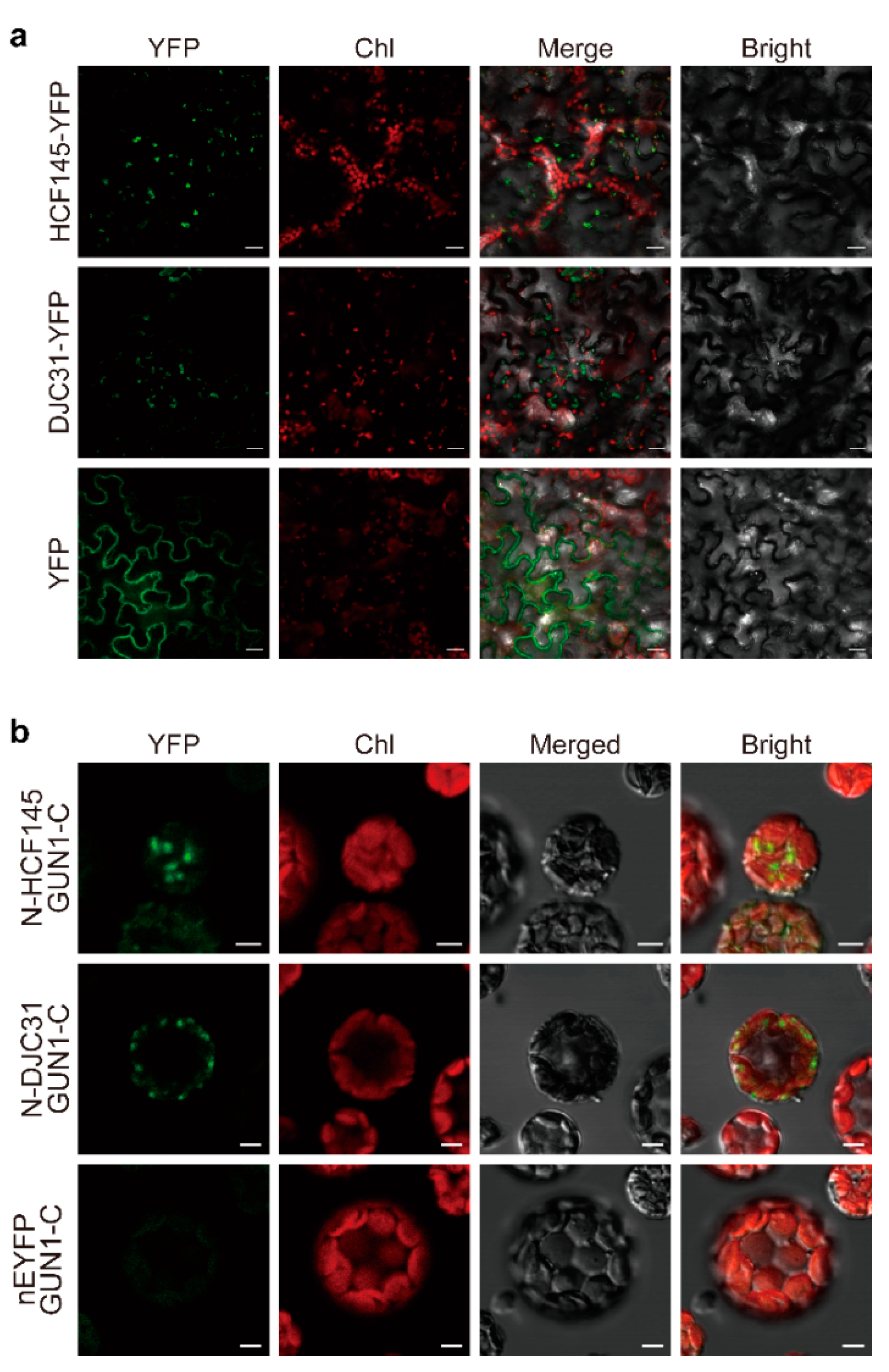
2.3. BiFC Assay
2.4. Expression of DJC31 and HCF145 in Response to NF and Lincomycin Treatments
3. Methods
3.1. Plant Materials and Treatments
3.2. Molecular Operation and Gene Expression Quantification
3.3. Y2H Assays
3.4. BiFC Assay
3.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nott, A.; Jung, H.-S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53. [Google Scholar] [CrossRef]
- Medina-Puche, L.; Tan, H.; Dogra, V.; Wu, M.; Rosas-Diaz, T.; Wang, L.; Ding, X.; Zhang, D.; Fu, X.; Kim, C.; et al. A defense pathway linking plasma membrane and chloroplasts and co-opted by pathogens. Cell 2020, 182, 1109–1124. [Google Scholar] [CrossRef]
- Woodson, J.D.; Chory, J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008, 9, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Susek, R.E.; Ausubel, F.M.; Chory, J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 1993, 74, 787–799. [Google Scholar] [CrossRef] [Green Version]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Woodson, J.D.; Perez-Ruiz, J.M.; Chory, J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 2011, 21, 897–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, N.; Tanaka, R.; Tanaka, A.; Masuda, T.; Nagatani, A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 15184–15189. [Google Scholar] [CrossRef] [Green Version]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef] [Green Version]
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Savchenko, T.; Baidoo, E.E.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corwin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 2012, 149, 1525–1535. [Google Scholar] [CrossRef] [Green Version]
- Pesaresi, P.; Kim, C. Current understanding of GUN1: A key mediator involved in biogenic retrograde signaling. Plant Cell Rep. 2019, 38, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, N.; Susek, R.; Chory, J. An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 1996, 112, 1465–1469. [Google Scholar] [CrossRef] [Green Version]
- Tadini, L.; Pesaresi, P.; Kleine, T.; Rossi, F.; Guljamow, A.; Sommer, F.; Mühlhaus, T.; Schroda, M.; Masiero, S.; Pribil, M.; et al. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 2016, 170, 1817–1830. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.Z.; Meyer, E.H.; Richter, A.S.; Schuster, M.; Ling, Q.H.; Schöttler, M.A.; Walther, D.; Zoschke, R.; Grimm, B.; Jarvis, R.P.; et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 2019, 5, 525–538. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, J.; Chory, J. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 10162–10167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.-Z.; Chalvin, C.; Hoelscher, M.P.; Meyer, E.H.; Wu, X.N.; Bock, R. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol. 2018, 176, 2472–2495. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.Q.; Wang, L.J.; Kong, M.J.; Huang, N.; Liu, X.Y.; Liang, H.Y.; Zhang, J.X.; Lu, S. At3g53630 encodes a GUN1-interacting protein under norflurazon treatment. Protoplasma 2021, 258, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Tadini, L.; Peracchio, C.; Ferrari, R.; Pesaresi, P. GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front. Plant Sci. 2016, 7, 1427. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.B.; Tian, H.Y.; Zhang, S.; Ding, Z.J.; Ma, C.L. GUN1-interacting proteins open the door for retrograde signaling. Trends Plant Sci. 2019, 24, 884–887. [Google Scholar] [CrossRef]
- Chiu, C.-C.; Chen, L.-J.; Su, P.-H.; Li, H.-M. Evolution of chloroplast J proteins. PLoS ONE 2013, 8, 70384. [Google Scholar] [CrossRef] [Green Version]
- Manavski, N.; Torabi, S.; Lezhneva, L.; Arif, M.A.; Frank, W.; Meurer, J. HIGH CHLOROPHYLL FLUORESCENCE145 binds to and stabilizes the psaA 5′ UTR via a newly defined repeat motif in Embryophyta. Plant Cell 2015, 27, 2600–2615. [Google Scholar] [CrossRef] [Green Version]
- Dittmer, S.; Kleine, T.; Schwenkert, S. The TPR and J-domain containing proteins DJC31 and DJC62 are involved in abiotic stress response in Arabidopsis thaliana. J. Cell Sci. 2021, 134, jcs259032. [Google Scholar] [CrossRef] [PubMed]
- Lezhneva, L.; Meurer, J. The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 2004, 38, 740–753. [Google Scholar] [CrossRef]
- Lu, S.; Van Eck, J.; Zhou, X.J.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Conlin, B.J.; Paolillo, D.J.; Garvin, D.F.; Vrebalov, J.; et al. The cauliflower Or gene encodes a cysteine-rich zinc finger domain-containing protein that mediates high-levels of β-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Zhou, F.; Huang, X.-Q.; Chen, W.-C.; Kong, M.-J.; Zhou, C.-F.; Zhuang, Z.; Li, L.; Lu, S. ORANGE represses chloroplast biogenesis in etiolated Arabidopsis cotyledons via interaction with TCP14. Plant Cell 2019, 31, 2996–3014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cottage, A.; Mott, E.K.; Kempster, J.A.; Gray, J.C. The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J. Exp. Bot. 2010, 61, 3773–3786. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Huang, J.; Chory, J. genome uncoupled1 mutants are hypersensitive to norflurazon and lincomycin. Plant Physiol. 2018, 178, 960–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Cao, T.-J.; Zheng, H.; Zhou, C.-F.; Wang, Z.; Wang, R.; Lu, S. Manipulation of carotenoid metabolic flux by lycopene cyclization in ripening red pepper (Capsicum annuum var. Conoides) fruits. J. Agric. Food. Chem. 2019, 67, 4300–4310. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Lee, L.Y.; Vyas, S.; Glick, E.; Chen, M.H.; Vainstein, A.; Gafni, Y.; Gelvin, S.B.; Tzfira, T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 2006, 362, 1120–1131. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Wang, C.-Y.; Gutensohn, M.; Jiang, L.; Zhang, P.; Zhang, D.; Dudareva, N.; Lu, S. A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc. Natl. Acad. Sci. USA 2017, 114, 6866–6871. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | AGI No. | Predicted Localization |
|---|---|---|
| BAG7 | AT5G62390 | endoplasmic reticulum |
| DJC31 * | AT5G12430 | chloroplast |
| DNAJ * | AT2G25560 | cytoplasm |
| EML3 * | AT5G13020 | nucleus |
| ERF74 * | AT1G53910 | nucleus |
| GATA * | AT1G28400 | extracellular |
| GGT1 | AT1G23310 | peroxisome |
| GNAT5 * | AT1G24040 | chloroplast |
| HAD | AT5G36790 | chloroplast |
| HCF145 * | AT5G08720 | chloroplast |
| HNI9 * | AT1G32130 | nucleus |
| KAC1 * | AT5G10470 | cytoplasm |
| MUSE1 * | AT3G58030 | nucleus |
| STO * | AT1G06040 | nucleus |
| TPR | AT5G28740 | nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Huang, X.; Li, K.; Song, S.; Jing, Y.; Lu, S. Screening and Identification of Candidate GUN1-Interacting Proteins in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 11364. https://doi.org/10.3390/ijms222111364
Wang L, Huang X, Li K, Song S, Jing Y, Lu S. Screening and Identification of Candidate GUN1-Interacting Proteins in Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(21):11364. https://doi.org/10.3390/ijms222111364
Chicago/Turabian StyleWang, Linjuan, Xingqi Huang, Kui Li, Shuyuan Song, Yunhe Jing, and Shan Lu. 2021. "Screening and Identification of Candidate GUN1-Interacting Proteins in Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 21: 11364. https://doi.org/10.3390/ijms222111364






