The Functional Association of ACQOS/VICTR with Salt Stress Resistance in Arabidopsis thaliana Was Confirmed by CRISPR-Mediated Mutagenesis
Abstract
:1. Introduction
2. Results
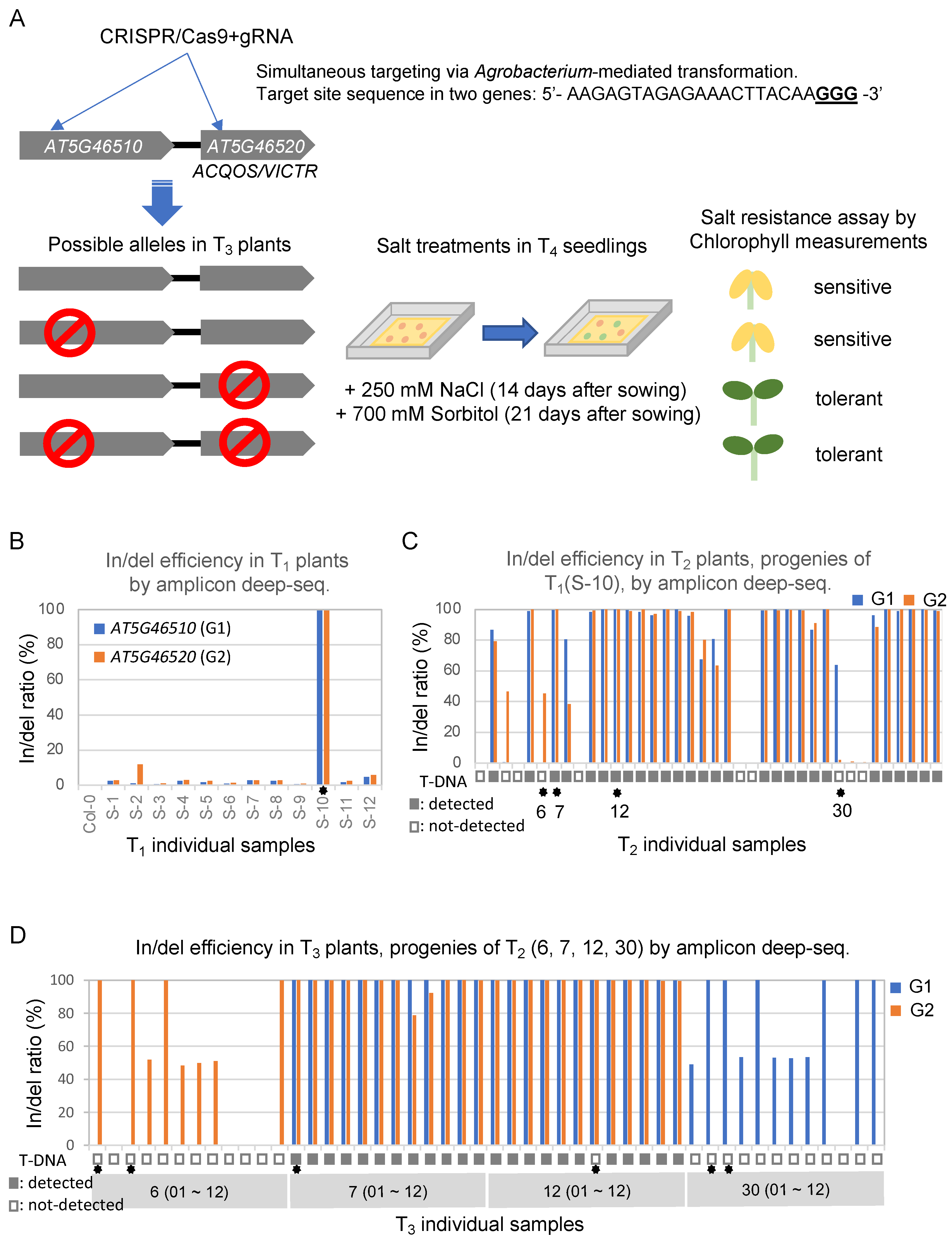
2.1. Generating Mutant Lines by Targeting Two Genes Using One Specific sgRNA for CRISPR-Mediated Mutagenesis in Arabidopsis
2.2. Salt Resistance Responses with Four Different Sets of Allelic Mutants
3. Discussion
4. Materials and Methods
4.1. Guide RNA Target Design
4.2. Vector Construction and gRNA Cloning
4.3. Agrobacterium-Mediated Transformation
4.4. Targeted Deep Sequencing and Mutation Pattern Analysis
4.5. Chlorophyll Isolation and Measurement to Test Osmotic Tolerance to Salt Stress
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.; Zhang, F. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas System. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.-L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, S.-T.; Ryu, J.; Choi, M.K.; Kweon, J.; Kang, B.; Ahn, H.; Bae, S.; Kim, J.; Kim, J.-S. A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J. Integr. Plant Biol. 2016, 58, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kim, J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grünewald, J.; Joung, J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353, aaf8729. [Google Scholar] [CrossRef]
- Hassan, M.M.; Zhang, Y.; Yuan, G.; De, K.; Chen, J.G.; Muchero, W.; Tuskan, G.A.; Qi, Y.; Yang, X. Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci. 2021, 26, 1133–1152. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evolut. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [Green Version]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Shen, L.; Fu, Y.; Yan, C.; Wang, K. A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genom. 2015, 42, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-P.; Xing, H.-L.; Dong, L.; Zhang, H.-Y.; Han, C.-Y.; Wang, X.-C.; Chen, Q.-J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [Green Version]
- Ordon, J.; Bressan, M.; Kretschmer, C.; Dall′Osto, L.; Marillonnet, S.; Bassi, R.; Stuttmann, J. Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Funct. Integr. Genom. 2020, 20, 151–162. [Google Scholar] [CrossRef]
- Doll, N.M.; Gilles, L.M.; Gérentes, M.-F.; Richard, C.; Just, J.; Fierlej, Y.; Borrelli, V.M.G.; Gendrot, G.; Ingram, G.C.; Rogowsky, P.M.; et al. Single and multiple gene knockouts by CRISPR–Cas9 in maize. Plant Cell Rep. 2019, 38, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Hsieh-Feng, V.; Yang, Y. Efficient expression of multiple guide RNAs for CRISPR/Cas genome editing. aBIOTECH 2020, 1, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W.; et al. 1135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef] [Green Version]
- Weigel, D.; Nordborg, M. Population genomics for understanding adaptation in wild plant species. Annu. Rev. Genet. 2015, 49, 315–338. [Google Scholar] [CrossRef] [Green Version]
- Exposito-Alonso, M.; Burbano, H.A.; Bossdorf, O.; Nielsen, R.; Weigel, D. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nat. Cell Biol. 2019, 573, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Katori, T.; Ikeda, A.; Iuchi, S.; Kobayashi, M.; Shinozaki, K.; Maehashi, K.; Sakata, Y.; Tanaka, S.; Taji, T. Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J. Exp. Bot. 2010, 61, 1125–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariga, H.; Katori, T.; Tsuchimatsu, T.; Hirase, T.; Tajima, Y.; Parker, J.E.; Alcazar, R.; Koornneef, M.; Hoekenga, O.; Lipka, A.E.; et al. NLR locus-mediated trade-off between abiotic and biotic stress adaptation in Arabidopsis. Nat. Plants 2017, 3, 17072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-H.; Kunz, H.-H.; Bhattacharjee, S.; Hauser, F.; Park, J.; Engineer, C.; Liu, A.; Ha, T.; Parker, J.E.; Gassmann, W.; et al. Natural variation in small molecule–induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 2013, 24, 5177–5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, B.-C.; Yun, J.-Y.; Kim, S.-T.; Shin, Y.; Ryu, J.; Choi, M.; Woo, J.W.; Kim, J.-S. Precision genome engineering through adenine base editing in plants. Nat. Plants 2018, 4, 427–431. [Google Scholar] [CrossRef]
- Choi, M.; Yun, J.-Y.; Kim, J.-H.; Kim, J.-S.; Kim, S.-T. The efficacy of CRISPR-mediated cytosine base editing with the RPS5a promoter in Arabidopsis thaliana. Sci. Rep. 2021, 11, 8087. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, Z.; Hua, K.; Gao, X.; Mao, Y.; Botella, J.R.; Zhu, J.-K. A highly efficient cell division-specific CRISPR/Cas9 system generates homozygous mutants for multiple genes in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3925. [Google Scholar] [CrossRef] [Green Version]
- Castel, B.; Tomlinson, L.; Locci, F.; Yang, Y.; Jones, J.D.G. Optimization of T-DNA architecture for Cas9-mediated mutagenesis in Arabidopsis. PLoS ONE 2019, 14, e0204778. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Lim, K.; Kim, J.-S.; Bae, S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2017, 33, 286–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronen, R.; Galun, M. Pigment extraction from lichens with dimethyl sulfoxide (DMSO) and estimation of chlorophyll degradation. Environ. Exp. Bot. 1984, 24, 239–245. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-T.; Choi, M.; Bae, S.-J.; Kim, J.-S. The Functional Association of ACQOS/VICTR with Salt Stress Resistance in Arabidopsis thaliana Was Confirmed by CRISPR-Mediated Mutagenesis. Int. J. Mol. Sci. 2021, 22, 11389. https://doi.org/10.3390/ijms222111389
Kim S-T, Choi M, Bae S-J, Kim J-S. The Functional Association of ACQOS/VICTR with Salt Stress Resistance in Arabidopsis thaliana Was Confirmed by CRISPR-Mediated Mutagenesis. International Journal of Molecular Sciences. 2021; 22(21):11389. https://doi.org/10.3390/ijms222111389
Chicago/Turabian StyleKim, Sang-Tae, Minkyung Choi, Su-Ji Bae, and Jin-Soo Kim. 2021. "The Functional Association of ACQOS/VICTR with Salt Stress Resistance in Arabidopsis thaliana Was Confirmed by CRISPR-Mediated Mutagenesis" International Journal of Molecular Sciences 22, no. 21: 11389. https://doi.org/10.3390/ijms222111389
APA StyleKim, S.-T., Choi, M., Bae, S.-J., & Kim, J.-S. (2021). The Functional Association of ACQOS/VICTR with Salt Stress Resistance in Arabidopsis thaliana Was Confirmed by CRISPR-Mediated Mutagenesis. International Journal of Molecular Sciences, 22(21), 11389. https://doi.org/10.3390/ijms222111389





