Pathogenic Variants in Selenoproteins and Selenocysteine Biosynthesis Machinery
Abstract
1. Introduction
2. Pathogenic Variants in Selenoproteins
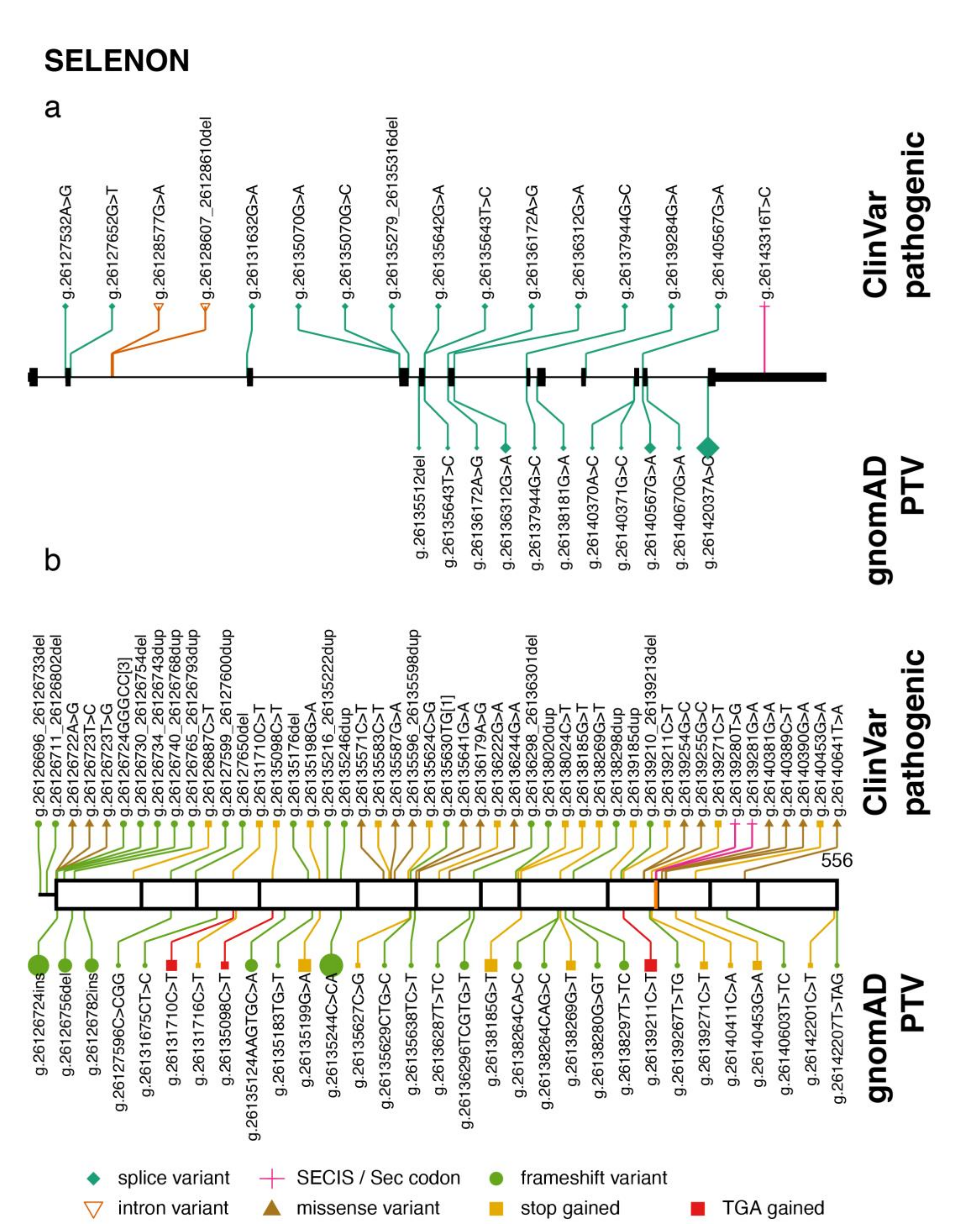
2.1. SELENON
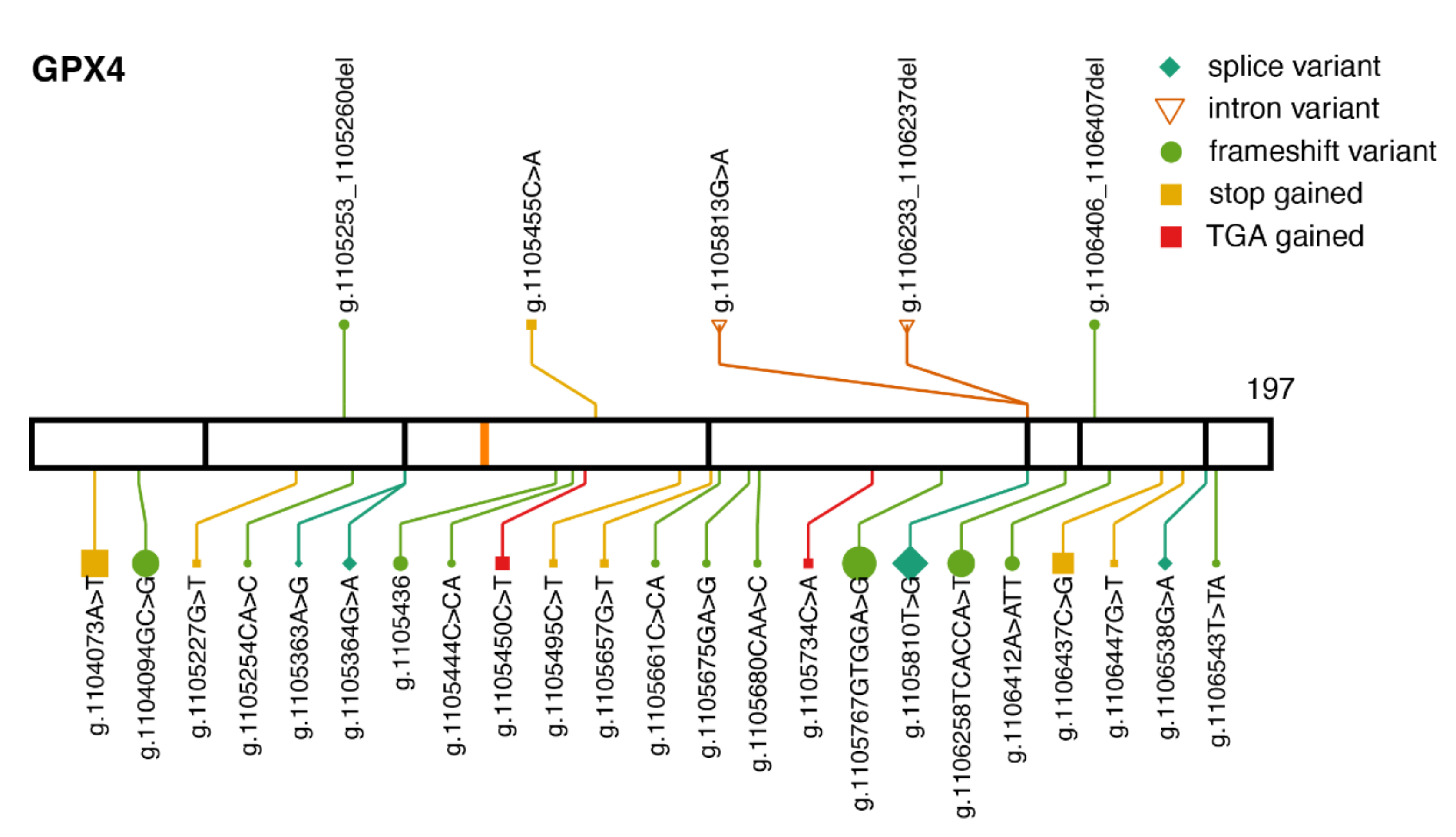
2.2. GPX4
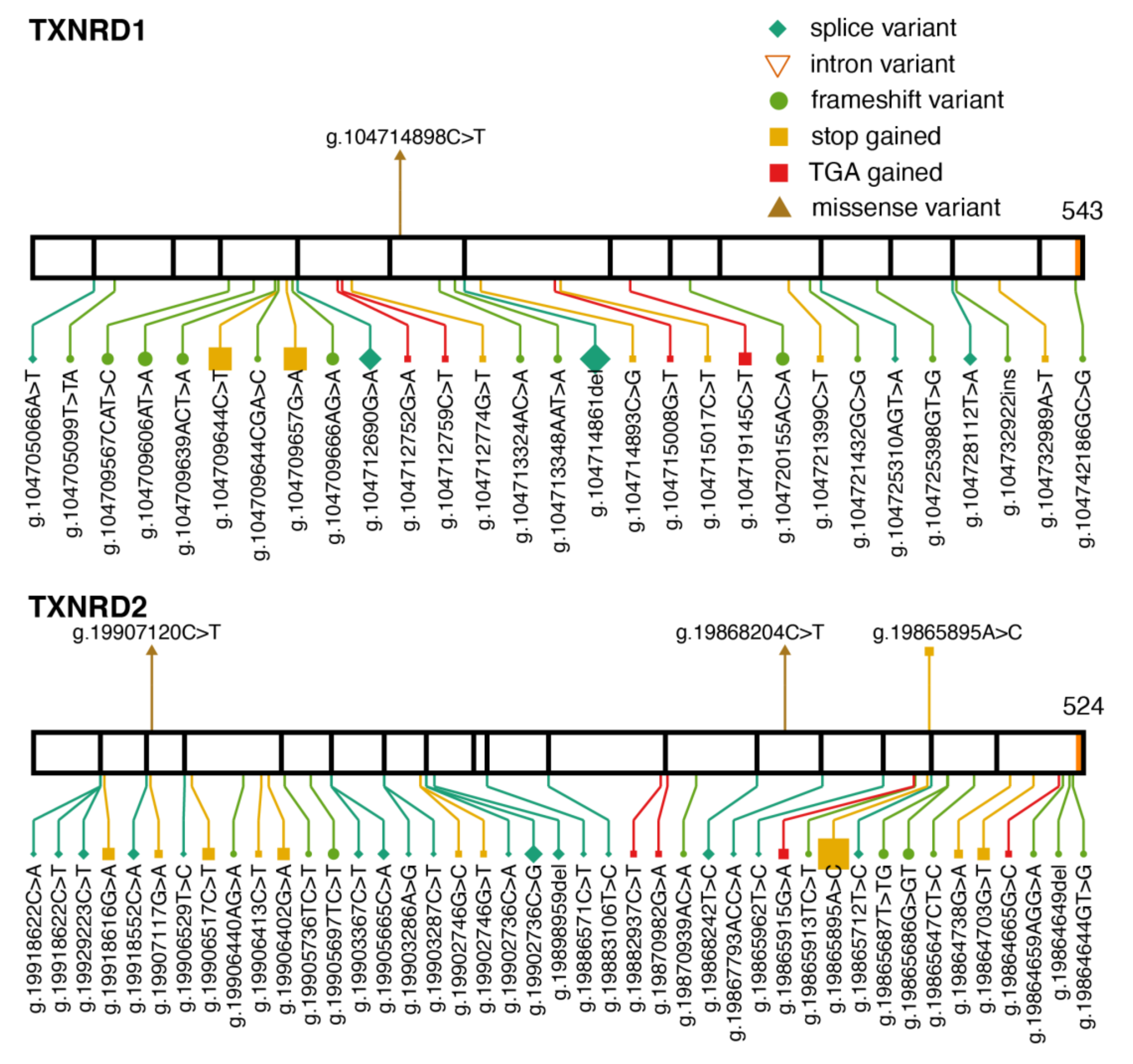
2.3. Thioredoxin Reductases
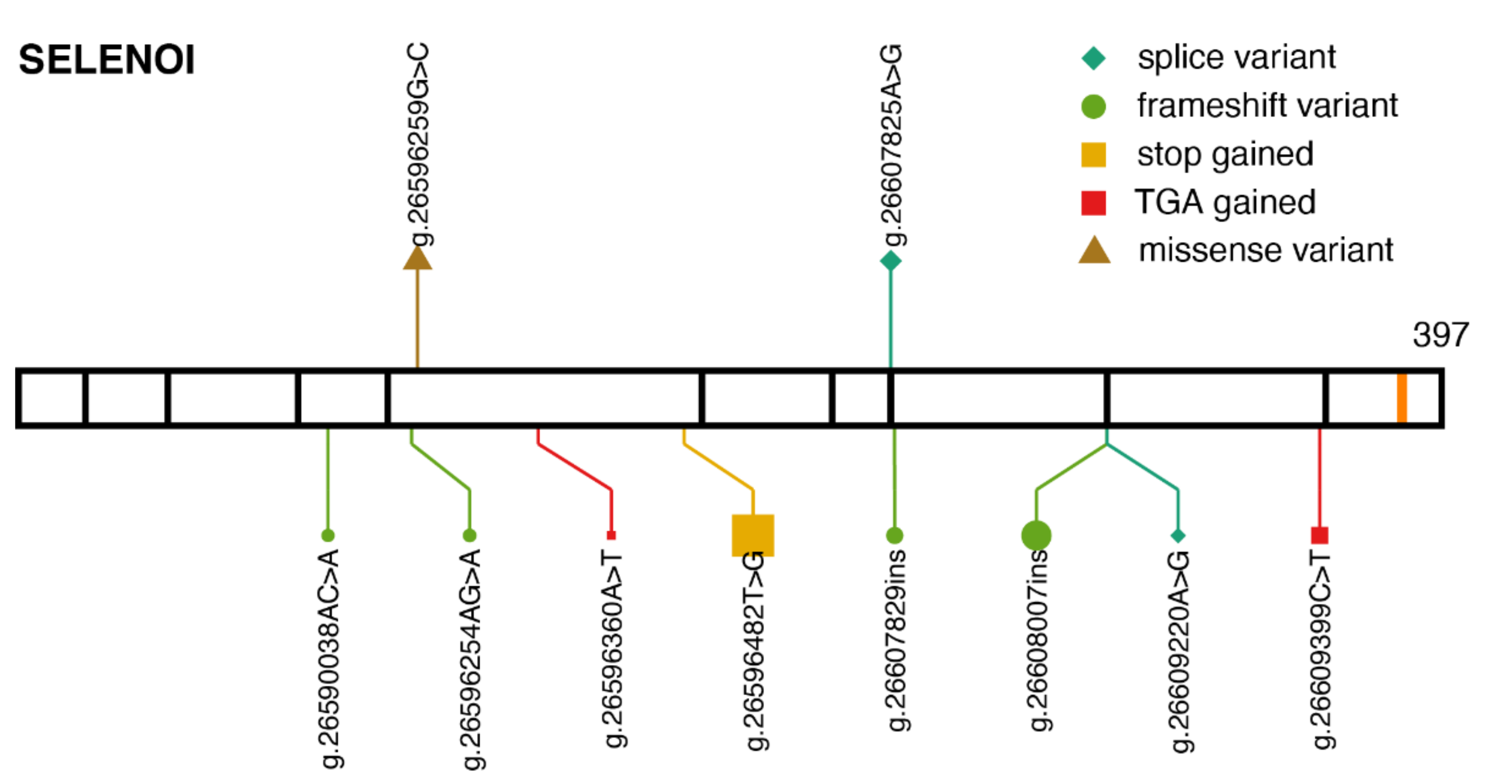
2.4. SELENOI
3. Pathogenic Variants Disrupting Selenoprotein Synthesis
3.1. TRU-TCA1-1
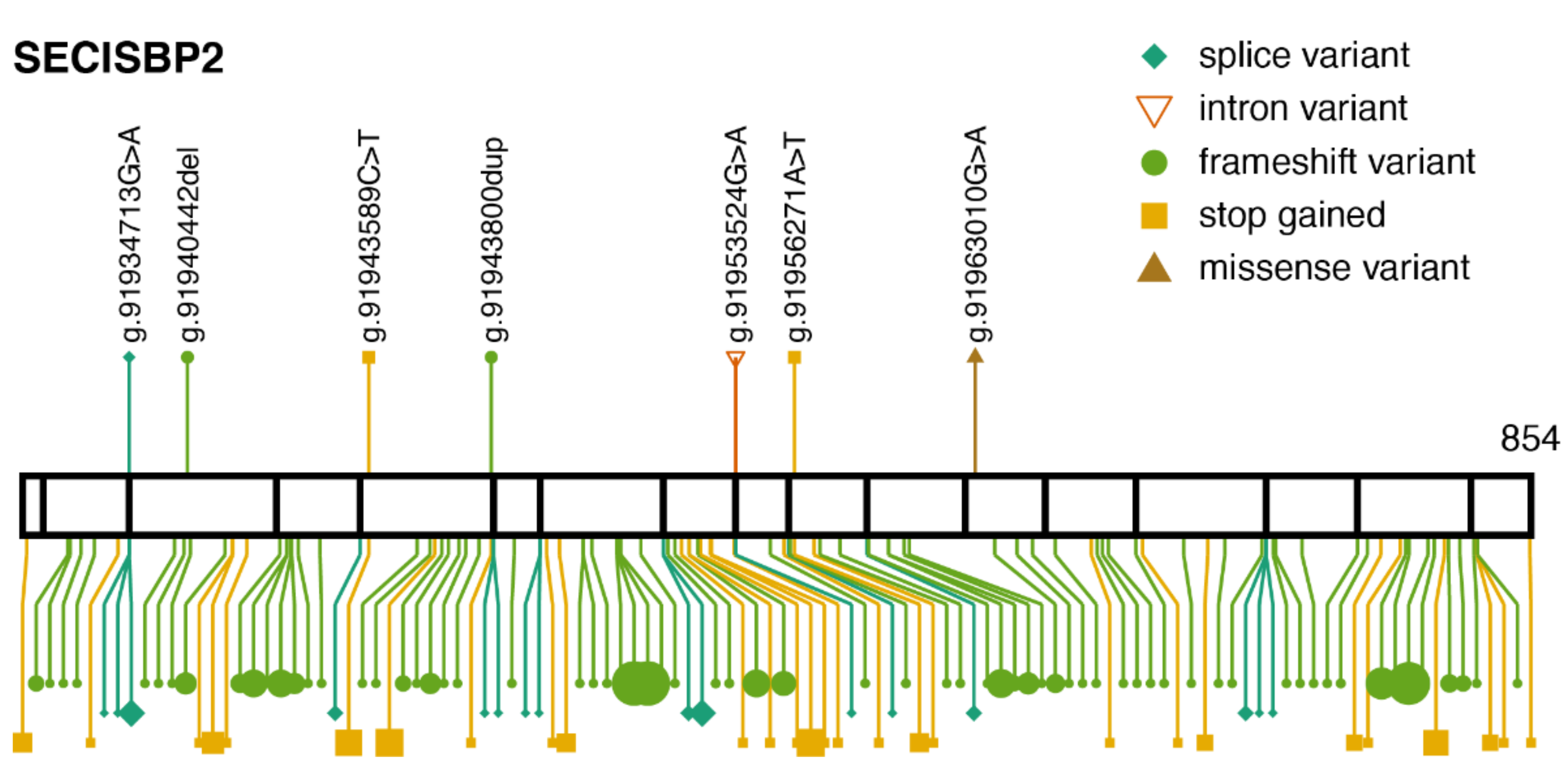
3.2. SECISBP2
3.3. SEPSECS
4. Common Genetic Variance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mariotti, M.; Ridge, P.G.; Zhang, Y.; Lobanov, A.V.; Pringle, T.H.; Guigo, R.; Hatfield, D.L.; Gladyshev, V.N. Composition and Evolution of the Vertebrate and Mammalian Selenoproteomes. PLoS ONE 2012, 7, e33066. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed]
- Allmang, C.; Wurth, L.; Krol, A. The Selenium to Selenoprotein Pathway in Eukaryotes: More Molecular Partners than Anticipated. Biochim. Biophys. Acta 2009, 1790, 1415–1423. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigó, R.; Gladyshev, V.N. Characterization of Mammalian Selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef]
- Conrad, M.; Schweizer, U. Mouse Models That Target Individual Selenoproteins. In Selenium; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 567–578. [Google Scholar] [CrossRef]
- Yant, L.J.; Ran, Q.; Rao, L.; Van Remmen, H.; Shibatani, T.; Belter, J.G.; Motta, L.; Richardson, A.; Prolla, T.A. The Selenoprotein GPX4 Is Essential for Mouse Development and Protects from Radiation and Oxidative Damage Insults. Free Radic. Biol. Med. 2003, 34, 496–502. [Google Scholar] [CrossRef]
- Imai, H.; Hirao, F.; Sakamoto, T.; Sekine, K.; Mizukura, Y.; Saito, M.; Kitamoto, T.; Hayasaka, M.; Hanaoka, K.; Nakagawa, Y. Early Embryonic Lethality Caused by Targeted Disruption of the Mouse PHGPx Gene. Biochem. Biophys. Res. Commun. 2003, 305, 278–286. [Google Scholar] [CrossRef]
- Jakupoglu, C.; Przemeck, G.K.H.; Schneider, M.; Moreno, S.G.; Mayr, N.; Hatzopoulos, A.K.; de Angelis, M.H.; Wurst, W.; Bornkamm, G.W.; Brielmeier, M.; et al. Cytoplasmic Thioredoxin Reductase Is Essential for Embryogenesis but Dispensable for Cardiac Development. Mol. Cell. Biol. 2005, 25, 1980–1988. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Jakupoglu, C.; Moreno, S.G.; Lippl, S.; Banjac, A.; Schneider, M.; Beck, H.; Hatzopoulos, A.K.; Just, U.; Sinowatz, F.; et al. Essential Role for Mitochondrial Thioredoxin Reductase in Hematopoiesis, Heart Development, and Heart Function. Mol. Cell. Biol. 2004, 24, 9414–9423. [Google Scholar] [CrossRef] [PubMed]
- Boukhzar, L.; Hamieh, A.; Cartier, D.; Tanguy, Y.; Alsharif, I.; Castex, M.; Arabo, A.; El Hajji, S.; Bonnet, J.-J.; Errami, M.; et al. Selenoprotein T Exerts an Essential Oxidoreductase Activity That Protects Dopaminergic Neurons in Mouse Models of Parkinson’s Disease. Antioxid. Redox Signal. 2016, 24, 557–574. [Google Scholar] [CrossRef]
- Avery, J.C.; Yamazaki, Y.; Hoffmann, F.W.; Folgelgren, B.; Hoffmann, P.R. Selenoprotein I Is Essential for Murine Embryogenesis. Arch. Biochem. Biophys. 2020, 689, 108444. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to Accessing Data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Schweizer, U.; Fradejas-Villar, N. Why 21? The Significance of Selenoproteins for Human Health Revealed by Inborn Errors of Metabolism. FASEB J. 2016, 30, 3669–3681. [Google Scholar] [CrossRef] [PubMed]
- Fradejas-Villar, N. Consequences of Mutations and Inborn Errors of Selenoprotein Biosynthesis and Functions. Free Radic. Biol. Med. 2018, 127, 206–214. [Google Scholar] [CrossRef]
- Schoenmakers, E.; Chatterjee, K. Human Disorders Affecting the Selenocysteine Incorporation Pathway Cause Systemic Selenoprotein Deficiency. Antioxid. Redox Signal. 2020, 33, 481–497. [Google Scholar] [CrossRef]
- Schweizer, U.; Bohleber, S.; Zhao, W.; Fradejas-Villar, N. The Neurobiology of Selenium: Looking Back and to the Future. Front. Neurosci. 2021, 15, 652099. [Google Scholar] [CrossRef]
- Castets, P.; Lescure, A.; Guicheney, P.; Allamand, V. Selenoprotein N in Skeletal Muscle: From Diseases to Function. J. Mol. Med. 2012, 90, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Lescure, A.; Gautheret, D.; Carbon, P.; Krol, A. Novel Selenoproteins Identified in Silico and in Vivo by Using a Conserved RNA Structural Motif. J. Biol. Chem. 1999, 274, 38147–38154. [Google Scholar] [CrossRef]
- Moghadaszadeh, B.; Petit, N.; Jaillard, C.; Brockington, M.; Quijano Roy, S.; Merlini, L.; Romero, N.; Estournet, B.; Desguerre, I.; Chaigne, D.; et al. Mutations in SEPN1 Cause Congenital Muscular Dystrophy with Spinal Rigidity and Restrictive Respiratory Syndrome. Nat. Genet. 2001, 29, 17–18. [Google Scholar] [CrossRef]
- Jurynec, M.J.; Xia, R.; Mackrill, J.J.; Gunther, D.; Crawford, T.; Flanigan, K.M.; Abramson, J.J.; Howard, M.T.; Grunwald, D.J. Selenoprotein N Is Required for Ryanodine Receptor Calcium Release Channel Activity in Human and Zebrafish Muscle. Proc. Natl. Acad. Sci. USA 2008, 105, 12485–12490. [Google Scholar] [CrossRef] [PubMed]
- Petit, N.; Lescure, A.; Rederstorff, M.; Krol, A.; Moghadaszadeh, B.; Wewer, U.M.; Guicheney, P. Selenoprotein N: An Endoplasmic Reticulum Glycoprotein with an Early Developmental Expression Pattern. Hum. Mol. Genet. 2003, 12, 1045–1053. [Google Scholar] [CrossRef]
- Chernorudskiy, A.; Varone, E.; Colombo, S.F.; Fumagalli, S.; Cagnotto, A.; Cattaneo, A.; Briens, M.; Baltzinger, M.; Kuhn, L.; Bachi, A.; et al. Selenoprotein N Is an Endoplasmic Reticulum Calcium Sensor That Links Luminal Calcium Levels to a Redox Activity. Proc. Natl. Acad. Sci. USA 2020, 117, 21288–21298. [Google Scholar] [CrossRef]
- Rederstorff, M.; Castets, P.; Arbogast, S.; Lainé, J.; Vassilopoulos, S.; Beuvin, M.; Dubourg, O.; Vignaud, A.; Ferry, A.; Krol, A.; et al. Increased Muscle Stress-Sensitivity Induced by Selenoprotein N Inactivation in Mouse: A Mammalian Model for SEPN1-Related Myopathy. PLoS ONE 2011, 6, e23094. [Google Scholar] [CrossRef] [PubMed]
- Castets, P.; Bertrand, A.T.; Beuvin, M.; Ferry, A.; Le Grand, F.; Castets, M.; Chazot, G.; Rederstorff, M.; Krol, A.; Lescure, A.; et al. Satellite Cell Loss and Impaired Muscle Regeneration in Selenoprotein N Deficiency. Hum. Mol. Genet. 2011, 20, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Moghadaszadeh, B.; Desguerre, I.; Topaloglu, H.; Muntoni, F.; Pavek, S.; Sewry, C.; Mayer, M.; Fardeau, M.; Tomé, F.M.; Guicheney, P. Identification of a New Locus for a Peculiar Form of Congenital Muscular Dystrophy with Early Rigidity of the Spine, on Chromosome 1p35-36. Am. J. Hum. Genet. 1998, 62, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Ferreiro, A.; Quijano-Roy, S.; Pichereau, C.; Moghadaszadeh, B.; Goemans, N.; Bönnemann, C.; Jungbluth, H.; Straub, V.; Villanova, M.; Leroy, J.-P.; et al. Mutations of the Selenoprotein N Gene, Which Is Implicated in Rigid Spine Muscular Dystrophy, Cause the Classical Phenotype of Multiminicore Disease: Reassessing the Nosology of Early-Onset Myopathies. Am. J. Hum. Genet. 2002, 71, 739–749. [Google Scholar] [CrossRef]
- Clarke, N.F.; Kidson, W.; Quijano-Roy, S.; Estournet, B.; Ferreiro, A.; Guicheney, P.; Manson, J.I.; Kornberg, A.J.; Shield, L.K.; North, K.N. SEPN1: Associated with Congenital Fiber-Type Disproportion and Insulin Resistance. Ann. Neurol. 2006, 59, 546–552. [Google Scholar] [CrossRef]
- Ferreiro, A.; Ceuterick-de Groote, C.; Marks, J.J.; Goemans, N.; Schreiber, G.; Hanefeld, F.; Fardeau, M.; Martin, J.-J.; Goebel, H.H.; Richard, P.; et al. Desmin-Related Myopathy with Mallory Body-like Inclusions Is Caused by Mutations of the Selenoprotein, N. Gene. Ann. Neurol. 2004, 55, 676–686. [Google Scholar] [CrossRef]
- Villar-Quiles, R.N.; von der Hagen, M.; Métay, C.; Gonzalez, V.; Donkervoort, S.; Bertini, E.; Castiglioni, C.; Chaigne, D.; Colomer, J.; Cuadrado, M.L.; et al. The Clinical, Histologic, and Genotypic Spectrum of SEPN1-Related Myopathy: A Case Series. Neurology 2020, 95, e1512–e1527. [Google Scholar] [CrossRef]
- Allamand, V.; Richard, P.; Lescure, A.; Ledeuil, C.; Desjardin, D.; Petit, N.; Gartioux, C.; Ferreiro, A.; Krol, A.; Pellegrini, N.; et al. A Single Homozygous Point Mutation in a 3’untranslated Region Motif of Selenoprotein N MRNA Causes SEPN1-Related Myopathy. EMBO Rep. 2006, 7, 450–454. [Google Scholar] [CrossRef]
- Maiti, B.; Arbogast, S.; Allamand, V.; Moyle, M.W.; Anderson, C.B.; Richard, P.; Guicheney, P.; Ferreiro, A.; Flanigan, K.M.; Howard, M.T. A Mutation in the SEPN1 Selenocysteine Redefinition Element (SRE) Reduces Selenocysteine Incorporation and Leads to SEPN1-Related Myopathy. Hum. Mutat. 2009, 30, 411–416. [Google Scholar] [CrossRef]
- Ursini, F.; Maiorino, M.; Brigelius-Flohé, R.; Aumann, K.D.; Roveri, A.; Schomburg, D.; Flohé, L. Diversity of Glutathione Peroxidases. Methods Enzymol. 1995, 252, 38–53. [Google Scholar] [CrossRef]
- Ursini, F.; Heim, S.; Kiess, M.; Maiorino, M.; Roveri, A.; Wissing, J.; Flohé, L. Dual Function of the Selenoprotein PHGPx during Sperm Maturation. Science 1999, 285, 1393–1396. [Google Scholar] [CrossRef] [PubMed]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Porto Freitas, F.; Seibt, T.; et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 2018, 172, 409–422.e21. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Hambright, W.S.; Fonseca, R.S.; Chen, L.; Na, R.; Ran, Q. Ablation of Ferroptosis Regulator Glutathione Peroxidase 4 in Forebrain Neurons Promotes Cognitive Impairment and Neurodegeneration. Redox Biol. 2017, 12, 8–17. [Google Scholar] [CrossRef]
- Sedaghatian, M.R. Congenital Lethal Metaphyseal Chondrodysplasia: A Newly Recognized Complex Autosomal Recessive Disorder. Am. J. Med. Genet. 1980, 6, 269–274. [Google Scholar] [CrossRef]
- Smith, A.C.; Mears, A.J.; Bunker, R.; Ahmed, A.; MacKenzie, M.; Schwartzentruber, J.A.; Beaulieu, C.L.; Ferretti, E.; FORGE Canada Consortium; Majewski, J.; et al. Mutations in the Enzyme Glutathione Peroxidase 4 Cause Sedaghatian-Type Spondylometaphyseal Dysplasia. J. Med. Genet. 2014, 51, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Aygun, C.; Celik, F.C.; Nural, M.S.; Azak, E.; Kucukoduk, S.; Ogur, G.; Incesu, L. Simplified Gyral Pattern with Cerebellar Hypoplasia in Sedaghatian Type Spondylometaphyseal Dysplasia: A Clinical Report and Review of the Literature. Am. J. Med. Genet. A 2012, 158A, 1400–1405. [Google Scholar] [CrossRef] [PubMed]
- Sibbing, D.; Pfeufer, A.; Perisic, T.; Mannes, A.M.; Fritz-Wolf, K.; Unwin, S.; Sinner, M.F.; Gieger, C.; Gloeckner, C.J.; Wichmann, H.-E.; et al. Mutations in the Mitochondrial Thioredoxin Reductase Gene TXNRD2 Cause Dilated Cardiomyopathy. Eur. Heart J. 2011, 32, 1121–1133. [Google Scholar] [CrossRef]
- Prasad, R.; Chan, L.F.; Hughes, C.R.; Kaski, J.P.; Kowalczyk, J.C.; Savage, M.O.; Peters, C.J.; Nathwani, N.; Clark, A.J.L.; Storr, H.L.; et al. Thioredoxin Reductase 2 (TXNRD2) Mutation Associated with Familial Glucocorticoid Deficiency (FGD). J. Clin. Endocrinol. Metab. 2014, 99, E1556–E1563. [Google Scholar] [CrossRef]
- Kudin, A.P.; Baron, G.; Zsurka, G.; Hampel, K.G.; Elger, C.E.; Grote, A.; Weber, Y.; Lerche, H.; Thiele, H.; Nürnberg, P.; et al. Homozygous Mutation in TXNRD1 Is Associated with Genetic Generalized Epilepsy. Free Radic. Biol. Med. 2017, 106, 270–277. [Google Scholar] [CrossRef]
- Rojnueangnit, K.; Sirichongkolthong, B.; Wongwandee, R.; Khetkham, T.; Noojarern, S.; Khongkraparn, A.; Wattanasirichaigoon, D. Identification of Gene Mutations in Primary Pediatric Cardiomyopathy by Whole Exome Sequencing. Pediatr. Cardiol. 2020, 41, 165–174. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Al-Khayat, A.; Al-Murshedi, F.; Al-Futaisi, A.; Chioza, B.A.; Fernandez-Murray, J.P.; Self, J.E.; Salter, C.G.; Harlalka, G.V.; Rawlins, L.E.; et al. A Mutation of EPT1 (SELENOI) Underlies a New Disorder of Kennedy Pathway Phospholipid Biosynthesis. Brain 2017, 140, 547–554. [Google Scholar] [CrossRef]
- Horibata, Y.; Elpeleg, O.; Eran, A.; Hirabayashi, Y.; Savitzki, D.; Tal, G.; Mandel, H.; Sugimoto, H. EPT1 (Selenoprotein I) Is Critical for the Neural Development and Maintenance of Plasmalogen in Humans. J. Lipid Res. 2018, 59, 1015–1026. [Google Scholar] [CrossRef]
- Santesmasses, D.; Mariotti, M.; Gladyshev, V.N. Tolerance to Selenoprotein Loss Differs between Human and Mouse. Mol. Biol. Evol. 2020, 37, 341–354. [Google Scholar] [CrossRef]
- Turanov, A.A.; Xu, X.-M.; Carlson, B.A.; Yoo, M.-H.; Gladyshev, V.N.; Hatfield, D.L. Biosynthesis of Selenocysteine, the 21st Amino Acid in the Genetic Code, and a Novel Pathway for Cysteine Biosynthesis. Adv. Nutr. 2011, 2, 122–128. [Google Scholar] [CrossRef]
- Diamond, A.; Dudock, B.; Hatfield, D. Structure and Properties of a Bovine Liver UGA Suppressor Serine TRNA with a Tryptophan Anticodon. Cell 1981, 25, 497–506. [Google Scholar] [CrossRef]
- Bösl, M.R.; Takaku, K.; Oshima, M.; Nishimura, S.; Taketo, M.M. Early Embryonic Lethality Caused by Targeted Disruption of the Mouse Selenocysteine TRNA Gene (Trsp). Proc. Natl. Acad. Sci. USA 1997, 94, 5531–5534. [Google Scholar] [CrossRef]
- Carlson, B.A. Selenocysteine TRNA[Ser]Sec Mouse Models for Elucidating Roles of Selenoproteins in Health and Development. In Selenium; Springer International Publishing: Cham, Switzerland, 2016; pp. 555–566. [Google Scholar] [CrossRef]
- Itoh, Y.; Chiba, S.; Sekine, S.-I.; Yokoyama, S. Crystal Structure of Human Selenocysteine TRNA. Nucleic Acids Res. 2009, 37, 6259–6268. [Google Scholar] [CrossRef]
- Carlson, B.A.; Lee, B.J.; Tsuji, P.A.; Tobe, R.; Park, J.M.; Schweizer, U.; Gladyshev, V.N.; Hatfield, D.L. Selenocysteine TRNA[Ser]Sec: From Nonsense Suppressor TRNA to the Quintessential Constituent in Selenoprotein Biosynthesis. In Selenium; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–12. [Google Scholar] [CrossRef]
- Diamond, A.M.; Choi, I.S.; Crain, P.F.; Hashizume, T.; Pomerantz, S.C.; Cruz, R.; Steer, C.J.; Hill, K.E.; Burk, R.F.; McCloskey, J.A.; et al. Dietary Selenium Affects Methylation of the Wobble Nucleoside in the Anticodon of Selenocysteine TRNA([Ser]Sec). J. Biol. Chem. 1993, 268, 14215–14223. [Google Scholar] [CrossRef]
- Carlson, B.A.; Moustafa, M.E.; Sengupta, A.; Schweizer, U.; Shrimali, R.; Rao, M.; Zhong, N.; Wang, S.; Feigenbaum, L.; Lee, B.J.; et al. Selective Restoration of the Selenoprotein Population in a Mouse Hepatocyte Selenoproteinless Background with Different Mutant Selenocysteine TRNAs Lacking Um34. J. Biol. Chem. 2007, 282, 32591–32602. [Google Scholar] [CrossRef]
- Schoenmakers, E.; Carlson, B.; Agostini, M.; Moran, C.; Rajanayagam, O.; Bochukova, E.; Tobe, R.; Peat, R.; Gevers, E.; Muntoni, F.; et al. Mutation in Human Selenocysteine Transfer RNA Selectively Disrupts Selenoprotein Synthesis. J. Clin. Investig. 2016, 126, 992–996. [Google Scholar] [CrossRef]
- Geslot, A.; Savagner, F.; Caron, P. Inherited Selenocysteine Transfer RNA Mutation: Clinical and Hormonal Evaluation of 2 Patients. Eur. Thyroid J. 2021, 1–6. [Google Scholar] [CrossRef]
- Palioura, S.; Sherrer, R.L.; Steitz, T.A.; Söll, D.; Simonovic, M. The Human SepSecS-TRNASec Complex Reveals the Mechanism of Selenocysteine Formation. Science 2009, 325, 321–325. [Google Scholar] [CrossRef]
- Copeland, P.R.; Fletcher, J.E.; Carlson, B.A.; Hatfield, D.L.; Driscoll, D.M. A Novel RNA Binding Protein, SBP2, Is Required for the Translation of Mammalian Selenoprotein MRNAs. EMBO J. 2000, 19, 306–314. [Google Scholar] [CrossRef]
- Seeher, S.; Atassi, T.; Mahdi, Y.; Carlson, B.A.; Braun, D.; Wirth, E.K.; Klein, M.O.; Reix, N.; Miniard, A.C.; Schomburg, L.; et al. Secisbp2 Is Essential for Embryonic Development and Enhances Selenoprotein Expression. Antioxid. Redox Signal. 2014, 21, 835–849. [Google Scholar] [CrossRef]
- Mehta, A.; Rebsch, C.M.; Kinzy, S.A.; Fletcher, J.E.; Copeland, P.R. Efficiency of Mammalian Selenocysteine Incorporation. J. Biol. Chem. 2004, 279, 37852–37859. [Google Scholar] [CrossRef] [PubMed]
- Copeland, P.R.; Stepanik, V.A.; Driscoll, D.M. Insight into Mammalian Selenocysteine Insertion: Domain Structure and Ribosome Binding Properties of Sec Insertion Sequence Binding Protein 2. Mol. Cell. Biol. 2001, 21, 1491–1498. [Google Scholar] [CrossRef]
- Allmang, C.; Carbon, P.; Krol, A. The SBP2 and 15.5 KD/Snu13p Proteins Share the Same RNA Binding Domain: Identification of SBP2 Amino Acids Important to SECIS RNA Binding. RNA 2002, 8, 1308–1318. [Google Scholar] [CrossRef][Green Version]
- Zhao, W.; Bohleber, S.; Schmidt, H.; Seeher, S.; Howard, M.T.; Braun, D.; Arndt, S.; Reuter, U.; Wende, H.; Birchmeier, C.; et al. Ribosome Profiling of Selenoproteins in Vivo Reveals Consequences of Pathogenic Secisbp2 Missense Mutations. J. Biol. Chem. 2019, jbc.RA119.009369. [Google Scholar] [CrossRef]
- Donovan, J.; Copeland, P.R. Selenocysteine Insertion Sequence Binding Protein 2L Is Implicated as a Novel Post-Transcriptional Regulator of Selenoprotein Expression. PLoS ONE 2012, 7, e35581. [Google Scholar] [CrossRef]
- Donovan, J.; Copeland, P.R. Evolutionary History of Selenocysteine Incorporation from the Perspective of SECIS Binding Proteins. BMC Evol. Biol. 2009, 9, 229. [Google Scholar] [CrossRef]
- Small-Howard, A.; Morozova, N.; Stoytcheva, Z.; Forry, E.P.; Mansell, J.B.; Harney, J.W.; Carlson, B.A.; Xu, X.-M.; Hatfield, D.L.; Berry, M.J. Supramolecular Complexes Mediate Selenocysteine Incorporation in Vivo. Mol. Cell. Biol. 2006, 26, 2337–2346. [Google Scholar] [CrossRef]
- Schoenmakers, E.; Schoenmakers, N.; Chatterjee, K. Mutations in Humans That Adversely Affect the Selenoprotein Synthesis Pathway. In Selenium. Its Molecular Biology and Role in Human Health; Hatfield, D.L., Berry, M.J., Gladyshev, N.V., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 523–538. [Google Scholar] [CrossRef]
- Anttonen, A.-K.; Hilander, T.; Linnankivi, T.; Isohanni, P.; French, R.L.; Liu, Y.; Simonović, M.; Söll, D.; Somer, M.; Muth-Pawlak, D.; et al. Selenoprotein Biosynthesis Defect Causes Progressive Encephalopathy with Elevated Lactate. Neurology 2015, 85, 306–315. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Mentuccia, D.; Proietti-Pannunzi, L.; Tanner, K.; Bacci, V.; Pollin, T.I.; Poehlman, E.T.; Shuldiner, A.R.; Celi, F.S. Association between a Novel Variant of the Human Type 2 Deiodinase Gene Thr92Ala and Insulin Resistance: Evidence of Interaction with the Trp64Arg Variant of the Beta-3-Adrenergic Receptor. Diabetes 2002, 51, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Castagna, M.G.; Dentice, M.; Cantara, S.; Ambrosio, R.; Maino, F.; Porcelli, T.; Marzocchi, C.; Garbi, C.; Pacini, F.; Salvatore, D. DIO2 Thr92Ala Reduces Deiodinase-2 Activity and Serum-T3 Levels in Thyroid-Deficient Patients. J. Clin. Endocrinol. Metab. 2017, 102, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Jung, J.; Araki, O.; Tsunekawa, K.; Park, S.Y.; Kim, J.; Murakami, M.; Jeong, S.-Y.; Lee, S. Concurrent TSHR Mutations and DIO2 T92A Polymorphism Result in Abnormal Thyroid Hormone Metabolism. Sci. Rep. 2018, 8, 10090. [Google Scholar] [CrossRef]
- Meulenbelt, I.; Min, J.L.; Bos, S.; Riyazi, N.; Houwing-Duistermaat, J.J.; van der Wijk, H.-J.; Kroon, H.M.; Nakajima, M.; Ikegawa, S.; Uitterlinden, A.G.; et al. Identification of DIO2 as a New Susceptibility Locus for Symptomatic Osteoarthritis. Hum. Mol. Genet. 2008, 17, 1867–1875. [Google Scholar] [CrossRef]
- Mez, J.; Chung, J.; Jun, G.; Kriegel, J.; Bourlas, A.P.; Sherva, R.; Logue, M.W.; Barnes, L.L.; Bennett, D.A.; Buxbaum, J.D.; et al. Two Novel Loci, COBL and SLC10A2, for Alzheimer’s Disease in African Americans. Alzheimers. Dement. 2017, 13, 119–129. [Google Scholar] [CrossRef]
- Sreelatha, A.; Yee, S.S.; Lopez, V.A.; Park, B.C.; Kinch, L.N.; Pilch, S.; Servage, K.A.; Zhang, J.; Jiou, J.; Karasiewicz-Urbańska, M.; et al. Protein AMPylation by an Evolutionarily Conserved Pseudokinase. Cell 2018, 175, 809–821.e19. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.M.; Zhu, G.; Dy, V.; Heath, A.C.; Madden, P.A.F.; Kemp, J.P.; McMahon, G.; St Pourcain, B.; Timpson, N.J.; Golding, J.; et al. Genome-Wide Association Study Identifies Loci Affecting Blood Copper, Selenium and Zinc. Hum. Mol. Genet. 2013, 22, 3998–4006. [Google Scholar] [CrossRef]
- Gong, J.; Hsu, L.; Harrison, T.; King, I.B.; Stürup, S.; Song, X.; Duggan, D.; Liu, Y.; Hutter, C.; Chanock, S.J.; et al. Genome-Wide Association Study of Serum Selenium Concentrations. Nutrients 2013, 5, 1706–1718. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; Fornage, M.; Foy, M.; Xun, P.; Gladyshev, V.N.; Morris, S.; Chasman, D.I.; Hu, F.B.; Rimm, E.B.; Kraft, P.; et al. Genome-Wide Association Study of Selenium Concentrations. Hum. Mol. Genet. 2015, 24, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Batai, K.; Trejo, M.J.; Chen, Y.; Kohler, L.N.; Lance, P.; Ellis, N.A.; Cornelis, M.C.; Chow, H.-H.S.; Hsu, C.-H.; Jacobs, E.T. Genome-Wide Association Study of Response to Selenium Supplementation and Circulating Selenium Concentrations in Adults of European Descent. J. Nutr. 2021, 151, 293–302. [Google Scholar] [CrossRef]
- Thomson, C.D.; Burton, C.E.; Robinson, M.F. On Supplementing the Selenium Intake of New Zealanders. 1. Short Experiments with Large Doses of Selenite or Selenomethionine. Br. J. Nutr. 1978, 39, 579–587. [Google Scholar] [CrossRef]
- Xia, Y.; Hill, K.E.; Byrne, D.W.; Xu, J.; Burk, R.F. Effectiveness of Selenium Supplements in a Low-Selenium Area of China. Am. J. Clin. Nutr. 2005, 81, 829–834. [Google Scholar] [CrossRef]
- White, L.; Romagné, F.; Müller, E.; Erlebach, E.; Weihmann, A.; Parra, G.; Andrés, A.M.; Castellano, S. Genetic Adaptation to Levels of Dietary Selenium in Recent Human History. Mol. Biol. Evol. 2015, 32, 1507–1518. [Google Scholar] [CrossRef]
- Canani, L.H.; Capp, C.; Dora, J.M.; Meyer, E.L.S.; Wagner, M.S.; Harney, J.W.; Larsen, P.R.; Gross, J.L.; Bianco, A.C.; Maia, A.L. The Type 2 Deiodinase A/G (Thr92Ala) Polymorphism Is Associated with Decreased Enzyme Velocity and Increased Insulin Resistance in Patients with Type 2 Diabetes Mellitus. J. Clin. Endocrinol. Metab. 2005, 90, 3472–3478. [Google Scholar] [CrossRef]
- Foster, C.B.; Aswath, K.; Chanock, S.J.; McKay, H.F.; Peters, U. Polymorphism Analysis of Six Selenoprotein Genes: Support for a Selective Sweep at the Glutathione Peroxidase 1 Locus (3p21) in Asian Populations. BMC Genet. 2006, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Engelken, J.; Espadas, G.; Mancuso, F.M.; Bonet, N.; Scherr, A.-L.; Jímenez-Álvarez, V.; Codina-Solà, M.; Medina-Stacey, D.; Spataro, N.; Stoneking, M.; et al. Signatures of Evolutionary Adaptation in Quantitative Trait Loci Influencing Trace Element Homeostasis in Liver. Mol. Biol. Evol. 2016, 33, 738–754. [Google Scholar] [CrossRef] [PubMed]
- Turanov, A.A.; Lobanov, A.V.; Hatfield, D.L.; Gladyshev, V.N. UGA Codon Position-Dependent Incorporation of Selenocysteine into Mammalian Selenoproteins. Nucleic Acids Res. 2013, 41, 6952–6959. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Weiss, S.L.; Sunde, R.A. UGA Codon Position Affects the Efficiency of Selenocysteine Incorporation into Glutathione Peroxidase-1. J. Biol. Chem. 1998, 273, 28533–28541. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santesmasses, D.; Gladyshev, V.N. Pathogenic Variants in Selenoproteins and Selenocysteine Biosynthesis Machinery. Int. J. Mol. Sci. 2021, 22, 11593. https://doi.org/10.3390/ijms222111593
Santesmasses D, Gladyshev VN. Pathogenic Variants in Selenoproteins and Selenocysteine Biosynthesis Machinery. International Journal of Molecular Sciences. 2021; 22(21):11593. https://doi.org/10.3390/ijms222111593
Chicago/Turabian StyleSantesmasses, Didac, and Vadim N. Gladyshev. 2021. "Pathogenic Variants in Selenoproteins and Selenocysteine Biosynthesis Machinery" International Journal of Molecular Sciences 22, no. 21: 11593. https://doi.org/10.3390/ijms222111593
APA StyleSantesmasses, D., & Gladyshev, V. N. (2021). Pathogenic Variants in Selenoproteins and Selenocysteine Biosynthesis Machinery. International Journal of Molecular Sciences, 22(21), 11593. https://doi.org/10.3390/ijms222111593





