Chronic Activation of AMPK Induces Mitochondrial Biogenesis through Differential Phosphorylation and Abundance of Mitochondrial Proteins in Dictyostelium discoideum
Abstract
:1. Introduction
2. Results
2.1. Identification of Phosphorylation Sites in Dictyostelium Mitochondrial Proteins
2.2. Protein Kinases Associated with Dictyostelium Mitochondria
2.3. Differential Phosphorylation Responses Caused by the Constitutive Hyperactivity of AMPKα
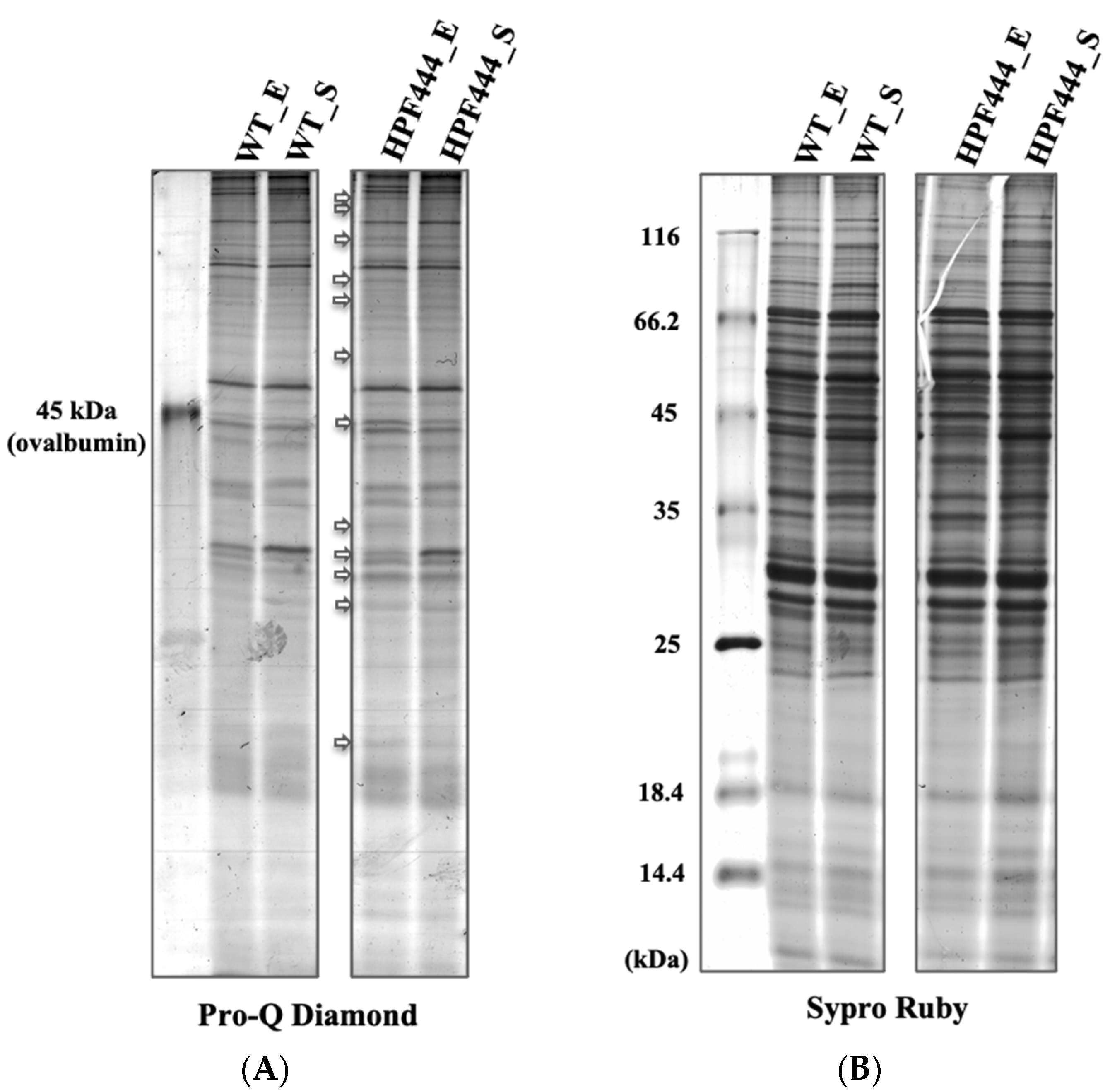
2.4. Effect of Chronically Elevated AMPKα Activity on the Abundance of the Mitochondrial Proteins
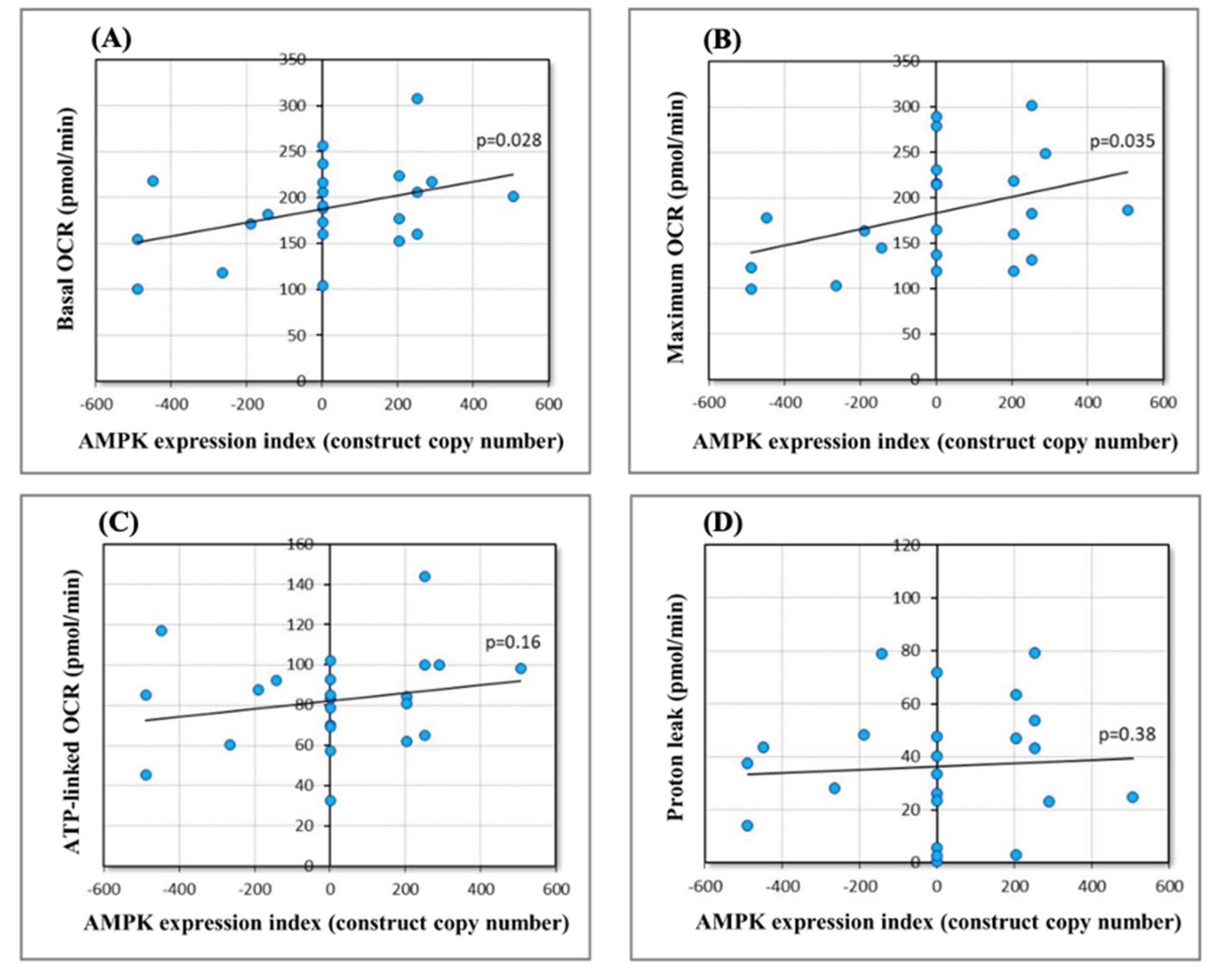
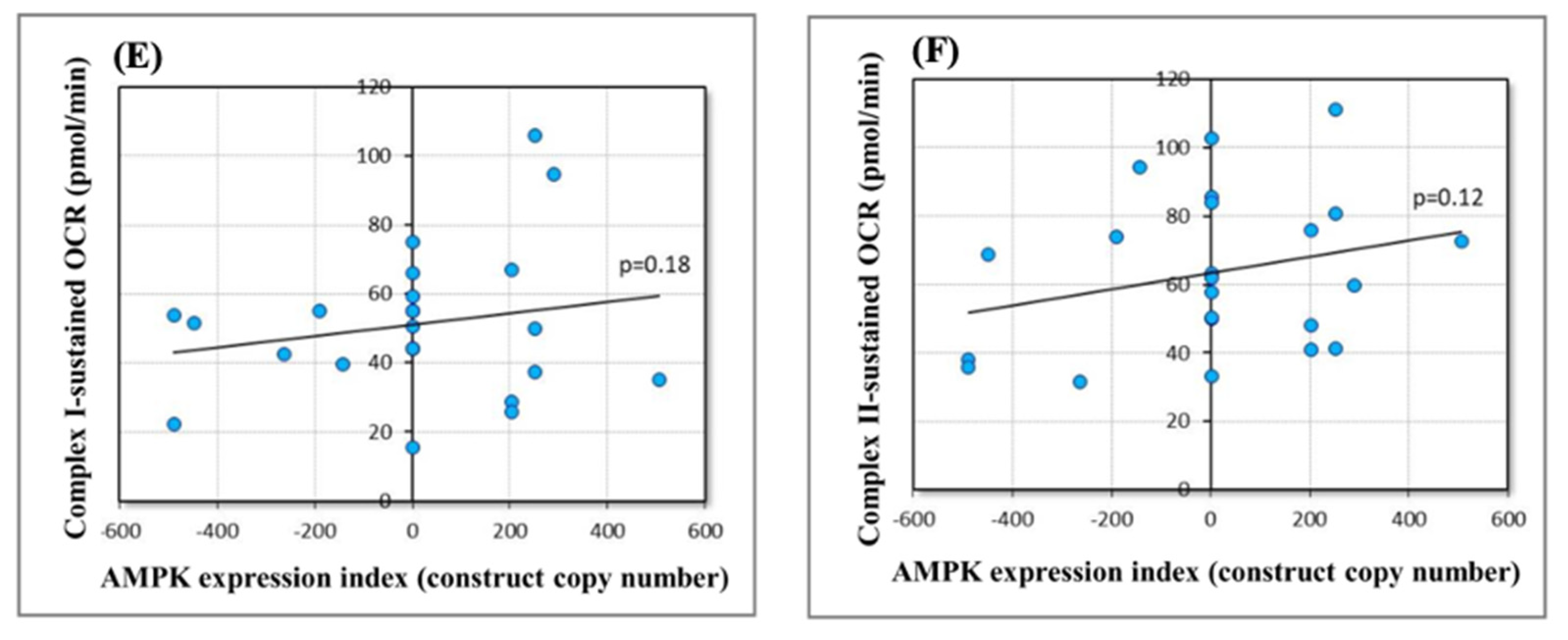
2.5. Effect of Chronically Elevated AMPKα Activity on Mitochondrial Respiratory Function
3. Discussion
4. Materials and Methods
4.1. D. discoideum Strains and Culture Growth Conditions
4.2. Mitochondria Isolation
4.3. Protein Sample Preparation for Phosphoproteome Identification with Mass Spectrometry
4.4. Phosphopeptide Enrichment
4.5. 1-D SDS-PAGE and Pro-Q Diamond Staining for Phosphoprotein Detection in Polyacrylamide Gels
4.6. Two-dimensional Polyacrylamide Gel Electrophoresis Analysis and Protein Identification
4.7. Seahorse Respirometry
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Xu, H. Translational regulation of mitochondrial biogenesis. Biochem. Soc. Trans. 2016, 44, 1717–1724. [Google Scholar] [CrossRef] [Green Version]
- Dominy, J.E.; Puigserver, P. Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a015008. [Google Scholar] [CrossRef]
- Williams, R.S.; Salmons, S.; Newsholme, E.A.; Kaufman, R.E.; Mellor, J. Regulation of nuclear and mitochondrial gene expression by contractile activity in skeletal muscle. J. Biol. Chem. 1986, 1, 376–380. [Google Scholar] [CrossRef]
- Hood, D.A. Invited review: Contractile activity-induced mitochondrial biogenesis in skeletal muscle. J. Appl. Physiol. 2001, 90, 1137–1157. [Google Scholar] [CrossRef] [PubMed]
- Zoladz, J.A.; Koziel, A.; Woyda-Ploszczyca, A.; Celichowski, J.; Jarmuszkiewicz, W. Endurance training increases the efficiency of rat skeletal mitochondria. Pflugers Archiv. Eur. J. Physiol. 2016, 468, 1709–1724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, H.; Ren, J.M.; Young, L.H.; Pypaert, M.; Mu, J.; Birnbaum, M.J.; Shulman, G.I. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc. Natl. Acad. Sci. USA 2002, 99, 15983–15987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Kanatous, S.B.; Thurmond, F.A.; Gallardo, T.; Isotani, E.; Bassel-Duby, R.; Williams, R.S. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 2002, 296, 349–352. [Google Scholar] [CrossRef]
- Thelander, M.; Olsson, T.; Ronne, H. Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 2004, 23, 1900–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baena-Gonzalez, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Bokko, P.B.; Francione, L.; Bandala-Sanchez, E.; Ahmed, A.U.; Annesley, S.J.; Huang, X.; Khurana, T.; Kimmel, A.R.; Fisher, P.R. Diverse cytopathologies in mitochondrial disease are caused by AMP-activated protein kinase signaling. Mol. Biol. Cell 2007, 18, 1874–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedbacker, K.; Carlson, M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008, 13, 2408–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narbonne, P.; Roy, R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 2009, 457, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.C.; Kazgan, N.; Bretz, C.A.; Forsberg, L.J.; Hector, C.E.; Worthen, R.J.; Onyenwoke, R.; Brenman, J.Y. Altered metabolism and persistent starvation behaviors caused by reduced AMPK function in Drosophila. PLoS ONE 2010, 5, e12799. [Google Scholar] [CrossRef] [Green Version]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vara-Ciruelos, D.; Dandapani, M.; Hardie, D.G. AMP-activated protein kinase: Friend or foe in cancer? Annu. Rev. Cancer Biol. 2020, 4, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L., Jr.; Loson, O.C.; Hellberg, K.; Young, N.; Chen, H.; Polleux, F.; Chan, D.C.; et al. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Ducommun, S.; Deak, M.; Sumpton, D.; Ford, R.J.; Galindo, A.N.; Kussmann, M.; Viollet, B.; Steinberg, G.R.; Foretz, M.; Dayon, L.; et al. Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: Identification of mitochondrial fission factor as a new AMPK substrate. Cell Signal. 2015, 27, 978–988. [Google Scholar] [CrossRef] [Green Version]
- Munday, M.R.; Campbell, D.G.; Carling, D.; Hardie, D.G. Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur. J. Biochem. 1988, 175, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, Z.-X.; Ding, Z.; Lu, Y.; Yu, Q.; Werle, K.D.; Zhou, G.; Park, Y.-Y.; Peng, G.; Gambello, M.J.; et al. Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat. Commun. 2015, 6, 7926. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Rho, E.; Sample, V.; Akano, H.; Magari, M.; Ueno, T.; Gorshkov, K.; Chen, M.; Tokumitsu, H.; Zhang, J.; et al. Compartmentalized AMPK signaling illuminated by genetically encoded molecular sensors and actuators. Cell Rep. 2015, 11, 657–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotsifas, M.; Barth, C.; De Lozanne, A.; Lay, S.T.; Fisher, P.R. Chaperonin 60 and mitochondrial disease in Dictyostelium. J. Muscle Res. Cell Motil. 2002, 23, 839–852. [Google Scholar] [CrossRef]
- Francione, L.; Fisher, P.R. Heteroplasmic mitochondrial disease in Dictyostelium discoideum. Biochem. Pharmacol. 2011, 82, 1510–1520. [Google Scholar] [CrossRef]
- Pearce, X.G.; Annesley, S.J.; Fisher, P.R. The Dictyostelium model for mitochondrial biology and disease. Int. J. Dev. Biol. 2019, 63, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Maurya, R.; Kumar, R.; Saran, S. Dictyostelium AMPKα regulates aggregate size and cell-type patterning. Open Biol. 2017, 7, 170055. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, T.H.; Agnew, B.J.; Gee, K.R.; Leung, W.-Y.; Goodman, T.; Schulenberg, B.; Hendrickson, J.; Beechem, J.M.; Haugland, R.P.; Patton, W.F. Global quantitative phosphoprotein analysis using multiplexed proteomics technology. Proteomics 2003, 3, 1128–1144. [Google Scholar] [CrossRef] [PubMed]
- Schulenberg, B.; Aggeler, R.; Beechem, J.M.; Capaldi, R.A.; Patton, W.F. Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J. Biol. Chem. 2003, 278, 27251–27255. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Bermudez, J.; Sanchez-Arago, M.; Soldevilla, B.; Del Arco, A.; Nuevo-Tapioles, C.; Cuezva, J.M. PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H-ATP synthase. Cell Rep. 2015, 12, 2143–2155. [Google Scholar] [CrossRef] [Green Version]
- Austin, S.; Nowikovsky, K. LETM1: Essential for mitochondrial biology and cation homeostasis? Trends Biochem. Sci. 2019, 44, 648–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, E.; Qu, D.; Huang, T.; Rizzi, N.; Boonying, W.; Krolak, D.; Ciana, P.; Woulfe, J.; Klein, C.; Slack, R.S.; et al. PINK1-mediated phosphorylation of LETM1 regulates mitochondrial calcium transport and protects neurons against mitochondrial stress. Nat. Commun. 2017, 8, 1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Landry, A.P.; Ding, X.H. The mitochondrial outer membrane protein mitoNEET is aredox enzyme catalyzing electron transfer from FMNH2 tooxygen or ubiquinone. J. Biol. Chem. 2017, 292, 10061–10067. [Google Scholar] [CrossRef] [Green Version]
- Mertins, P.; Yang, F.; Liu, T.; Mani, D.R.; Petyuk, V.A.; Gillette, M.A.; Clauser, K.R.; Qiao, J.W.; Gritsenko, M.A.; Moore, R.J.; et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteom. 2014, 13, 1690–1704. [Google Scholar] [CrossRef] [Green Version]
- Schweppe, D.K.; Rigas, J.R.; Gerber, S.A. Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J. Proteom. 2013, 91, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Di Palma, S.; Preisinger, C.; Peng, M.; Polat, A.N.; Heck, A.J.; Mohammed, S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013, 12, 260–271. [Google Scholar] [CrossRef]
- Shah, G.N.; Rubbelke, T.S.; Hendin, J.; Nguyen, H.; Waheed, A.; Shoemaker, J.D.; Sly, W.S. Targeted mutagenesis of mitochondrial carbonic anhydrases VA and VB implicates both enzymes in ammonia detoxification and glucose metabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 7423–7428. [Google Scholar] [CrossRef] [Green Version]
- Fiore, A.D.; Supuran, C.T.; Scalonic, A.; De Simonea, G. Human carbonic anhydrases and post-translational modifications: A hidden world possibly affecting protein properties and functions. J. Enzyme Inhib. Med. Chem. 2020, 35, 1450–1461. [Google Scholar] [CrossRef] [PubMed]
- Kawata, T.; Shevchenko, A.; Fukuzawa, M.; Jermyn, K.A.; Totty, N.F.; Zhukovskaya, N.V.; Sterling, A.E.; Mann, M.; Williams, J.G. SH2 signaling in a lower eukaryote: A STAT protein that regulates stalk cell differentiation in Dictyostelium. Cell 1997, 89, 909–916. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, J.M.; Manning, G.; Liu, A.; Fey, P.; Pilcher, K.E.; Xu, Y.; Smith, J.L. The Dictyostelium kinome-analysis of the protein kinases from a simple model organism. PLoS Genet. 2006, 2, e38. [Google Scholar] [CrossRef]
- Padrao, A.I.; Vitorino, R.; Duarte, J.A.; Ferreira, R.; Amado, F. Unraveling the phosphoproteome dynamics in mammal mitochondria from a network perspective. J. Proteome Res. 2013, 12, 4257–4267. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.M.E.; Paschke, P.; Peak-Chew, S.; Williams, T.D.; Tweedy, L.; Skehel, M.; Stephens, E.; Chubb, J.R.; Kay, R.R. The atypical MAP kinase ErkB transmits distinct chemotactic signals through a core signaling module. Dev. Cell 2019, 48, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Sugden, C.; Urbaniak, M.D.; Araki, T.; Williams, J.G. The Dictyostelium prestalk inducer differentiation-inducing factor-1 (DIF-1) triggers unexpectedly complex global phosphorylation changes. Mol. Biol. Cell 2015, 26, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Kim, L.; Harwood, A.; Kimmel, A.R. Receptor-dependent and tyrosine phosphatase-mediated inhibition of GSK3 regulates cell fate choice. Dev. Cell 2002, 3, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, S.K.; Blau-Wasser, R.; Rohlfs, M.; Gallinger, C.; Schleicher, M.; Noegel, A.A. The centrosomal component CEP161 of Dictyostelium discoideum interacts with the Hippo signaling pathway. Cell Cycle 2015, 14, 1024–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichinger, L.; Bähler, M.; Dietz, M.; Eckerskorn, C.; Schleicher, M. Characterization and cloning of a Dictyostelium Ste20-like protein kinase that phosphorylates the actin-binding protein severin. J. Biol. Chem. 1998, 273, 12952–12959. [Google Scholar] [CrossRef] [Green Version]
- Bergeron, R.; Ren, J.M.; Cadman, K.S.; Moore, I.K.; Perret, P.; Pypaert, M.; Young, L.H.; Semenkovich, C.F.; Shulman, G.I. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am. J. Physiol. Endocrinol. Metab. 2001, 281, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Nowikovsky, K.; Froschauer, E.M.; Zsurka, G.; Samaj, J.; Reipert, S.; Kolisek, M.; Wiesenberger, G.; Schweyen, R.J. The LETM1/YOL027 gene family encodes a factor of the mitochondrial K+ homeostasis with a potential role in the Wolf–Hirschhorn syndrome. J. Biol. Chem. 2004, 279, 30307–30315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamai, S.; Iida, H.; Yokota, S.; Sayano, T.; Kiguchiya, S.; Ishihara, N.; Hayashi, J.-I.; Mihara, K.; Oka, T. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J. Cell Sci. 2008, 121, 2588–2600. [Google Scholar] [CrossRef] [Green Version]
- Jiang, D.; Zhao, L.; Clapham, D.E. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science 2009, 326, 144–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McQuibban, A.G.; Joza, N.; Megighian, A.; Scorzeto, M.; Zanini, D.; Reipert, S.; Richter, C.; Schweyen, R.J.; Nowikovsky, K. A Drosophila mutant of LETM1, a candidate gene for seizures in Wolf–Hirschhorn syndrome. Hum. Mol. Genet. 2010, 19, 987–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doonan, P.J.; Chandramoorthy, H.C.; Hoffman, N.E.; Zhang, X.; Cárdenas, C.; Shanmughapriya, S.; Rajan, S.; Vallem, S.; Chen, X.; Foskett, J.K.; et al. LETM1-dependent mitochondrial Ca2 flux modulates cellular bioenergetics and proliferation. FASEB J. 2014, 28, 4936–4949. [Google Scholar] [CrossRef] [Green Version]
- Shao, J.; Fu, Z.; Ji, Y.; Guan, X.; Guo, S.; Ding, Z.; Yang, X.; Cong, Y.; Shen, Y. Leucine zipper-EF-hand containing trans- membrane protein 1 (LETM1) forms a Ca2+/H+ antiporter. Sci. Rep. 2016, 6, 34174. [Google Scholar] [CrossRef] [Green Version]
- Frazier, A.E.; Taylor, R.D.; Mick, D.U.; Warscheid, B.; Stoepel, N.; Meyer, H.; Ryan, M.T.; Guiard, B.; Rehling, P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 2006, 172, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Bauerschmitt, H.; Mick, D.U.; Deckers, M.; Vollmer, C.; Funes, S.; Kehrein, K.; Ott, M.; Rehling, P.; Herrmann, J. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol. Biol. Cell 2010, 21, 1937–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durigon, R.; Mitchell, A.L.; Jones, A.W.E.; Manole, A.; Mennuni, M.; Hirst, E.M.A.; Houlden, H.; Maragni, G.; Lattante, S.; Doronzio, P.N.; et al. LETM1 couples mitochondrial DNA metabolism and nutrient preference. EMBO Mol. Med. 2018, 10, e8550. [Google Scholar] [CrossRef]
- Wiley, S.E.; Murphy, A.N.; Ross, S.A.; van der Geer, P.; Dixon, J.E. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. USA 2007, 104, 5318–5323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Ruan, Y.; Zhang, K.; Jian, F.; Hu, C.; Miao, L.; Gong, L.; Sun, L.; Zhang, X.; Chen, S.; et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016, 23, 380–392. [Google Scholar] [CrossRef] [Green Version]
- Meeusen, S.; Tieu, Q.; Wong, E.; Weiss, E.; Schieltz, D.; Yates, J.R.; Nunnari, J. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 1999, 145, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Lay, S.; Sanislav, O.; Annesley, S.J.; Fisher, P.R. Mitochondrial stress tests using Seahorse respirometry on intact Dictyostelium discoideum cells. Methods Mol. Biol. 2016, 1407, 41–61. [Google Scholar] [CrossRef]
- Gnad, F.; Forner, F.; Zielinska, D.F.; Birney, E.; Gunawardena, J.; Mann, M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol. Cell. Proteom. 2010, 9, 2642–2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotrasova, V.; Keresztesova, B.; Ondrovicova, G.; Bauer, J.A.; Havalova, H.; Pevala, V.; Kutejova, E.; Kunova, N. Mitochondrial kinases and the role of mitochondrial protein phosphorylation in health and disease. Life 2021, 11, 82. [Google Scholar] [CrossRef] [PubMed]
- Frankovsky, J.; Vozarikova, V.; Nosek, J.; Tomaska, L. Mitochondrial protein phosphorylation in yeast revisited. Mitochondrion 2021, 57, 148–162. [Google Scholar] [CrossRef]
- Castellanos, E.; Lanning, N.J. Phosphorylation of OXPHOS machinery subunits: Functional implications in cell biology and disease. Yale J. Biol. Med. 2019, 92, 523–531. [Google Scholar]
- Hojlund, K.; Wrzesinski, K.; Larsen, P.M.; Fey, S.J.; Roepstorff, P.; Handberg, A.; Dela, F.; Vinten, J.; McCormack, J.G.; Reynet, C.; et al. Proteome analysis reveals phosphorylation of ATP synthase beta -subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J. Biol. Chem. 2003, 278, 10436–10442. [Google Scholar] [CrossRef] [Green Version]
- Hojlund, K.; Yi, Z.; Lefort, N.; Langlais, P.; Bowen, B.; Levin, K.; Beck-Nielsen, H.; Mandarino, L.J. Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia 2010, 53, 541–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Leon, I.R.; Bak, S.; Mogensen, M.; Wrzesinski, K.; Hojlund, K.; Jensen, O.N. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol. Cell. Proteom. 2011, 10, M110.000299. [Google Scholar] [CrossRef] [Green Version]
- Kane, L.A.; Youngmas, M.J.; Jensen, R.E.; Van Eyk, J.E. Phosphorylation of the F1F0 ATP synthase β subunit: Functional and structural consequences in a model system. Circ. Res. 2010, 106, 504–513. [Google Scholar] [CrossRef] [Green Version]
- Strauss, M.; Hofhaus, G.; Schröder, R.R.; Kühlbrandt, W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008, 27, 1154–1160. [Google Scholar] [CrossRef] [Green Version]
- Bakhtiari, N.; Lai-Zhang, J.; Yao, B.; Mueller, D.M. Structure/function of the beta-barrel domain of F1-ATPase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 16363–16369. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Ford, H.C.; Carroll, J.; Douglas, C.; Gonzales, E.; Ding, S.; Fearnley, I.M.; Walker, J.E. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. USA 2018, 115, 2988–2993. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Godwin, M.L.; Nowak, G. Protein kinase C-alpha inhibits the repair of oxidative phosphorylation after S-(1,2-dichlorovinyl)-L-cysteine injury in renal cells. Am. J. Physiol. Renal Physiol. 2004, 287, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Di Pancrazio, F.; Bisetto, E.; Alverdi, V.; Mavelli, I.; Esposito, G.; Professor, G.L. Differential steady-state tyrosine phosphorylation of two oligomeric forms of mitochondrial F0F1 ATP synthase: A structural proteomic analysis. Proteomics 2006, 3, 921–926. [Google Scholar] [CrossRef]
- Bak, S.; Leon, I.; Jensen, O.N.; Hojlund, K. Tissue specific phosphorylation of mitochondrial proteins isolated from rat liver, heart muscle, and skeletal muscle. J. Proteome Res. 2013, 12, 4327–4339. [Google Scholar] [CrossRef]
- Yang, K.; Chen, Z.; Gao, J.; Shi, W.; Li, L.; Jiang, S.; Hu, H.; Liu, Z.; Xu, D.; Wu, L. The key roles of GSK-3beta in regulating mitochondrial activity. Cell Physiol. Biochem. 2017, 44, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Hindi, S.M.; Sato, S.; Xiong, G.; Bohnert, K.R.; Gibb, A.A.; Gallot, Y.S.; McMillan, J.D.; Hill, B.G.; Uchida, S.; Kumar, A. TAK1 regulates skeletal muscle mass and mitochondrial function. JCI Insight 2018, 3, e98441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momcilovic, M.; Hong, S.-P.; Carlson, M. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 2006, 281, 25336–25343. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Bissa, B.; Brecht, L.; Allers, L.; Choi, S.W.; Gu, Y.; Zbinden, M.; Burge, M.R.; Timmins, G.; Hallows, K.; et al. AMPK, a regulator of metabolism and autophagy, is activated by lysosomal damage via a novel galectin-directed ubiquitin signal transduction system. Mol. Cell 2020, 77, 951–969. [Google Scholar] [CrossRef]
- Mia, S.; Castor, T.; Musculus, K.; Voelkl, J.; Alesutan, I.; Lang, F. Role of AMP-activated protein kinase α1 in angiotensin-II-induced renal Tgfβ-activated kinase 1 activation. Biochem. Biophys. Res. Commun. 2016, 476, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Jeong, S.; Jung, E.; Baik, K.-H.; Chang, M.H.; Kim, S.A.; Shim, J.-H.; Chun, E.; Lee, K.-Y. AMP-activated protein kinase-α1 as an activating kinase of TGF-β-activated kinase 1 has a key role in inflammatory signals. Cell Death Dis. 2012, 3, e357. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, G.; Varughese, A.; Ji, Q.; Lee, I.; Liu, J.; Vaishnav, A.; Sinkler, C.; Kapralov, A.A.; Moraes, C.T.; Sanderson, T.H.; et al. Phosphorylation of cytochrome c threonine 28 regulates electron transport chain activity in kidney: Implications for Amp kinase. J. Biol. Chem. 2017, 292, 64–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardie, D.G.; Schaffer, B.E.; Brunet, A. AMPK: An energy-sensing pathway with multiple inputs. Trends Cell Biol. 2016, 26, 190–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, N.J.; Parker, B.L.; Chaudhuri, R.; Fisher-Wellman, K.H.; Kleinert, M.; Humphrey, S.J.; Yang, P.; Holliday, M.; Trefely, S.; Fazakerley, D.J.; et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 2015, 22, 922–935. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Jin, M.; Wang, Y.; Zhu, J.; Tan, R.; Zhao, J.; Ji, X.; Jin, C.; Jia, Y.; Ren, T.; et al. MCU-induced mitochondrial calcium uptake promotes mitochondrial biogenesis and colorectal cancer growth. Sig. Transduct. Target Ther. 2020, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, Y.; Takata, N.; Suzuki-Karasaki, M.; Yoshida, Y.; Tokuhashi, Y.; Suzuki-Karasaki, Y. Disrupting mitochondrial Ca2+ homeostasis causes tumor-selective TRAIL sensitization through mitochondrial network abnormalities. Int. J. Oncol. 2017, 51, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Thompson, C.B. Nutrient acquisition strategies of mammalian cells. Nature 2017, 546, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Hall, M.N.; Lin, S.-C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of cellular nutrient sensing and growth control. Cell Metab. 2020, 31, 472–492. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensi- tive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Jaiswal, P.; Majithia, A.R.; Rosel, D.; Liao, X.-H.; Khurana, T.; Kimmel, A.R. Integrated actions of mTOR complexes 1 and 2 for growth and development of Dictyostelium. Int. J. Dev. Biol. 2019, 63, 521–527. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Comer, F.I.; Sasaki, A.; McLeod, I.X.; Duong, Y.; Okumura, K.; Yates, J.R., III; Parent, C.A.; Firtel, R.A. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 2005, 16, 4572–4583. [Google Scholar] [CrossRef] [Green Version]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic check-point. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nada, S.; Hondo, A.; Kasai, A.; Koike, M.; Saito, K.; Uchiyama, Y.; Okada, M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009, 28, 477–489. [Google Scholar] [CrossRef] [Green Version]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal sur- face and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.-S.; Jiang, B.; Li, M.; Zhu, M.; Peng, Y.; Zhang, Y.-L.; Wu, Y.-Q.; Li, T.Y.; Liang, Y.; Lu, Z.; et al. The Lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014, 20, 526–540. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.D.; Zhang, Y.; Wie, Y.; Cho, J.-H.; Morris, L.E.; Wang, H.-Y.; Zheng, X.F.S. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell 2014, 26, 754–769. [Google Scholar] [CrossRef] [Green Version]
- Levin, R.S.; Hertz, N.T.; Burlingame, A.L.; Shokat, K.M.; Mukherjee, S. Innate immunity kinate TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc. Natl. Acad. Sci. USA 2016, 113, 4776–4783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, P.; Kimmel, A.R. mTORC1/AMPK responses define a core gene set for developmental cell fate switching. BMC Biol. 2019, 17, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Missailidis, D.; Annesley, S.J.; Allan, C.Y.; Sanislav, O.; Lidbury, B.A.; Lewis, D.P.; Fisher, P.R. An isolated Complex V inefficiency and dysregulated mitochondrial function in immortalized lymphocytes from ME/CFS patients. Int. J. Mol. Sci. 2020, 21, 1074. [Google Scholar] [CrossRef] [Green Version]
- Missailidis, D.; Sanislav, O.; Allan, C.Y.; Smith, P.K.; Annesley, S.J.; Fisher, P.R. Dysregulated provision of oxidisable substrates to the mitochondria in ME/CFS lymphoblasts. Int. J. Mol. Sci. 2021, 22, 2046. [Google Scholar] [CrossRef]
- Watts, D.J.; Ashworth, J.M. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 1970, 119, 171–174. [Google Scholar] [CrossRef] [Green Version]
- Lowry, O.H.; Rosenbrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Boersema, P.J.; Mohammed, S.; Heck, A.J. Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass Spectrom. 2009, 44, 861–878. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ye, M.; Dong, J.; Corradini, E.; Cristobal, A.; Heck, A.J.; Zou, H.; Mohammed, S. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 2013, 8, 461–480. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Czarna, M.; Mathy, G.; Mac’Cord, A.; Dobson, R.; Jarmuszkiewicz, W.; Sluse-Goffart, C.M.; Leprince, P.; De Pauw, E.; Sluse, F.E. Dynamics of the Dictyostelium discoideum mitochondrial proteome during vegetative growth, starvation and early stages of development. Proteomics 2010, 10, 6–22. [Google Scholar] [CrossRef]
- Dyballa, N.; Metzger, S. Fast and sensitive colloidal Coomassie G-250 staining for proteins in polyacrylamide gels. J. Vis. Exp. 2009, 30, e1431. [Google Scholar] [CrossRef] [PubMed]




| UniProt | DictyBase | Protein | Phosphopeptide(s) | Phosphorylation Site |
|---|---|---|---|---|
| OXPHOS | ||||
| Q9U3X4 | DDB0214886 | Succinate dehydrogenase (sdhA) | gEGGYLLNSsGER | Ser-300 |
| Q54D07 | DDB0238603 | Cytochrome c1, heme protein (cyc1) | amAADtTVmDGPDSEGDmFER amAADTTVmDGPDsEGDmFER | Thr-104 Ser-112 |
| Q1ZXP3 | DDB0233077 | Cytochrome b-c1 complex, subunit 6 (uqcrh) | gPIQEGcAsGcEk vkGPIQEGcAsGcEk | Ser-18 |
| Q54V76 | DDB0267111 | Cytochrome b-c1 complex, subunit 8 (uqcrq) | qITYSVsPFQQk | Ser-17 |
| Q54NW9 | DDB0238608 | Cytochrome b-c1 complex, subunit Rieske (ucr) | fITsDkIIVGDE | Ser-206 |
| P30815 | DDB0214995 | Cytochrome c oxidase, subunit 4 (cxdA) | vGsPEFDk | Ser-55 |
| P29505 | DDB0191104 | Cytochrome c oxidase, subunit 5 (cxeA) | hISsEGEVmYY | Ser-113 |
| A9CLV8 | DDB0350620 | ATP synthase, subunit 4 (atp4) | dLsSDEITNSSTk dLSsDEITNSSTk dLSSDEItNSSTk | Ser-108 Ser-109 Thr-113 |
| Q54DF1 | DDB0237782 | ATP synthase, subunit gamma (atp5C1) | vLGVVETADAFNTAtEPIEDR | Thr-85 |
| Q54RA8 | DDB0266798 | ATP synthase, subunit O (OSCP) (atp5O) | tALEGDIDNsEk yLtLDESkN | Ser-133 Thr-290 |
| Q55CS9 | DDB0233951 | ATP synthase, subunit beta (atp5B) | sLLDkESNEESTEVDYSk sLLDkEsNEESTEVDYSk sLLDkESNEEsTEVDYSk sLLDkESNEEStEVDYSk vIEDLNNPsLk aPPPFADLAPSAsILETGIk | Ser-58 Ser-64 Ser-68 Thr-69 Ser-128 Ser-231 |
| OXPHOS complex regulation and assembly | ||||
| Q54ID0 | DDB0305161 | Cytochrome c oxidase copper chaperone (cox17) | sIAETNTTTEVAAPk | Ser-2 |
| Q9GSE7 | DDB0216175 | F1F0-ATPase putative regulatory protein (if1) | kAGSQPTPNASSSANNsk | Ser-81 |
| Genome repair & maintenance | ||||
| Q8MYF0 | DDB0304673 | Mitochondrial genome maintenance protein (mgm101) | iTEQQDDsEDIDIDDVVPPQLk | Ser-315 |
| Protein synthesis, folding & stabilization | ||||
| Q54WN8 | DDB0304956 | Uncharacterized protein; probable mitochondrial small ribosomal subunit (DDB_G0279527) | eIIDQNPNDStDmPITR | Thr-846 |
| Q54CA5 | DDB0306787 | S5 DRBM domain-containing protein, mitochondrial small ribosomal subunit | tIFEDGELDTPsVNSIR tIFEDGELDTPSVNsIR lGTVEHEELtFDHEk rFDEDYAEEsEILSQFSk eLsEYEEVIHQR | Ser-802 Ser-805 Thr-915 Ser-1006 Ser-1587 |
| Q55GH1 | DDB0306542 | Protein similar to yeast tRNA threonylcarbamoyladenosine dehydratase 2 (DDB_G0268496) | sLNGGGGGGDDDGDNNNSsPSNQHIDk | Ser-51 |
| Q54F93 | DDB0232199 | Mitochondrial-processing peptidase subunit alpha-2 (mppA) | vTFGNDESSTSIsNETAQYIGGESLk vTFGNDESSTSISNETAQYIGGESLkYSSGNsk | Ser-235 Ser-254 |
| Q8I0H7 | DDB0215366 | Heat shock 70 kDa protein, mitochondrial (mhsp70) | dNTTEAEFtEkk | Thr-655 |
| Q8MPA5 | DDB0232124 | Heat shock protein, Hsp20 domain-containing protein (hspG7) | sSTsPSSSTLDSk | Ser-96 |
| C7G004 | DDB0304476 | Heat shock protein, DnaJ family protein (DDB_G0304475) | yIDNLIIPSSSSsDSGSGSGGSk yIDNLIIPSSSSSDsGSGSGGSk yIDNLIIPSSSSSDSGSGSGGsk | Ser-213 Ser-215 Ser-222 |
| Q54Q31 | DDB0232062 | Prohibitin-2 (phbB) | sIsSLTGSk | Ser-91 |
| Transport | ||||
| Q01501 | DDB0185213 | Mitochondrial outer membrane protein porin (porA) | yGsIVAVTDIk qILLSTLYTATsk | Ser-47 Ser-191 |
| O97470 | DDB0201558 | Mitochondrial substrate carrier family protein ANT; ADP/ATP carrier protein (ancA) | dsLIGGTAGGVSk lLLQVQsASTQIAADk lLLQVQSAsTQIAADk lLLQVQSAStQIAADk lAADVGtGSAR lAADVGTGsAR lmGFEGGVGsE | Ser-16 Ser-44 Ser-46 Thr-47 Thr-150 Ser-152 Ser-308 |
| Q54BF6 | DDB0233888 | Mitochondrial substrate carrier family protein N (mcfN) | aGDLtPSLFLk | Thr-6 |
| Q54H87 | DDB0346938 | Uncharacterized protein (similar to S. cerevisiae mitochondrial phosphate transport protein PHO88m) (DDB_G0289621) | vTEINEsESSSEESEkEEk | Ser-216 |
| Q86AV5 | DDB0234131 | Mitochondrial substrate carrier family protein X (mcfX) | gLSsNLIGIIPEk | Ser-87 |
| Q54Y17 | DDB0306880 | LETM1 and EF-hand domain-containing protein (DDB_G0278471) | sGQtVTSDEVLk | Thr-282 |
| Metabolism | ||||
| Q55GD7 | DDB0306487 | CDGSH iron-sulfur domain-containing protein (ortholog of human CISD3), similar to mitoNEET-related protein 2 (DDB_G0267712) | yNEETGLNDsPLk yNEETGLNDsPLkVEk kYNEETGLNDsPLk | Ser-75 |
| Q555A3 | DDB0203193 | Carbonic anhydrase (DDB_G0274643) | lkENISLSTsN lkENISLStSN | Ser-273 Thr-272 |
| Q55BA8 | DDB0215348 | Probable calnexin (cnxA) | eSVSIQDkPTIEsEESDESDEDNETTkk eSVSIQDkPTIESEEsDESDEDNETTk eSVSIQDkPTIESEESDEsDEDNETTk | Ser-510 Ser-513 Ser-516 |
| Signalling | ||||
| P51136 | DDB0185150 | Glycogen synthase kinase-3 (gskA) | gETNVsYIcSR gETNVSyIcSR iLIkGETNVSyIcSR | Ser-213 Tyr-214 |
| O61122 | DDB0191176 | Severin kinase (svkA) | sLsNSSQTTPVk | Ser-375 |
| Q54RB7 | DDB0191149 | Dual specificity protein kinase SHKA (shkA) | aQLSGyIN | Tyr-525 |
| Q54U31 | DDB0230122 | Dual specificity protein kinase SHKD (shkD) | fTQETFNPyDPYTN | Tyr-739 |
| O00910 | DDB0215388 | Signal transducer and activator of transcription A (dstA) | rTAPVPVGGyEPLNS | Tyr-702 |
| Q1ZXA8 | DDB0231625 | Protein similar to human Ragulator complex protein LAMTOR1 (DDB_G0292160) | nQASSSQQPSSSQtPSk nQASSSQQPSSSQTPsk dSEEQPQEVSYsQmR ecGELVVFFGNsLk | Thr-34 Ser-36 Ser-56 Ser-172 |
| P34139 | DDB0191476 | Ras-related protein Rab1A (rab1A) | tITSsYYR | Ser-76 |
| UniProt | DictyBase | Protein | Kinase Group | Mitochondrial Preparations |
|---|---|---|---|---|
| P51136 | DDB0185150 | Glycogen synthase kinase-3 | CMGC kinases | 2 |
| O61122 | DDB0191176 | Severin kinase | STE kinases | 2 |
| Q54RB7 | DDB0191149 | Dual specificity protein kinase SHKA | TKL kinases | 3 |
| Q54U31 | DDB0230122 | Dual specificity protein kinase SHKD | TKL kinases | 1 |
| DictyBase | Protein | P-Site |
|---|---|---|
| Proteins with up-regulated phosphorylation | ||
| DDB0306880 | LETM1 and EF-hand domain-containing protein | Thr-282 |
| DDB0306787 | S5 DRBM domain-containing protein | Ser-1587 |
| DDB0306113 | Uncharacterized protein | Thr-65 |
| DDB0306695 | Uncharacterized protein | Thr-367 |
| Proteins with down-regulated phosphorylation | ||
| DDB0238603 | Cytochrome c1, heme protein | Ser-112 |
| DDB0237782 | ATP synthase, subunit gamma | Thr-85 |
| DDB0233951 | ATP synthase, subunit beta | Thr-69 |
| DDB0215366 | Heat shock 70 kDa protein, mitochondrial | Thr-655 |
| DDB0304476 | Heat shock protein, DnaJ family protein | Ser-213 |
| DDB0201558 | Mitochondrial substrate carrier family protein ANT | Ser-152, 308 |
| DDB0234131 | Mitochondrial substrate carrier family protein X | Ser-87 |
| DDB0306487 | Similar to mitoNEET-related protein 2 | Ser-75 |
| DDB0215348 | Probable calnexin | Ser-513 |
| DDB0191149 | Dual specificity protein kinase SHKA | Tyr-525 |
| DDB0231625 | Protein similar to human Ragulator complex protein LAMTOR1 | Thr-34, Ser-56, 172 |
| DDB0191476 | Ras-related protein Rab-1A | Ser-76 |
| DDB0304899 | Uncharacterized protein | Ser-290 |
| DDB0347754 | Uncharacterized protein | Ser-106, 107 |
| DDB0305799 | Uncharacterized protein | Ser-61, 536 |
| DDB0302557 | Uncharacterized protein | Thr-52 |
| DDB0307109 | Uncharacterized protein | Ser-647, 648, 650 |
| DDB0306527 | Uncharacterized protein | Ser-314 |
| DDB0346952 | Uncharacterized protein | Ser-230 |
| DDB0232257 | Uncharacterized protein | Ser-473 |
| DDB0306348 | Uncharacterized protein | Ser-128, Thr-130 |
| DDB0346952 | Uncharacterized protein | Ser-230 |
| DDB0307677 | Uncharacterized protein | Ser-301 |
| DDB0306179 | Uncharacterized protein | Ser-333 |
| DDB0191444 | Myosin-2 heavy chain | Ser-1636, 1637 |
| DDB0191351 | Myosin IB heavy chain | Tyr-334 |
| DDB0185146 | Myosin regulatory light chain | Ser-14 |
| DDB0214946 | Probable ATPase, P-type ATPase | Ser-22 |
| DDB0232252 | Crt homolog 3 | Ser-438 |
| DDB0191505 | Vacuolin-A | Ser-14 |
| DDB0234063 | Phospholipid-translocating ATPase | Ser-86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heidorn-Czarna, M.; Heidorn, H.-M.; Fernando, S.; Sanislav, O.; Jarmuszkiewicz, W.; Mutzel, R.; Fisher, P.R. Chronic Activation of AMPK Induces Mitochondrial Biogenesis through Differential Phosphorylation and Abundance of Mitochondrial Proteins in Dictyostelium discoideum. Int. J. Mol. Sci. 2021, 22, 11675. https://doi.org/10.3390/ijms222111675
Heidorn-Czarna M, Heidorn H-M, Fernando S, Sanislav O, Jarmuszkiewicz W, Mutzel R, Fisher PR. Chronic Activation of AMPK Induces Mitochondrial Biogenesis through Differential Phosphorylation and Abundance of Mitochondrial Proteins in Dictyostelium discoideum. International Journal of Molecular Sciences. 2021; 22(21):11675. https://doi.org/10.3390/ijms222111675
Chicago/Turabian StyleHeidorn-Czarna, Malgorzata, Herbert-Michael Heidorn, Sanjanie Fernando, Oana Sanislav, Wieslawa Jarmuszkiewicz, Rupert Mutzel, and Paul R. Fisher. 2021. "Chronic Activation of AMPK Induces Mitochondrial Biogenesis through Differential Phosphorylation and Abundance of Mitochondrial Proteins in Dictyostelium discoideum" International Journal of Molecular Sciences 22, no. 21: 11675. https://doi.org/10.3390/ijms222111675
APA StyleHeidorn-Czarna, M., Heidorn, H.-M., Fernando, S., Sanislav, O., Jarmuszkiewicz, W., Mutzel, R., & Fisher, P. R. (2021). Chronic Activation of AMPK Induces Mitochondrial Biogenesis through Differential Phosphorylation and Abundance of Mitochondrial Proteins in Dictyostelium discoideum. International Journal of Molecular Sciences, 22(21), 11675. https://doi.org/10.3390/ijms222111675







