Antimicrobial Peptides: An Update on Classifications and Databases
Abstract
:1. Introduction
2. Sources of AMPs
2.1. Bacteriophage/Viral AMPs
2.2. Bacterial AMPs
2.2.1. AMPs from Gram-Positive Bacteria
2.2.2. AMPs from Gram-Negative Bacteria
2.3. Fungal AMPs
2.4. Plant Derived AMPs
2.5. Animal Derived AMPs
2.5.1. Invertebrates
2.5.2. Fish and Amphibian AMPs
2.5.3. Reptile- and Avian-Derived Peptides
2.5.4. Mammalian-Derived AMPs
3. Structural and Physicochemical Properties of AMPs
3.1. Sequence Based Classification
3.1.1. UCLL/Class L
3.1.2. UCSS/Class S
3.1.3. UCSB/Class P
3.1.4. UCBB/Class O
3.2. Structure-Based Classification
3.2.1. α-helix AMPs
3.2.2. β-sheet AMPs
3.2.3. αβ AMPs
3.2.4. Non-αβ AMPs
3.2.5. Cyclic and Unusual or Complex AMPs
4. Diverse Activities and Modes of Action
4.1. Antiviral AMPs
4.2. Antibacterial AMPs
4.2.1. Membrane Targeting AMPs
4.2.2. Non-Membrane Targeting/Intracellular AMPs
4.3. Antifungal AMPs
5. AMP Databases
5.1. APD3
5.2. CAMPR3
5.3. dbAMP
5.4. DBAASP
5.5. LAMP2
6. Prediction Functionality in AMP Databases
6.1. Sequence Alignment
6.2. Pattern-Matching
6.3. Profile-HMM
6.4. Machine Learning and Deep Learning
6.5. Molecular Dynamics (MD) Simulations
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO Antimicrobial Resistance. Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar] [CrossRef]
- Upton, M.; Cotter, P.; Tagg, J. Antimicrobial peptides as therapeutic agents. Int. J. Microbiol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, R.J. Studies on a bactericidal agent extracted from a soil bacillus: I. Preparation of the agent. Its activity in vitro. J. Exp. Med. 1939, 70, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, R.J. Studies on a bactericidal agent extracted from a soil bacillus: II. Protective effect of the bactericidal agent against experimental pieuococcus infections in mice. J. Exp. Med. 1939, 70, 11–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012, 32, 143–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.H.; Lu, T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chromek, M.; Slamová, Z.; Bergman, P.; Kovács, L.; Podracká, L.; Ehrén, I.; Hökfelt, T.; Gudmundsson, G.H.; Gallo, R.L.; Agerberth, B.; et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006, 12, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Pretzel, J.; Mohring, F.; Rahlfs, S.; Becker, K. Antiparasitic peptides. Adv. Biochem. Eng. Biotechnol. 2013, 135, 157–192. [Google Scholar] [CrossRef]
- De Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; De Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; Van Der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinstraesser, L.; Kraneburg, U.; Jacobsen, F.; Al-Benna, S. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology 2011, 216, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Nijnik, A.; Philpott, D.J. Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 2012, 10, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.K.; Craik, D.J. Designing macrocyclic disulfide-rich peptides for biotechnological applications perspective. Nat. Chem. Biol. 2018, 14, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Lawrence, N.; Chaousis, S.; Ravipati, A.S.; Cheneval, O.; Benfield, A.H.; Elliott, A.G.; Kavanagh, A.M.; Cooper, M.A.; Chan, L.Y.; et al. Redesigned Spider Peptide with Improved Antimicrobial and Anticancer Properties. ACS Chem. Biol. 2017, 12, 2324–2334. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; Rodríguez, A.; García, P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013, 39, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Mao, J.; Xie, J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs 2014, 28, 265–274. [Google Scholar] [CrossRef]
- Parisien, A.; Allain, B.; Zhang, J.; Mandeville, R.; Lan, C.Q. Novel alternatives to antibiotics: Bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 2008, 104, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens virulent bacteriophage CPS2 and its thermostable endolysin lysCPS2. Viruses 2018, 10, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotka, M.; Kapusta, M.; Dorawa, S.; Kaczorowska, A.K.; Kaczorowski, T. Ts2631 endolysin from the extremophilic thermus scotoductus bacteriophage vB_Tsc2631 as an antimicrobial agent against gram-negative multidrug-resistant bacteria. Viruses 2019, 11, 657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastagia, M.; Schuch, R.; Fischetti, V.A.; Huang, D.B. Lysins: The arrival of pathogen-directed anti-infectives. J. Med. Microbiol. 2013, 62, 1506–1516. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Ross, R.P.; Coffey, A. Bacteriophage and their lysins for elimination of infectious bacteria: Review article. FEMS Microbiol. Rev. 2009, 33, 801–819. [Google Scholar] [CrossRef] [Green Version]
- Salas, M.; Wernecki, M.; Fernández, L.; Iglesias, B.; Gutiérrez, D.; Álvarez, A.; García, L.; Prieto, E.; García, P.; Rodríguez, A. Characterization of clinical MRSA isolates from Northern Spain and assessment of their susceptibility to phage-derived antimicrobials. Antibiotics 2020, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.Y.; You, R.I.; Lai, M.J.; Lin, N.T.; Chen, L.K.; Chang, K.C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Wu, J.; Liu, H.; Yang, H.; Rong, M.; Li, D.; Zhang, P.; Han, J.; Lai, R. A mycobacteriophage-derived trehalose-6,6=-dimycolatebinding peptide containing both antimycobacterial and anti-inflammatory abilities. FASEB J. 2013, 27, 3067–3077. [Google Scholar] [CrossRef]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 2004, 186, 4808–4812. [Google Scholar] [CrossRef] [Green Version]
- Hermoso, J.A.; García, J.L.; García, P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007, 10, 461–472. [Google Scholar] [CrossRef]
- Zimmer, M.; Vukov, N.; Scherer, S.; Loessner, M.J. The murein hydrolase of the bacteriophage φ3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl. Environ. Microbiol. 2002, 68, 5311–5317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Donovan, D.M.; García, P. Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiiPla88 HydH5 Virion-associated peptidoglycan hydrolase: Fusions, deletions, and synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, L.; Martínez, B.; Zhou, Y.; Rodríguez, A.; Donovan, D.M.; García, P. Lytic activity of the virion-associated peptidoglycan hydrolase HydH5 of Staphylococcus aureus bacteriophage vB-SauS-phiIPLA88. BMC Microbiol. 2011, 11, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moak, M.; Molineux, I.J. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 2004, 51, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molineux, I.J. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 2001, 40, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Moak, M.; Molineux, I.J. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 2000, 37, 345–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldentey, J.; Bamford, D.H. The lytic enzyme of the Pseudomonas phage φ6. Purification and biochemical characterization. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1992, 1159, 44–50. [Google Scholar] [CrossRef]
- Weng, S.F.; Fu, Y.C.; Lin, J.W.; Tseng, T.T. Identification of a Broad-Spectrum Peptidoglycan Hydrolase Associated with the Particle of Xanthomonas oryzae Phage Xop411. J. Mol. Microbiol. Biotechnol. 2018, 28, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Molineux, I.J. Evolution of a new enzyme activity from the same motif fold. Mol. Microbiol. 2008, 69, 287–290. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.J.; Lin, T.L.; Lin, Y.T.; Su, P.A.; Chen, C.T.; Hsieh, P.F.; Hsu, C.R.; Chen, C.C.; Hsieh, Y.C.; Wang, J.T. Identification of capsular types in carbapenem-resistant klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.-J.; Lin, T.-L.; Chen, C.-C.; Tsai, Y.-T.; Cheng, Y.-H.; Chen, Y.-Y.; Hsieh, P.-F.; Lin, Y.-T.; Wang, J.-T. Klebsiella Phage ΦK64-1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Cornelissen, A.; Ceyssens, P.J.; Krylov, V.N.; Noben, J.P.; Volckaert, G.; Lavigne, R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 2012, 434, 251–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carson, L.; Gorman, S.P.; Gilmore, B.F. The use of lytic bacteriophages in the prevention and eradication of biofilms of Proteus mirabilis and Escherichia coli. FEMS Immunol. Med. Microbiol. 2010, 59, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Shi, Y.; Ji, W.; Meng, X.; Zhang, J.; Wang, H.; Lu, C.; Sun, J.; Yan, Y. Application of a bacteriophage lysin to disrupt biofilms formed by the animal pathogen Streptococcus suis. Appl. Environ. Microbiol. 2011, 77, 8272–8279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chibeu, A.; Lingohr, E.J.; Masson, L.; Manges, A.; Harel, J.; Ackermann, H.W.; Kropinski, A.M.; Boerlin, P. Bacteriophages with the ability to degrade uropathogenic Escherichia Coli biofilms. Viruses 2012, 4, 471–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, R.; Bläsi, U. Holins: Form and function in bacteriophage lysis. FEMS Microbiol. Rev. 1995, 17, 191–205. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage holins: Deadly diversity. J. Mol. Microbiol. Biotechnol. 2002, 4, 21–36. [Google Scholar] [PubMed]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N.; Deaton, J.; Young, R. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J. Bacteriol. 2003, 185, 779–787. [Google Scholar] [CrossRef] [Green Version]
- Krupovič, M.; Bamford, D.H. Holin of bacteriophage lambda: Structural insights into a membrane lesion. Mol. Microbiol. 2008, 69, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.; Fleming, T.C.; Pogliano, K.; Young, R. Visualization of pinholin lesions in vivo. Proc. Natl. Acad. Sci. USA 2013, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Niu, W.; Wu, R.; Wang, J.; Lei, L.; Han, W.; Gu, J. The Phage Holin HolGH15 Exhibits Potential As an Antibacterial Agent to Control Listeria monocytogenes. Foodborne Pathog. Dis. 2020. [Google Scholar] [CrossRef]
- Song, J.; Xia, F.; Jiang, H.; Li, X.; Hu, L.; Gong, P.; Lei, L.; Feng, X.; Sun, C.; Gu, J.; et al. Identification and characterization of HolGH15: The Holin of Staphylococcus Aureus bacteriophage GH15. J. Gen. Virol. 2016, 97, 1272–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, N.; Sun, Y.; Wang, Q.; Qiu, Y.; Chen, Z.; Wen, Y.; Wang, S.; Song, Y. Cloning and characterization of endolysin and holin from Streptomyces avermitilis bacteriophage phiSASD1 as potential novel antibiotic candidates. Int. J. Biol. Macromol. 2020, 147, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, M.; Karimi, A.; Fallah, F.; Akhavan, M.M. Overview of ribosomal and non-ribosomal antimicrobial peptides produced by Gram positive bacteria. Cell. Mol. Biol. 2017, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Diep, D.; Nes, I. Ribosomally Synthesized Antibacterial Peptides in Gram Positive Bacteria. Curr. Drug Targets 2005, 3, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.; Draper, L.; Ross, R.; Cotter, P. Lantibiotic Immunity. Curr. Protein Pept. Sci. 2008, 9, 39–49. [Google Scholar] [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Bierbaum, G.; Szekat, C.; Josten, M.; Heidrich, C.; Kempter, C.; Jung, G.; Sahl, H.G. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol. 1996, 62, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Sahl, H.-G.; Jack, R.W.; Bierbaum, G. Biosynthesis and Biological Activities of Lantibiotics with Unique Post-Translational Modifications. Eur. J. Biochem. 1995, 230, 827–853. [Google Scholar] [CrossRef]
- Nishie, M.; Nagao, J.I.; Sonomoto, K. Antibacterial peptides “bacteriocins”: An overview of their diverse characteristics and applications. Biocontrol Sci. 2012, 17, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willey, J.M.; Van Der Donk, W.A. Lantibiotics: Peptides of diverse structure and function. Annu. Rev. Microbiol. 2007, 61, 477–501. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Van Der Donk, W.A. Post-translational modifications during lantibiotic biosynthesis. Curr. Opin. Chem. Biol. 2004, 8, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Guder, A.; Wiedemann, I.; Sahl, H. Posttranslationally modified bacteriocins—the lantibiotics. Biopolymers 2000, 55, 62–73. [Google Scholar] [CrossRef]
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Hasper, H.E.; Kramer, N.E.; Smith, J.L.; Hillman, J.D.; Zachariah, C.; Kuipers, O.P.; De Kruijff, B.; Breukink, E. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 2006, 313, 1636–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, S.T.D.; Breukink, E.; Tischenko, E.; Lutters, M.A.G.; De Kruijff, B.; Kaptein, R.; Bonvin, A.M.J.J.; Van Nuland, N.A.J. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 2004, 11, 963–967. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Ulm, H.; Reder-Christ, K.; Sahl, H.G.; Schneider, T. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb. Drug Resist. 2012, 18, 261–270. [Google Scholar] [CrossRef]
- Wiedemann, I.; Breukink, E.; Van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; De Kruijff, B.; Sahl, H.G. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbour, A.; Philip, K.; Muniandy, S. Enhanced Production, Purification, Characterization and Mechanism of Action of Salivaricin 9 Lantibiotic Produced by Streptococcus salivarius NU10. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Geng, M.; Austin, F.; Shin, R.; Smith, L. Covalent structure and bioactivity of the type All lantibiotic salivaricin A2. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbour, A.; Tagg, J.; Abou-Zied, O.K.; Philip, K. New insights into the mode of action of the lantibiotic salivaricin B. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zitzer, A.; Palmer, M.; Weller, U.; Wassenaar, T.; Biermann, C.; Tranum-Jensen, J.; Bhakdi, S. Mode of primary binding to target membranes and pore formation induced by Vibrio cholerae cytolysin (hemolysin). Eur. J. Biochem. 1997, 247, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Kumar Rai, A.; Chattopadhyay, K. Vibrio cholerae cytolysin: Structure–function mechanism of an atypical β-barrel pore- forming toxin. Adv. Exp. Med. Biol. 2015, 842, 109–125. [Google Scholar] [CrossRef]
- Jack, R.W.; Tagg, J.R.; Ray, B. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 1995, 59, 171–200. [Google Scholar] [CrossRef] [PubMed]
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2010, 2, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oscáriz, J.C.; Pisabarro, A.G. Classification and mode of action of membrane-active bacteriocins produced by gram-positive bacteria. Int. Microbiol. 2001, 4, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Fimland, G.; Johnsen, L.; Dalhus, B.; Nissen-Meyer, J. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: Biosynthesis, structure, and mode of action. J. Pept. Sci. 2005, 11, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Hill, C.; Ross, R.P. Food microbiology: Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Green, G.; Dicks, L.M.T.; Bruggeman, G.; Vandamme, E.J.; Chikindas, M.L. Pediocin PD-1, a bactericidal antimicrobial peptide from Pediococcus damnosus NCFB 1832. J. Appl. Microbiol. 1997, 83, 127–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devi, S.M.; Halami, P.M. Detection and characterization of pediocin PA-1/AcH like bacteriocin producing lactic acid bacteria. Curr. Microbiol. 2011, 63, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernández, P.E.; Nes, I.F. Biochemical and genetic characterization of enterocin P, a novel sec- dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nissen-Meyer, J.; Rogne, P.; Oppegard, C.; Haugen, H.; Kristiansen, P. Structure-Function Relationships of the Non-Lanthionine-Containing Peptide (class II) Bacteriocins Produced by Gram-Positive Bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; Garcia-Garcera, M.J.; Driessen, A.J.M.; Ledeboer, A.M.; Nissen- Meyer, J.; Nes, I.F.; Abee, T.; Konings, W.N.; Venema, G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 1993, 59, 3577–3584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Héchard, Y.; Sahl, H.G. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 2002, 84, 545–557. [Google Scholar] [CrossRef]
- Abee, T.; Klaenhammer, T.R.; Letellier, L. Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration complexes in the cytoplasmic membrane. Appl. Environ. Microbiol. 1994, 60, 1006–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moll, G.; Ubbink-Kok, T.; Hildeng-Hauge, H.; Nissen-Meyer, J.; Nes, I.F.; Konings, W.N.; Driessen, A.J.M. Lactococcin G is a potassium ion-conducting, two-component bacteriocin. J. Bacteriol. 1996, 178, 600–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moll, G.; Hildeng-hauge, H.; Nissen-meyer, J.; Nes, I.F.; Konings, W.N.; Driessen, A.J.M. Mechanistic properties of the two-component bacteriocin lactococcin G. J. Bacteriol. 1998, 180, 96–99. [Google Scholar] [CrossRef] [Green Version]
- Rea, M.C.; Ross, R.P.; Cotter, P.D.; Hill, C. Classification of Bacteriocins from Gram-Positive Bacteria. In Prokaryotic Antimicrobial Peptides; Springer: New York, NY, USA, 2011; pp. 29–53. [Google Scholar]
- Zendo, T. Screening and characterization of novel bacteriocins from lactic acid bacteria. Biosci. Biotechnol. Biochem. 2013, 77, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Structure and genetics of circular bacteriocins. Trends Microbiol. 2011, 19, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, E.E.; Daly, C.; Fitzgerald, G.F. Identification and characterization of helveticin V-1829, a bacteriocin produced by Lactobacillus helveticus 1829. J. Appl. Bacteriol. 1992, 73, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Heng, N.C.K.; Tagg, J.R. What’s in a name? Class distinction for bacteriocins. Nat. Rev. Microbiol. 2006, 4, 160. [Google Scholar] [CrossRef]
- Heng, N.C.K.; Wescombe, P.A.; Burton, J.P.; Jack, R.W.; Tagg, J.R. The Diversity of Bacteriocins in Gram-Positive Bacteria. Bacteriocins 2007, 45–92. [Google Scholar] [CrossRef]
- da Silva Sabo, S.; Vitolo, M.; González, J.M.D.; de Souza Oliveira, R.P. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 2014, 64, 527–536. [Google Scholar] [CrossRef] [PubMed]
- De Martinis, E.C.P.; Alves, V.F.; Franco, B.D.G.M. Fundamentals and perspectives for the use of bacteriocins produced by lactic acid bacteria in meat products. Food Rev. Int. 2002, 18, 191–208. [Google Scholar] [CrossRef]
- Benedict, R.G.; Langlykke, A.F. Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 1947, 54, 24. [Google Scholar] [PubMed]
- Stansly, P.G.; Shepherd, R.G.; White, H.J. Polymyxin: A new chemotherapeutic agent. Bull. Johns Hopkins Hosp. 1947, 81, 43–54. [Google Scholar] [PubMed]
- Velkov, T.; Thompson, P.E.; Azad, M.A.K.; Roberts, K.D.; Bergen, P.J. History, Chemistry and Antibacterial Spectrum. Adv. Exp. Med. Biol. 2019, 1145, 15–36. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. JMS J. Med. Soc. 2018, 32, 76–77. [Google Scholar] [CrossRef]
- Deris, Z.Z.; Akter, J.; Sivanesan, S.; Roberts, K.D.; Thompson, P.E.; Nation, R.L.; Li, J.; Velkov, T. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J. Antibiot. 2014, 67, 147–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampson, T.R.; Liu, X.; Schroeder, M.R.; Kraft, C.S.; Burd, E.M.; Weiss, D.S. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 2012, 56, 5642–5649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, M.; Chavan, M. (Eds.) Bacteriocins: Ecology and Evolution; Springer: Berlin/Heidelberg, Germany, 2007; p. 150. [Google Scholar]
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubès, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin Biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillor, O.; Kirkup, B.C.; Riley, M.A. Colicins and microcins: The next generation antimicrobials. Adv. Appl. Microbiol. 2004, 54, 129–146. [Google Scholar] [CrossRef] [PubMed]
- Papadakos, G.; Wojdyla, J.A.; Kleanthous, C. Nuclease colicins and their immunity proteins. Q. Rev. Biophys. 2012, 45, 57–103. [Google Scholar] [CrossRef] [PubMed]
- Parret, A.H.A.; De Mot, R. Bacteria killing their own kind: Novel bacteriocins of Pseudomonas and other γ-proteobacteria. Trends Microbiol. 2002, 10, 107–112. [Google Scholar] [CrossRef]
- Denkovskienė, E.; Paškevičius, Š.; Misiūnas, A.; Stočkūnaitė, B.; Starkevič, U.; Vitkauskienė, A.; Hahn-Löbmann, S.; Schulz, S.; Giritch, A.; Gleba, Y.; et al. Broad and Efficient Control of Klebsiella Pathogens by Peptidoglycan-Degrading and Pore-Forming Bacteriocins Klebicins. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel-Briand, Y.; Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 2002, 84, 499–510. [Google Scholar] [CrossRef]
- Asensio, C.; Pérez-Díaz, J.C.; Martínez, M.C.; Baquero, F. A new family of low molecular weight antibiotics from enterobacteria. Biochem. Biophys. Res. Commun. 1976, 69, 7–14. [Google Scholar] [CrossRef]
- Jack, R.W.; Jung, G. Lantibiotics and microcins: Polypeptides with unusual chemical diversity. Curr. Opin. Chem. Biol. 2000, 4, 310–317. [Google Scholar] [CrossRef]
- Duquesne, S.; Destoumieux-Garzón, D.; Peduzzi, J.; Rebuffat, S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 2007, 24, 708–734. [Google Scholar] [CrossRef] [PubMed]
- Pons, A.M.; Lanneluc, I.; Cottenceau, G.; Sable, S. New developments in non-post translationally modified microcins. Biochimie 2002, 84, 531–537. [Google Scholar] [CrossRef]
- Braun, V.; Patzer, S.I.; Hantke, K. Ton-dependent colicins and microcins: Modular design and evolution. Biochimie 2002, 84, 365–380. [Google Scholar] [CrossRef]
- Herrero, M.; Moreno, F. Microcin B17 blocks DNA replication and induces the SOS system in Escherichia coli. J. Gen. Microbiol. 1986, 132, 393–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adelman, K.; Yuzenkova, J.; La Porta, A.; Zenkin, N.; Lee, J.; Lis, J.T.; Borukhov, S.; Wang, M.D.; Severinov, K. Molecular mechanism of transcription inhibition by peptide antibiotic Microcin J25. Mol. Cell 2004, 14, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Rintoul, M.R.; De Arcuri, B.F.; Morero, R.D. Effects of the antibiotic peptide microcin J25 on liposomes: Role of acyl chain length and negatively charged phospholipid. Biochim. Biophys. Acta Biomembr. 2000, 1509, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Scholl, D. Phage Tail-Like Bacteriocins. Annu. Rev. Virol. 2017, 4, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Daw, M.A.; Falkiner, F.R. Bacteriocins: Nature, function and structure. Micron 1996, 27, 467–479. [Google Scholar] [CrossRef]
- Morse, S.A.; Jones, B.V.; Lysko, P.G. Pyocin inhibition of Neisseria gonorrhoea: Mechanism of action. Antimicrob. Agents Chemother. 1980, 18, 416–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.; Chakraborty, U.; Gebhart, D.; Govoni, G.R.; Zhou, Z.H.; Scholl, D. F-type bacteriocins of Listeria monocytogenes: A new class of phage tail-like structures reveals broad parallel coevolution between tailed bacteriophages and high-molecular-weight bacteriocins. J. Bacteriol. 2016, 198, 2784–2793. [Google Scholar] [CrossRef] [Green Version]
- Duclohier, H. Antimicrobial Peptides and Peptaibols, Substitutes for Conventional Antibiotics. Curr. Pharm. Des. 2010, 16, 3212–3223. [Google Scholar] [CrossRef]
- Wu, J.; Gao, B.; Zhu, S. The fungal defensin family enlarged. Pharmaceuticals 2014, 7, 866–880. [Google Scholar] [CrossRef]
- Bissett, J.; Gams, W.; Jaklitsch, W.; Samuels, G.J. Accepted Trichoderma names in the year 2015. IMA Fungus 2015, 6, 263–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leitgeb, B.; Szekeres, A.; Manczinger, L.; Vágvölgyi, C.; Kredics, L. The history of Alamethicin: A review of the most extensively studied peptaibol. Chem. Biodivers. 2007, 4, 1027–1051. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, E.; Bavoso, A.; Di Blasio, B.; Pavone, V.; Pedone, C.; Toniolo, C.; Bonora, G.M. Peptaibol antibiotics: A study on the helical structure of the 2-9 sequence of emerimicins III and IV. Proc. Natl. Acad. Sci. USA 1982, 79, 7951–7954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitmore, L.; Wallace, B.A. The Peptaibol Database: A database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004, 32. [Google Scholar] [CrossRef] [Green Version]
- Meyer, C.E.; Reusser, F. A polypeptide antibacterial agent isolated from Trichoderma viride. Experientia 1967, 23, 85–86. [Google Scholar] [CrossRef] [PubMed]
- De Zotti, M.; Biondi, B.; Formaggio, F.; Toniolo, C.; Stella, L.; Park, Y.; Hahm, K.S. Trichogin GA IV: An antibacterial and protease-resistant peptide. J. Pept. Sci. 2009, 15, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Rebuffat, S.; Prigent, Y.; Auvin-Guette, C.; Bodo, B. Tricholongins BI and BII, 19-residue peptaibols from Trichoderma longibrachiatum Solution structure from two-dimensional NMR spectroscopy. Eur. J. Biochem. 1991, 201, 661–674. [Google Scholar] [CrossRef]
- Rebuffat, S.; Conraux, L.; Massias, M.; Auvin-Guette, C.; Bodo, B. Sequence and solution conformation of the 20-residue peptaibols, saturnisporins SA II and SA IV. Int. J. Pept. Protein Res. 1993, 41, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wu, J.; Somkuti, G.A.; Watson, J.T. Determination of the disulfide structure of sillucin, a highly knotted, cysteine-rich peptide, by cyanylation/cleavage mass mapping. Biochemistry 2001, 40, 4531–4538. [Google Scholar] [CrossRef]
- Bradley, W.A.; Somkuti, G.A. The primary structure of sillucin and antimicrobial peptide from mucor pusillus. FEBS Lett. 1979, 97, 81–83. [Google Scholar] [CrossRef] [Green Version]
- Ramachander Turaga, V.N. Peptaibols: Antimicrobial peptides from fungi. In Bioactive Natural Products in Drug Discovery; Springer: Singapore, 2020; pp. 713–730. ISBN 9789811513947. [Google Scholar]
- Wallace, B.A.; Chugh, J. Peptaibols: Models for Ion Channels. Biochem. Soc. Trans. 2001, 29, A54. [Google Scholar] [CrossRef]
- Chugh, J.K.; Brückner, H.; Wallace, B.A. Model for a helical bundle channel based on the high-resolution crystal structure of trichotoxin_A50E. Biochemistry 2002, 41, 12934–12941. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Janso, J.E.; Yang, H.Y.; Bernan, V.S.; Lin, S.L.; Yu, K. Culicinin D, an antitumor peptaibol produced by the fungus Culicinomyces clavisporus, strain LL-12I252. J. Nat. Prod. 2006, 69, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Csermely, P.; Radics, L.; Rossi, C.; Szamel, M.; Ricci, M.; Mihály, K.; Somogyi, J. The nonapeptide leucinostatin A acts as a weak ionophore and as an immunosuppressant on T lymphocytes. BBA Mol. Cell Res. 1994, 1221, 125–132. [Google Scholar] [CrossRef]
- Grishin, D.V.; Sokolov, N.N. Defensins are natural peptide antibiotics of higher eukaryotes. Biochem. Suppl. Ser. B Biomed. Chem. 2014, 8, 11–18. [Google Scholar] [CrossRef]
- Zhu, S. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSαβ defensins. Mol. Immunol. 2008, 45, 828–838. [Google Scholar] [CrossRef]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S.; et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mygind, P.H.; Fischer, R.L.; Schnorr, K.M.; Hansen, M.T.; Sönksen, C.P.; Ludvigsen, S.; Raventós, D.; Buskov, S.; Christensen, B.; De Maria, L.; et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Essig, A.; Hofmann, D.; Münch, D.; Gayathri, S.; Künzler, M.; Kallio, P.T.; Sahl, H.G.; Wider, G.; Schneider, T.; Aebi, M. Copsin, a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. J. Biol. Chem. 2014, 289, 34953–34964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Gao, B.; Harvey, P.J.; Craik, D.J. Dermatophytic defensin with antiinfective potential. Proc. Natl. Acad. Sci. USA 2012, 109, 8495–8500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Ben Hamida, J.; Vergoten, G.; Fliss, I. PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stec, B. Plant thionins—The structural perspective. Cell. Mol. Life Sci. 2006, 63, 1370–1385. [Google Scholar] [CrossRef]
- Pelegrini, P.B.; Franco, O.L. Plant γ-thionins: Novel insights on the mechanism of action of a multi-functional class of defense proteins. Int. J. Biochem. Cell Biol. 2005, 37, 2239–2253. [Google Scholar] [CrossRef]
- Fujimura, M.; Minami, Y.; Watanabe, K.; Tadera, K. Purification, characterization, and sequencing of a novel type of antimicrobial peptides, fa-amp1 and fa-amp2, from seeds of buckwheat (fagopyrum esculentum moench.). Biosci. Biotechnol. Biochem. 2003, 67, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Beintema, J.J. Structural features of plant chitinases and chitin-binding proteins. FEBS Lett. 1994, 350, 159–163. [Google Scholar] [CrossRef] [Green Version]
- Asensio, J.L.; Siebert, H.C.; von der Lieth, C.W.; Laynez, J.; Bruix, M.; Soedjanaamadja, U.M.; Beintema, J.J.; Cañada, F.J.; Gabius, H.J.; Jiménez-Barbero, J. NMR investigations of protein-carbohydrate interactions: Studies on the relevance of Trp/Tyr variations in lectin binding sites as deduced from titration microcalorimetry and NMR studies on hevein domains. Determination of the NMR structure of the complex. Proteins Struct. Funct. Genet. 2000, 40, 218–236. [Google Scholar] [CrossRef]
- Pallaghy, P.K.; Norton, R.S.; Nielsen, K.J.; Craik, D.J. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994, 3, 1833–1839. [Google Scholar] [CrossRef]
- Duvick, J.P.; Rood, T.; Rao, A.G.; Marshak, D.R. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J. Biol. Chem. 1992, 267, 18814–18820. [Google Scholar] [CrossRef]
- Marcus, J.P.; Green, J.L.; Goulter, K.C.; Manners, J.M. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 1999, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.M.; Yang, Y.; Sze, K.H.; Zhang, X.; Zheng, Y.T.; Shaw, P.C. Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrica). J. Struct. Biol. 2011, 174, 164–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira Carvalho, A.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology-A concise review. Peptides 2007, 28, 1144–1153. [Google Scholar] [CrossRef] [PubMed]
- Sels, J.; Mathys, J.; De Coninck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Oliveira-Lima, M.; Benko-Iseppon, A.; Neto, J.; Rodriguez-Decuadro, S.; Kido, E.; Crovella, S.; Pandolfi, V. Snakin: Structure, Roles and Applications of a Plant Antimicrobial Peptide. Curr. Protein Pept. Sci. 2016, 18, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and biological properties of snakins: The foremost cysteine-rich plant host Defense peptides. J. Fungi 2020, 6, 220. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.S.; Rettore, J.V.; Freitas, R.M.; Porto, W.F.; Duque, A.P.D.N.; Singulani, J.D.L.; Silva, O.N.; Detoni, M.D.L.; Vasconcelos, E.G.; Dias, S.C.; et al. Antimicrobial activity of recombinant Pg-AMP1, a glycine-rich peptide from guava seeds. Peptides 2012, 37, 294–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.J.; Park, C.B.; Hong, S.S.; Lee, H.S.; Lee, S.Y.; Kim, S.C. Characterization and cDNA cloning of two glycine- and histidine-rich antimicrobial peptides from the roots of shepherd’s purse, Capsella bursa-pastoris. Plant Mol. Biol. 2000, 44, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L. Antibacterial peptides isolated from insects. J. Pept. Sci. 2000, 6, 497–511. [Google Scholar] [CrossRef]
- Tincu, J.A.; Taylor, S.W. Antimicrobial peptides from marine invertebrates. Antimicrob. Agents Chemother. 2004, 48, 3645–3654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulet, P.; Stöcklin, R.; Menin, L. Anti-microbial peptides: From invertebrates to vertebrates. Immunol. Rev. 2004, 198, 169–184. [Google Scholar] [CrossRef]
- Shafee, T.M.A.; Lay, F.T.; Hulett, M.D.; Anderson, M.A. The Defensins Consist of Two Independent, Convergent Protein Superfamilies. Mol. Biol. Evol. 2016. [Google Scholar] [CrossRef] [Green Version]
- Shafee, T.M.A.; Lay, F.T.; Phan, T.K.; Anderson, M.A.; Hulett, M.D. Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 2017, 74, 663–682. [Google Scholar] [CrossRef] [PubMed]
- Tassanakajon, A.; Somboonwiwat, K.; Amparyup, P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev. Comp. Immunol. 2015, 48, 324–341. [Google Scholar] [CrossRef]
- Froy, O. Convergent evolution of invertebrate defensins and nematode antibacterial factors. Trends Microbiol. 2005, 13, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Sperstad, S.V.; Haug, T.; Blencke, H.M.; Styrvold, O.B.; Li, C.; Stensvåg, K. Antimicrobial peptides from marine invertebrates: Challenges and perspectives in marine antimicrobial peptide discovery. Biotechnol. Adv. 2011, 29, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Cociancich, S.; Ghazi, A.; Hetru, C.; Hoffmann, J.A.; Letellier, L. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus. J. Biol. Chem. 1993, 268, 19239–19245. [Google Scholar] [CrossRef]
- Matsuyama, K.; Natori, S. Mode of action of sapecin, a novel antibacterial protein of Sarcophaga peregrina (flesh fly). J. Biochem. 1990, 108, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, Q.; Wang, Q.; Chen, L.; Liu, Y.; Cong, M.; Wu, H.; Li, F.; Ji, C.; Zhao, J. A defensin-like antimicrobial peptide from the manila clam Ruditapes philippinarum: Investigation of the antibacterial activities and mode of action. Fish Shellfish Immunol. 2018, 80, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Takahashi, H.; Sugai, M.; Iwai, H.; Kohno, T.; Sekimizu, K.; Natori, S.; Shimada, I. Channel-forming membrane permeabilization by an antibacterial protein, sapecin. Determination of membrane-buried and oligomerization surfaces by NMR. J. Biol. Chem. 2004, 279, 4981–4987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornet, B.; Bonmatin, J.M.; Hetru, C.; Hoffmann, J.A.; Ptak, M.; Vovelle, F. Refined three-dimensional solution structure of insect defensin A. Structure 1995, 3, 435–448. [Google Scholar] [CrossRef] [Green Version]
- Saito, T.; Kawabata, S.I.; Shigenaga, T.; Takayenoki, Y.; Cho, J.; Nakajima, H.; Hirata, M.; Iwanaga, S. A novel big defensin identified in horseshoe crab hemocytes: Isolation, amino acid sequence, and antibacterial activity. J. Biochem. 1995, 117, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, S.I. Immunocompetent molecules and their response network in horseshoe crabs. Adv. Exp. Med. Biol. 2010, 708, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Song, L.; Li, C.; Ni, D.; Wu, L.; Zhu, L.; Wang, H.; Xu, W. Molecular cloning, expression of a big defensin gene from bay scallop Argopecten irradians and the antimicrobial activity of its recombinant protein. Mol. Immunol. 2007, 44, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Rosa, R.D.; Santini, A.; Fievet, J.; Bulet, P.; Destoumieux-Garzón, D.; Bachère, E. Big defensins, a diverse family of antimicrobial peptides that follows different patterns of expression in hemocytes of the oyster crassostrea gigas. PloS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; De Moro, G.; Manfrin, C.; Venier, P.; Pallavicini, A. Big defensins and mytimacins, new AMP families of the Mediterranean mussel Mytilus galloprovincialis. Dev. Comp. Immunol. 2012, 36, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Pisuttharachai, D.; Yasuike, M.; Aono, H.; Yano, Y.; Murakami, K.; Kondo, H.; Aoki, T.; Hirono, I. Characterization of two isoforms of Japanese spiny lobster Panulirus japonicus defensin cDNA. Dev. Comp. Immunol. 2009, 33, 434–438. [Google Scholar] [CrossRef]
- Montero-Alejo, V.; Acosta-Alba, J.; Perdomo-Morales, R.; Perera, E.; Hernández-Rodríguez, E.W.; Estrada, M.P.; Porto-Verdecia, M. Defensin like peptide from Panulirus argus relates structurally with beta defensin from vertebrates. Fish Shellfish Immunol. 2012, 33, 872–879. [Google Scholar] [CrossRef]
- Cheng, T.; Zhao, P.; Liu, C.; Xu, P.; Gao, Z.; Xia, Q.; Xiang, Z. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics 2006, 87, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E.; Shai, Y.; Lee, W.J.; Brey, P.T. Mode of Action of the Antibacterial Cecropin B2: A Spectrofluorometric Study. Biochemistry 1994, 33, 10681–10692. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E.; Miller, I.R.; Biggin, P.C.; Sansom, M.S.P.; Shai, Y. Structure and orientation of the mammalian antibacterial peptide cecropin P1 within phospholipid membranes. J. Mol. Biol. 1996, 258, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Jeong, K.W.; Lee, J.; Shin, A.; Kim, J.K.; Lee, J.; Lee, D.G.; Kim, Y. Structure-activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep. 2013, 46, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.J.; Beazley, W.D.; Bibby, M.C.; Devine, D.A. Antimicrobial activity of cecropins. J. Antimicrob. Chemother. 1996, 37, 1077–1089. [Google Scholar] [CrossRef] [Green Version]
- Hultmark, D.; Engström, Å.; Bennich, H.; Kapur, R.; Boman, H.G. Insect Immunity: Isolation and Structure of Cecropin D and Four Minor Antibacterial Components from Cecropia Pupae. Eur. J. Biochem. 1982, 127, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Ponnuvel, K.M.; Subhasri, N.; Sirigineedi, S.; Murthy, G.N.; Vijayaprakash, N.B. Molecular evolution of the cecropin multigene family in silkworm Bombyx mori. Bioinformation 2010, 5, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efimova, S.S.; Schagina, L.V.; Ostroumova, O.S. Channel-forming activity of cecropins in lipid bilayers: Effect of agents modifying the membrane dipole potential. Langmuir 2014, 30, 7884–7892. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Garzón, J.; Manrique-Moreno, M.; Trier, S.; Leidy, C.; Torres, R.; Patiño, E. Antimicrobial activity and interactions of cationic peptides derived from Galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. 2017, 70, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Chan, S.C.; Lee, J.C.; Chang, C.H.; Murugan, M.; Jack, R.W. Transmission Electron Microscopic Observations of Membrane Effects of Antibiotic Cecropin B on Escherichia coli. Microsc. Res. Tech. 2003, 62, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Simpson, K.J.; Shaw, D.C.; Nicholas, K.R. The whey acidic protein family: A new signature motif and three-dimensional structure by comparative modeling. J. Mol. Graph. Model. 1999, 17, 106–113. [Google Scholar] [CrossRef]
- Relf, J.M.; Chisholm, J.R.S.; Kemp, G.D.; Smith, V.J. Purification and characterization of a cysteine-rich 11.5-kDa antibacterial protein from the granular haemocytes of the shore crab, Carcinus maenas. Eur. J. Biochem. 1999, 264, 350–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, V.J.; Fernandes, J.M.O.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef] [Green Version]
- Smith, V.J. Phylogeny of whey acidic protein (WAP) four-disulfide core proteins and their role in lower vertebrates and invertebrates. Biochem. Soc. Trans. 2011, 39, 1403–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, A.E.; Rus, S.; Goiney, C.C.; Smith, C.M.; Towle, D.W.; Dickinson, P.S. Identification and characterization of a cDNA encoding a crustin-like, putative antibacterial protein from the American lobster Homarus americanus. Mol. Immunol. 2007, 44, 3333–3337. [Google Scholar] [CrossRef] [PubMed]
- Brockton, V.; Hammond, J.A.; Smith, V.J. Gene characterisation, isoforms and recombinant expression of carcinin, an antibacterial protein from the shore crab, Carcinus maenas. Mol. Immunol. 2007, 44, 943–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiravanichpaisal, P.; Lee, S.Y.; Kim, Y.A.; Andrén, T.; Söderhäll, I. Antibacterial peptides in hemocytes and hematopoietic tissue from freshwater crayfish Pacifastacus leniusculus: Characterization and expression pattern. Dev. Comp. Immunol. 2007, 31, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Amparyup, P.; Kondo, H.; Hirono, I.; Aoki, T.; Tassanakajon, A. Molecular cloning, genomic organization and recombinant expression of a crustin-like antimicrobial peptide from black tiger shrimp Penaeus monodon. Mol. Immunol. 2008, 45, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Krusong, K.; Poolpipat, P.; Supungul, P.; Tassanakajon, A. A comparative study of antimicrobial properties of crustinPm1 and crustinPm7 from the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 2012, 36, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, S. Comparative genomics analysis of five families of antimicrobial peptide-like genes in seven ant species. Dev. Comp. Immunol. 2012, 38, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Avila, E.E. Functions of Antimicrobial Peptides in Vertebrates. Curr. Protein Pept. Sci. 2016, 18. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [Green Version]
- Shinnar, A.E.; Butler, K.L.; Park, H.J. Cathelicidin family of antimicrobial peptides: Proteolytic processing and protease resistance. Bioorg. Chem. 2003, 31, 425–436. [Google Scholar] [CrossRef]
- Shinnar, A.; Uzzell, T.; Rao, M.; Spooner, E.; Lane, W.; Zasloff, M. New family of linear antimicrobial peptides from hagfish intestine contains bromo-tryptophan as novel amino acid; Peptides: Chemistry and Biology. In Proceedings of the 14th American Peptide Symposium, Columbus, OH, USA, 18–23 June 1995; Mayflower Scientific Ltd: Columbus, OH, USA, 1996; Volume 14, pp. 189–191. [Google Scholar]
- Basañez, G.; Shinnar, A.E.; Zimmerberg, J. Interaction of hagfish cathelicidin antimicrobial peptides with model lipid membranes. FEBS Lett. 2002, 532, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Broekman, D.C.; Zenz, A.; Gudmundsdottir, B.K.; Lohner, K.; Maier, V.H.; Gudmundsson, G.H. Functional characterization of codCath, the mature cathelicidin antimicrobial peptide from Atlantic cod (Gadus morhua). Peptides 2011, 32, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.I.; Zhang, Y.A.; Zou, J.; Nie, P.; Secombes, C.J. Two cathelicidin genes are present in both rainbow trout (Oncorhynchus mykiss) and atlantic salmon (Salmo salar). Antimicrob. Agents Chemother. 2006, 50, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.I.; Pleguezuelos, O.; Zhang, Y.A.; Zou, J.; Secombes, C.J. Identification of a novel cathelicidin gene in the rainbow trout, Oncorhynchus mykiss. Infect. Immun. 2005, 73, 5053–5064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuesta, A.; Meseguer, J.; Esteban, M.Á. Molecular and functional characterization of the gilthead seabream β-defensin demonstrate its chemotactic and antimicrobial activity. Mol. Immunol. 2011, 48, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Mercier, C.; Koussounadis, A.; Secombes, C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol. Immunol. 2007, 44, 638–647. [Google Scholar] [CrossRef]
- Wang, G.; Li, J.; Zou, P.; Xie, H.; Huang, B.; Nie, P.; Chang, M. Expression pattern, promoter activity and bactericidal property of β-defensin from the mandarin fish Siniperca chuatsi. Fish Shellfish Immunol. 2012, 33, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Falco, A.; Chico, V.; Marroquí, L.; Perez, L.; Coll, J.M.; Estepa, A. Expression and antiviral activity of a β-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008, 45, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Casadei, E.; Wang, T.; Zou, J.; González Vecino, J.L.; Wadsworth, S.; Secombes, C.J. Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 2009, 46, 3358–3366. [Google Scholar] [CrossRef] [PubMed]
- Nam, B.H.; Moon, J.Y.; Kim, Y.O.; Kong, H.J.; Kim, W.J.; Lee, S.J.; Kim, K.K. Multiple β-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol. 2010, 28, 267–274. [Google Scholar] [CrossRef]
- Guo, M.; Wei, J.; Huang, X.; Huang, Y.; Qin, Q. Antiviral effects of β-defensin derived from orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2012, 32, 828–838. [Google Scholar] [CrossRef]
- Jin, J.Y.; Zhou, L.; Wang, Y.; Li, Z.; Zhao, J.G.; Zhang, Q.Y.; Gui, J.F. Antibacterial and antiviral roles of a fish β-defensin expressed both in pituitary and testis. PloS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Ruangsri, J.; Kitani, Y.; Kiron, V.; Lokesh, J.; Brinchmann, M.F.; Karlsen, B.O.; Fernandes, J.M.O. A Novel Beta-Defensin Antimicrobial Peptide in Atlantic Cod with Stimulatory Effect on Phagocytic Activity. PloS ONE 2013, 8, e62302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.H.; Chen, J.Y.; Kuo, C.M. Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 2007, 44, 1922–1934. [Google Scholar] [CrossRef] [PubMed]
- Hunter, H.N.; Bruce Fulton, D.; Ganz, T.; Vogel, H.J. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 2002, 277, 37597–37603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Sun, Y.; Shi, G.; Wang, R. Miiuy croaker hepcidin gene and comparative analyses reveal evidence for positive selection. PloS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masso-Silva, J.; Diamond, G.; Macias-Rodriguez, M.; Ascencio, F. Genomic organization and tissue-specific expression of hepcidin in the pacific mutton hamlet, Alphestes immaculatus (Breder, 1936). Fish Shellfish Immunol. 2011, 31, 1297–1302. [Google Scholar] [CrossRef] [PubMed]
- Hilton, K.B.; Lambert, L.A. Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene 2008, 415, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Ma, L.; Yu, Y.; Yu, H.; Mohammed, S.; Chu, G.; Mu, L.; Zhang, Q. Identification and characterization of a hepcidin from half-smooth tongue sole Cynoglossus semilaevis. Fish Shellfish Immunol. 2012, 33, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Cai, J.J.; Liu, H.P.; Fan, D.Q.; Peng, H.; Wang, K.J. Recombinant medaka (Oryzias melastigmus) pro-hepcidin: Multifunctional characterization. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 161, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.O.; Ruangsri, J.; Kiron, V. Atlantic cod piscidin and its diversification through positive selection. PloS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raju, S.V.; Sarkar, P.; Kumar, P.; Arockiaraj, J. Piscidin, Fish Antimicrobial Peptide: Structure, Classification, Properties, Mechanism, Gene Regulation and Therapeutical Importance. Int. J. Pept. Res. Ther. 2021, 27, 91–107. [Google Scholar] [CrossRef]
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 1997, 272, 12008–12013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Lai, R. The chemistry and biological activities of peptides from amphibian skin secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Maisetta, G.; Di Luca, M.; Gaddi, L.M.H.; Esin, S.; Florio, W.; Brancatisano, F.L.; Barra, D.; Campa, M.; Batoni, G. Comparative analysis of the bactericidal activities of amphibian peptide analogues against multidrug-resistant nosocomial bacterial strains. Antimicrob. Agents Chemother. 2008, 52, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmaco, M.; Kreil, G.; Barra, D. Bombinins, antimicrobial peptides from Bombina species. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1551–1555. [Google Scholar] [CrossRef] [Green Version]
- Hale, J.D.F.; Hancock, R.E.W. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti. Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef]
- Cho, J.H.; Sung, B.H.; Kim, S.C. Buforins: Histone H2A-derived antimicrobial peptides from toad stomach. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1564–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, J.; Chen, X.; Ye, T.; Zeng, B.; Zeng, Q.; Wu, J.; Kascakova, B.; Martins, L.A.; Prudnikova, T.; Smatanova, I.K.; et al. Characterization and functional analysis of cathelicidin-mh, a novel frog-derived peptide with anti-septicemic properties. Elife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lin, Y.F.; Chen, J.H.; Chen, X.; Lin, Z.H. Molecular characterization of cathelicidin in tiger frog (Hoplobatrachus rugulosus): Antimicrobial activity and immunomodulatory activity. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021, 247. [Google Scholar] [CrossRef] [PubMed]
- Marcos, J.F.; Gandía, M.; Harries, E.; Carmona, L.; Muñoz, A. Antifungal peptides: Exploiting non-lytic mechanisms and cell penetration properties. ACS Symp. Ser. 2012, 1095, 337–357. [Google Scholar] [CrossRef]
- Huang, L.; Chen, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Dermaseptin-PH: A novel peptide with antimicrobial and anticancer activities from the skin secretion of the south American orange-legged leaf frog, pithecopus (phyllomedusa) hypochondrialis. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Belmadani, A.; Semlali, A.; Rouabhia, M. Dermaseptin-S1 decreases Candida albicans growth, biofilm formation and the expression of hyphal wall protein 1 and aspartic protease genes. J. Appl. Microbiol. 2018, 125, 72–83. [Google Scholar] [CrossRef]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell. Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef] [PubMed]
- Luca, V.; Olivi, M.; Di Grazia, A.; Palleschi, C.; Uccelletti, D.; Mangoni, M.L. Anti-Candida activity of 1-18 fragment of the frog skin peptide esculentin-1b: In vitro and in vivo studies in a Caenorhabditis elegans infection model. Cell. Mol. Life Sci. 2014, 71, 2535–2546. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M. A proposed nomenclature for antimicrobial peptides from frogs of the genus Leptodactylus. Peptides 2008, 29, 1631–1632. [Google Scholar] [CrossRef]
- Oliveira, M.; Gomes-Alves, A.G.; Sousa, C.; Mirta Marani, M.; Plácido, A.; Vale, N.; Delerue-Matos, C.; Gameiro, P.; Kückelhaus, S.A.S.; Tomas, A.M.; et al. Ocellatin-PT antimicrobial peptides: High-resolution microscopy studies in antileishmania models and interactions with mimetic membrane systems. Biopolymers 2016, 873–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, R.; Zheng, Y.T.; Shen, J.H.; Liu, G.J.; Liu, H.; Lee, W.H.; Tang, S.Z.; Zhang, Y. Antimicrobial peptides from skin secretions of Chinese red belly toad Bombina maxima. Peptides 2002, 23, 427–435. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, J.; Shen, J.H.; Jin, Y.; Lee, W.H.; Zhang, Y. Maximins S, a novel group of antimicrobial peptides from toad Bombina maxima. Biochem. Biophys. Res. Commun. 2005, 327, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kang, N.; Ko, S.J.; Park, J.; Park, E.; Shin, D.W.; Kim, S.H.; Lee, S.A.; Lee, J.I.; Lee, S.H.; et al. Antibacterial and antibiofilm activity and mode of action of magainin 2 against drug-resistant acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, J.; Retz, M.; Sidhu, S.S.; Suttmann, H.; Sell, M.; Paulsen, F.; Harder, J.; Unteregger, G.; Stöckle, M. Antitumor Activity of the Antimicrobial Peptide Magainin II against Bladder Cancer Cell Lines. Eur. Urol. 2006, 50, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Lee, D.G.; Hahm, K.S. HP(2-9)-magainin 2(1-12), a synthetic hybrid peptide, exerts its antifungal effect on Candida albicans by damaging the plasma membrane. J. Pept. Sci. 2004, 10, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Amiche, M.; Ladram, A.; Nicolas, P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides 2008, 29, 2074–2082. [Google Scholar] [CrossRef] [PubMed]
- El Amri, C.; Lacombe, C.; Zimmerman, K.; Ladram, A.; Amiche, M.; Nicolas, P.; Bruston, F. The plasticins: Membrane adsorption, lipid disorders, and biological activity. Biochemistry 2006, 45, 14285–14297. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.H. Insights into the Antimicrobial Activities of Unusual Antimicrobial Peptide Families from Amphibian Skin. J. Clin. Toxicol. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Basir, Y.J.; Knoop, F.C.; Dulka, J.; Conlon, J.M. Multiple antimicrobial peptides and peptides related to bradykinin and neuromedin N isolated from skin secretions of the pickerel frog, Rana palustris. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000, 1543, 95–105. [Google Scholar] [CrossRef]
- Pierre, T.N.; Seon, A.A.; Amiche, M.; Nicolas, P. Phylloxin, a novel peptide antibiotic of the dermaseptin family of antimicrobial/opioid peptide precursors. Eur. J. Biochem. 2000, 267, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Pinto, E.G.; Pimenta, D.C.; Antoniazzi, M.M.; Jared, C.; Tempone, A.G. Antimicrobial peptides isolated from Phyllomedusa nordestina (Amphibia) alter the permeability of plasma membrane of Leishmania and Trypanosoma cruzi. Exp. Parasitol. 2013, 135, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, Q.; Li, L.; Xi, X.; Wu, D.; Zhou, M.; Chen, T.; Shaw, C.; Wang, L. Discovery of phylloseptins that defense against gram-positive bacteria and inhibit the proliferation of the non-small cell lung cancer cell line, from the skin secretions of phyllomedusa frogs. Molecules 2017, 22, 1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, L.; Soto, A.M.; Knoop, F.C.; Conlon, J.M. Pseudin-2: An antimicrobial peptide with low hemolytic activity from the skin of the paradoxical frog. Biochem. Biophys. Res. Commun. 2001, 288, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kim, H.; Kim, J.Y.; Kim, H.; Cheong, G.W.; Lee, J.R.; Jang, M.K. Improved cell selectivity of pseudin-2 via substitution in the leucine-zipper motif: In vitro and in vivo antifungal activity. Antibiotics 2020, 9, 921. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Ma, C.; Zhang, Y.; Xi, X.; Wang, L.; Zhou, M.; Burrows, J.F.; Chen, T. A novel antimicrobial peptide, Ranatuerin-2PLx, showing therapeutic potential in inhibiting proliferation of cancer cells. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, D.P.; Durell, S.; Maloy, W.L.; Zasloff, M. Renalexin. A novel antimicrobial peptide from bullfrog (Rana catesbeiana) skin, structurally related to the bacterial antibiotic, polymyxin. J. Biol. Chem. 1994, 269, 10849–10855. [Google Scholar] [CrossRef]
- Domhan, C.; Uhl, P.; Meinhardt, A.; Zimmermann, S.; Kleist, C.; Lindner, T.; Leotta, K.; Mier, W.; Wink, M. A novel tool against multiresistant bacterial pathogens: Lipopeptide modification of the natural antimicrobial peptide ranalexin for enhanced antimicrobial activity and improved pharmacokinetics. Int. J. Antimicrob. Agents 2018, 52, 52–62. [Google Scholar] [CrossRef] [PubMed]
- van Hoek, M.L. Antimicrobial peptides in reptiles. Pharmaceuticals 2014, 7, 723–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Prickett, M.D.; Gutowska, W.; Kuo, R.; Belov, K.; Burt, D.W. Evolution of the avian β-defensin and cathelicidin genes. BMC Evol. Biol. 2015, 15, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Gan, T.X.; Liu, X.D.; Jin, Y.; Lee, W.H.; Shen, J.H.; Zhang, Y. Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides 2008, 29, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hong, J.; Liu, X.; Yang, H.; Liu, R.; Wu, J.; Wang, A.; Lin, D.; Lai, R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PloS ONE 2008, 3, e3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.W.; Beach, G.G.; Wunderlich, J.; Harmon, B.G. Isolation of antimicrobial peptides from avian heterophils. J. Leukoc. Biol. 1994, 56, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006, 281, 2858–2867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Herrera, A.I.; Bommineni, Y.R.; Soulages, J.L.; Prakash, O.; Zhang, G. The central kink region of fowlicidin-2, an α-helical host defense peptide, is critically involved in bacterial killing and endotoxin neutralization. J. Innate Immun. 2009, 1, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Gao, W.; Chen, H.; Li, D.; Yang, N.; Zhu, J.; Feng, X.; Liu, C.; Li, Z. The central hinge link truncation of the antimicrobial peptide fowlicidin-3 enhances its cell selectivity without antibacterial activity loss. Antimicrob. Agents Chemother. 2016, 60, 2798–2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goitsuka, R.; Chen, C.L.H.; Benyon, L.; Asano, Y.; Kitamura, D.; Cooper, M.D. Chicken cathelicidin-B1, an antimicrobial guardian at the mucosal M cell gateway. Proc. Natl. Acad. Sci. USA 2007, 104, 15063–15068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, F.; Chen, C.; Zhu, W.; He, W.; Guang, H.; Li, Z.; Wang, D.; Liu, J.; Chen, M.; Wang, Y.; et al. Gene cloning, expression and characterization of avian cathelicidin orthologs, Cc-CATHs, from Coturnix coturnix. FEBS J. 2011, 278, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Ishige, T.; Hara, H.; Hirano, T.; Kono, T.; Hanzawa, K. Characterization of the cathelicidin cluster in the Japanese quail (Coturnix japonica). Anim. Sci. J. 2017, 88, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lu, Y.; Qiao, X.; Wei, L.; Fu, T.; Cai, S.; Wang, C.; Liu, X.; Zhong, S.; Wang, Y. Novel Cathelicidins from Pigeon Highlights Evolutionary Convergence in Avain Cathelicidins and Functions in Modulation of Innate Immunity. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Xing, L.; Qu, P.; Tan, T.; Yang, N.; Li, D.; Chen, H.; Feng, X. Identification of a novel cathelicidin antimicrobial peptide from ducks and determination of its functional activity and antibacterial mechanism. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lu, Z.; Feng, F.; Zhu, W.; Guang, H.; Liu, J.; He, W.; Chi, L.; Li, Z.; Yu, H. Molecular cloning and characterization of novel cathelicidin-derived myeloid antimicrobial peptide from Phasianus colchicus. Dev. Comp. Immunol. 2011, 35, 314–322. [Google Scholar] [CrossRef]
- Yacoub, H.A.; Elazzazy, A.M.; Mahmoud, M.M.; Baeshen, M.N.; Al-Maghrabi, O.A.; Alkarim, S.; Ahmed, E.S.; Almehdar, H.A.; Uversky, V.N. Chicken cathelicidins as potent intrinsically disordered biocides with antimicrobial activity against infectious pathogens. Dev. Comp. Immunol. 2016, 65, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Hamad, S.K.; Kim, S.; El-Kadi, S.W.; Wong, E.A.; Dalloul, R.A. Comparative expression of host defense peptides in turkey poults. Poult. Sci. 2017, 96, 2083–2090. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, C.; Kolobov, A.; Leonova, Y.F.; Knappe, D.; Shamova, O.; Ovchinnikova, T.V.; Kokryakov, V.N.; Hoffmann, R. Isolation, purification and de novo sequencing of TBD-1, the first beta-defensin from leukocytes of reptiles. Proteomics 2009, 9, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Yount, N.Y.; Kupferwasser, D.; Spisni, A.; Dutz, S.M.; Ramjan, Z.H.; Sharma, S.; Waring, A.J.; Yeaman, M.R. Selective reciprocity in antimicrobial activity versus cytotoxicity of hBD-2 and crotamine. Proc. Natl. Acad. Sci. USA 2009, 106, 14972–14977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshminarayanan, R.; Vivekanandan, S.; Samy, R.P.; Banerjee, Y.; Chi-Jin, E.O.; Kay, W.T.; Jois, S.D.S.; Kini, R.M.; Valiyaveettil, S. Structure, self-assembly, and dual role of a β-defensin-like peptide from the Chinese soft-shelled turtle eggshell matrix. J. Am. Chem. Soc. 2008, 130, 4660–4668. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Sinha, N.K.; Banerjee, S.; Roy, D.; Chattopadhyay, D.; Roy, S. Small cationic protein from a marine turtle has β-defensin-like fold and antibacterial and antiviral activity. Proteins Struct. Funct. Genet. 2006, 64, 524–531. [Google Scholar] [CrossRef]
- van Dijk, A.; Veldhuizen, E.J.A.; Haagsman, H.P. Avian defensins. Vet. Immunol. Immunopathol. 2008, 124, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sugiarto, H.; Yu, P.L. Identification of three novel ostricacins: An update on the phylogenetic perspective of β-defensins. Int. J. Antimicrob. Agents 2006, 27, 229–235. [Google Scholar] [CrossRef]
- Sugiarto, H.; Yu, P.L. Avian antimicrobial peptides: The defense role of β-defensins. Biochem. Biophys. Res. Commun. 2004, 323, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Nguyen, T.; Liu, L.; Sacco, R.E.; Brogden, K.A.; Lehrer, R.I. Gallinacin-3, an inducible epithelial β-defensin in the chicken. Infect. Immun. 2001, 69, 2684–2691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thouzeau, C.; Le Maho, Y.; Froget, G.; Sabatier, L.; Le Bohec, C.; Hoffmann, J.A.; Bulet, P. Spheniscins, Avian β-Defensins in Preserved Stomach Contents of the King Penguin, Aptenodytes patagonicus. J. Biol. Chem. 2003, 278, 51053–51058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Tomasinsig, L.; Zanetti, M. The Cathelicidins—Structure, Function and Evolution. Curr. Protein Pept. Sci. 2005, 6, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duplantier, A.J.; van Hoek, M.L. The human cathelicidin antimicrobial peptide LL-37 as a potential treatment for polymicrobial infected wounds. Front. Immunol. 2013, 4, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, S.N.; Bishop, B.M.; Van Hoek, M.L. Susceptibility of Pseudomonas aeruginosa biofilm to alpha-helical peptides: D-enantiomer of LL-37. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011, 11, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.W.; Rehm, B.H.A.; Hancock, R.E.W. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sochacki, K.A.; Barns, K.J.; Bucki, R.; Weisshaar, J.C. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. USA 2011, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barns, K.J.; Weisshaar, J.C. Real-time attack of LL-37 on single Bacillus subtilis cells. Biochim. Biophys. Acta Biomembr. 2013, 1828, 1511–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neville, F.; Hodges, C.S.; Liu, C.; Konovalov, O.; Gidalevitz, D. In situ characterization of lipid A interaction with antimicrobial peptides using surface X-ray scattering. Biochim. Biophys. Acta Biomembr. 2006, 1758, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Henzler-Wildman, K.A.; Martinez, G.V.; Brown, M.F.; Ramamoorthy, A. Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry 2004, 43, 8459–8469. [Google Scholar] [CrossRef] [PubMed]
- Brahma, B.; Patra, M.C.; Karri, S.; Chopra, M.; Mishra, P.; De, B.C.; Kumar, S.; Mahanty, S.; Thakur, K.; Poluri, K.M.; et al. Diversity, antimicrobial action and structure- activity relationship of buffalo cathelicidins. PloS ONE 2015, 10, e0144741. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.H.; Chen, C.; Jou, M.L.; Lee, A.Y.L.; Lin, Y.C.; Yu, Y.P.; Huang, W.T.; Wu, S.H. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: Evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 2005, 33, 4053–4064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kokryakov, V.N.; Harwig, S.S.L.; Panyutich, E.A.; Shevchenko, A.A.; Aleshina, G.M.; Shamova, O.V.; Korneva, H.A.; Lehrer, R.I. Protegrins: Leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993, 327, 231–236. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Liu, L.; Lehrer, R.I. Identification of a new member of the protegrin family by cDNA cloning. FEBS Lett. 1994, 346, 285–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Ganz, T.; Lehrer, R.I. The structure of porcine protegrin genes. FEBS Lett. 1995, 368, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, D.A.; Hurst, M.A.; Fujii, C.A.; Kung, A.H.C.; Ho, J.F.; Cheng, F.C.; Loury, D.J.; Fiddes, J.C. Protegrin-1: A broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 1997, 41, 1738–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gour, S.; Kumar, V.; Singh, A.; Gadhave, K.; Goyal, P.; Pandey, J.; Giri, R.; Yadav, J.K. Mammalian antimicrobial peptide protegrin-4 self assembles and forms amyloid-like aggregates: Assessment of its functional relevance. J. Pept. Sci. 2019, 25. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A.; Ackermann, M.; McCray, P.B.; Tack, B.F. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 2003, 22, 465–478. [Google Scholar] [CrossRef] [Green Version]
- Radermacher, S.W.; Schoop, V.M.; Schluesener, H.J. Bactenecin, a leukocytic antimicrobial peptide, is cytotoxic to neuronal and glial cells. J. Neurosci. Res. 1993, 36, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Skerlavaj, B.; Romeo, D.; Gennaro, R. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect. Immun. 1990, 58, 3724–3730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skerlavaj, B.; Scocchi, M.; Gennaro, R.; Risso, A.; Zanetti, M. Structural and functional analysis of horse cathelicidin peptides. Antimicrob. Agents Chemother. 2001, 45, 715–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef]
- Ayabe, T.; Satchell, D.P.; Wilson, C.L.; Parks, W.C.; Selsted, M.E.; Ouellette, A.J. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000, 1, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Saito, K. Purification, primary structure, and biological activity of guinea pig neutrophil cationic peptides. Infect. Immun. 1989, 57, 2405–2409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selsted, M.E.; Szklarek, D.; Lehrer, R.I. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect. Immun. 1984, 45, 150–154. [Google Scholar] [CrossRef] [Green Version]
- Ganz, T.; Selsted, M.E.; Szklarek, D.; Harwig, S.S.; Daher, K.; Bainton, D.F.; Lehrer, R.I. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 1985, 76, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Wilde, C.G.; Griffith, J.E.; Marra, M.N.; Snable, J.L.; Scott, R.W. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J. Biol. Chem. 1989, 264, 11200–11203. [Google Scholar] [CrossRef]
- Jones, D.E.; Bevins, C.L. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 1992, 267, 23216–23225. [Google Scholar] [CrossRef]
- Jones, D.E.; Bevins, C.L. Defensin-6 mRNA in human Paneth cells: Implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993, 315, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Ehmann, D.; Wendler, J.; Koeninger, L.; Larsen, I.S.; Klag, T.; Berger, J.; Marette, A.; Schaller, M.; Stange, E.F.; Malek, N.P.; et al. Paneth cell α-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 3746–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, K.; Contreras, R. Human antimicrobial peptides: Defensins, cathelicidins and histatins. Biotechnol. Lett. 2005, 27, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Barton, A.; Daher, K.A.; Harwig, S.S.L.; Ganz, T.; Selsted, M.E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Invest. 1989, 84, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.J.; Unholzer, A.; Schaller, M.; Schäfer-Korting, M.; Korting, H.C. Human defensins. J. Mol. Med. 2005, 83, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Diamond, G.; Zasloff, M.; Eck, H.; Brasseur, M.; Lee Maloy, W.; Bevins, C.L. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: Peptide isolation and cloning of a cDNA. Proc. Natl. Acad. Sci. USA 1991, 88, 3952–3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonwetter, B.S.; Stolzenberg, E.D.; Zasloff, M.A. Epithelial antibiotics induced at sites of inflammation. Science 1995, 267, 1645–1648. [Google Scholar] [CrossRef] [PubMed]
- Rieg, S.; Meier, B.; Fähnrich, E.; Huth, A.; Wagner, D.; Kern, W.V.; Kalbacher, H. Differential activity of innate defense antimicrobial peptides against Nocardia species. BMC Microbiol. 2010, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selsted, M.E.; Tang, Y.Q.; Morris, W.L.; McGuire, P.A.; Novotny, M.J.; Smith, W.; Henschen, A.H.; Cullor, J.S. Purification, primary structures, and antibacterial activities of β- defensins, a new family of antimicrobial peptides from bovine neutrophils. J. Biol. Chem. 1993, 268, 6641–6648. [Google Scholar] [CrossRef]
- Pazgier, M.; Hoover, D.M.; Yang, D.; Lu, W.; Lubkowski, J. Human β-defensins. Cell. Mol. Life Sci. 2006, 63, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- McCray, P.B.; Bentley, L. Human Airway Epithelia Express a β-defensin. Am. J. Respir. Cell Mol. Biol. 1997, 16, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Frye, M. 186. Differential expression of human $alpha;- and $beta;-defensins mRNA in gastrointestinal epithelia. Neth. J. Med. 1999, 54, S71. [Google Scholar] [CrossRef]
- Valore, E.V.; Park, C.H.; Quayle, A.J.; Wiles, K.R.; McCray, P.B.; Ganz, T. Human β-defensin-1: An antimicrobial peptide of urogenital tissues. J. Clin. Invest. 1998, 101, 1633–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, J.-R.C.; Krause, A.; Schulz, S.; Rodríguez-Jiménez, F.-J.; Klüver, E.; Adermann, K.; Forssmann, U.; Frimpong-Boateng, A.; Bals, R.; Forssmann, W.-G. Human β-defensin 4: A novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001, 15, 1819–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harder, J.; Meyer-Hoffert, U.; Wehkamp, K.; Schwichtenberg, L.; Schröder, J.M. Differential gene induction of human β-defensins (hBD-1, -2, -3, and -4) in keratinocytes is inhibited by retinoic acid. J. Invest. Dermatol. 2004, 123, 522–529. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.X.; Cole, A.M.; Lehrer, R.I. Evolution of primate θ-defensins: A serpentine path to a sweet tooth. Peptides 2003, 24, 1647–1654. [Google Scholar] [CrossRef] [PubMed]
- Hazlett, L.; Wu, M. Defensins in innate immunity. Cell Tissue Res. 2011, 343, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, M.; Stockem, M.; Bierbaum, G.; Schlag, M.; Götz, F.; Tran, D.Q.; Schaal, J.B.; Ouellette, A.J.; Selsted, M.E.; Sahl, H.G. Killing of staphylococci by θ-defensins involves membrane impairment and activation of autolytic enzymes. Antibiotics 2014, 3, 617–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welkos, S.; Cote, C.K.; Hahn, U.; Shastak, O.; Jedermann, J.; Bozue, J.; Jung, G.; Ruchala, P.; Pratikhya, P.; Tang, T.; et al. Humanized θ-defensins (retrocyclins) enhance macrophage performance and protect mice from experimental anthrax infections. Antimicrob. Agents Chemother. 2011, 55, 4238–4250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, V.; Garcia, A.; Tran, D.Q.; Schaal, J.B.; Tran, P.; Ngole, D.; Aqeel, Y.; Tongaonkar, P.; Ouellette, A.J.; Selsteda, M.E. Fungicidal Potency and Mechanisms of –Defensins against Multidrug-Resistant Candida Species. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maróti Gergely, G.; Kereszt, A.; Kondorosi, É.; Mergaert, P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 2011, 162, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Guo, H.; Chen, F.; Zhao, L.; He, L.; Ou, Y.; Huang, M.; Zhang, Y.; Guo, B.; Cao, Y.; et al. Antibacterial Effects of a Cell-Penetrating Peptide Isolated from Kefir. J. Agric. Food Chem. 2016, 64, 3234–3242. [Google Scholar] [CrossRef] [PubMed]
- Tareq, F.S.; Lee, M.A.; Lee, H.S.; Lee, Y.J.; Lee, J.S.; Hasan, C.M.; Islam, M.T.; Shin, H.J. Gageotetrins A-C, noncytotoxic antimicrobial linear lipopeptides from a marine bacterium bacillus subtilis. Org. Lett. 2014, 16, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Brady, A.; Young, A.; Rasimick, B.; Chen, K.; Zhou, C.; Kallenbach, N.R. Length effects in antimicrobial peptides of the (RW)n series. Antimicrob. Agents Chemother. 2007, 51, 597–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringstad, L.; Schmidtchen, A.; Malmsten, M. Effect of peptide length on the interaction between consensus peptides and DOPC/DOPA bilayers. Langmuir 2006, 22, 5042–5050. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Phadke, S.M.; Lazarevic, V.; Cascio, M.; Islam, K.; Montelaro, R.C.; Mietzner, T.A. De novo generation of cationic antimicrobial peptides: Influence of length and tryptophan substitution on antimicrobial activity. Antimicrob. Agents Chemother. 2005, 49, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Maier, E.; Benz, R.; Hancock, R.E.W. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 1999, 38, 7235–7242. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, B. Detergent-like properties of magainin antibiotic peptides: A 31P solid-state NMR spectroscopy study. Biochim. Biophys. Acta Biomembr. 2005, 1712, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pouny, Y.; Rapaport, D.; Shai, Y.; Mor, A.; Nicolas, P. Interaction of Antimicrobial Dermaseptin and its Fluorescently Labeled Analogs with Phospholipid Membranes. Biochemistry 1992, 31, 12416–12423. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, K.; Tazume, T.; Uji, K.; Tanaka, M.; Kumura, H.; Mikawa, K.; Shimo-Oka, T. Properties of a Heparin-binding Peptide Derived from Bovine Lactoferrin. J. Dairy Sci. 1998, 81, 2841–2849. [Google Scholar] [CrossRef]
- Fernandes, J.M.O.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]
- Anderson, R.C.; Yu, P.L. Isolation and characterisation of proline/arginine-rich cathelicidin peptides from ovine neutrophils. Biochem. Biophys. Res. Commun. 2003, 312, 1139–1146. [Google Scholar] [CrossRef]
- Taylor, S.W.; Sun, C.; Hsieh, A.; Andon, N.L.; Ghosh, S.S. A sulfated, phosphorylated 7 kDa secreted peptide characterized by direct analysis of cell culture media. J. Proteome Res. 2008, 7, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H.; Miao, V.; Wrigley, S.K. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 2005, 22, 717–741. [Google Scholar] [CrossRef] [PubMed]
- Paulmann, M.; Arnold, T.; Linke, D.; Özdirekcan, S.; Kopp, A.; Gutsmann, T.; Kalbacher, H.; Wanke, I.; Schuenemann, V.J.; Habeck, M.; et al. Structure-activity analysis of the dermcidin-derived peptide DCD-1L, an anionic antimicrobial peptide present in human sweat. J. Biol. Chem. 2012, 287, 8434–8443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffen, H.; Rieg, S.; Wiedemann, I.; Kalbacher, H.; Deeg, M.; Sahl, H.G.; Peschel, A.; Götz, F.; Garbe, C.; Schittek, B. Naturally processed dermcidin-derived peptides do not permeabilize bacterial membranes and kill microorganisms irrespective of their charge. Antimicrob. Agents Chemother. 2006, 50, 2608–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Wang, G. Antimicrobial peptides: Discovery, design, and novel therapeutic strategies. In Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; Cabi: Wallingford, UK, 2017; ISBN 9781845936570. [Google Scholar]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringstad, L.; Protopapa, E.; Lindholm-Sethson, B.; Schmidtchen, A.; Nelson, A.; Malmsten, M. An electrochemical study into the interaction between complement-derived peptides and DOPC mono- and bilayers. Langmuir 2008, 24, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Post-Translational Modifications of Natural Antimicrobial Peptides and Strategies for Peptide Engineering. Curr. Biotechnol. 2011, 1, 72–79. [Google Scholar] [CrossRef]
- Taylor, S.W.; Craig, A.G.; Fischer, W.H.; Park, M.; Lehrer, R.I. Styelin D, an extensively modified antimicrobial peptide from ascidian hemocytes. J. Biol. Chem. 2000, 275, 38417–38426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unger, T.; Oren, Z.; Shai, Y. The effect of cyclization of magainin 2 and melittin analogues on structure, function, and model membrane interactions: Implication to their mode of action. Biochemistry 2001, 40, 6388–6397. [Google Scholar] [CrossRef]
- Koziel, J.; Bryzek, D.; Sroka, A.; Maresz, K.; Glowczyk, I.; Bielecka, E.; Kantyka, T.; Pyrć, K.; Svoboda, P.; Pohl, J.; et al. Citrullination Alters Immunomodulatory Function of LL-37 Essential for Prevention of Endotoxin-Induced Sepsis. J. Immunol. 2014, 192, 5363–5372. [Google Scholar] [CrossRef] [Green Version]
- Picchianti, M.; Russo, C.; Castagnini, M.; Biagini, M.; Soldaini, E.; Balducci, E. NAD-dependent ADP-ribosylation of the human antimicrobial and immune-modulatory peptide LL-37 by ADP-ribosyltransferase-1. Innate Immun. 2015, 21, 314–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koro, C.; Hellvard, A.; Delaleu, N.; Binder, V.; Scavenius, C.; Bergum, B.; Gówczyk, I.; Roberts, H.M.; Chapple, I.L.C.; Grant, M.M.; et al. Carbamylated LL-37 as a modulator of the immune response. Innate Immun. 2016, 22, 218–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G. Improved methods for classification, Prediction, And design of antimicrobial peptides. Methods Mol. Biol. 2015, 1268, 43–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nar, H.; Schmid, A.; Puder, C.; Potterat, O. High-resolution crystal structure of a lasso peptide. ChemMedChem 2010, 5, 1689–1692. [Google Scholar] [CrossRef] [PubMed]
- Rosengren, K.J.; Clark, R.J.; Daly, N.L.; Göransson, U.; Jones, A.; Craik, D.J. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 2003, 125, 12464–12474. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, J.; Sun, L.C.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q.Y. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial peptides: Interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uggerhøj, L.E.; Poulsen, T.J.; Munk, J.K.; Fredborg, M.; Sondergaard, T.E.; Frimodt-Moller, N.; Hansen, P.R.; Wimmer, R. Rational design of alpha-helical antimicrobial peptides: Do’s and don’ts. ChemBioChem 2015, 16, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Mohanram, H.; Bhattacharjya, S. Cysteine deleted protegrin-1 (CDP-1): Anti-bacterial activity, outer-membrane disruption and selectivity. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 3006–3016. [Google Scholar] [CrossRef] [PubMed]
- Dhople, V.; Krukemeyer, A.; Ramamoorthy, A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1499–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodziewicz-Motowidło, S.; Mickiewicz, B.; Greber, K.; Sikorska, E.; Szultka, Ł.; Kamysz, E.; Kamysz, W. Antimicrobial and conformational studies of the active and inactive analogues of the protegrin-1 peptide. FEBS J. 2010, 277, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Mandard, N.; Sodano, P.; Labbe, H.; Bonmatin, J.M.; Bulet, P.; Hetru, C.; Ptak, M.; Vovelle, F. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. Eur. J. Biochem. 1998, 256, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Laederach, A.; Andreotti, A.H.; Fulton, D.B. Solution and micelle-bound structures of tachyplesin I and its active aromatic linear derivatives. Biochemistry 2002, 41, 12359–12368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, J.P.S.; Rozek, A.; Hancock, R.E.W. Structure-activity relationships for the β-hairpin cationic antimicrobial peptide polyphemusin I. Biochim. Biophys. Acta Proteins Proteomics 2004, 1698, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Mandard, N.; Bulet, P.; Caille, A.; Daffre, S.; Vovelle, F. The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider. Eur. J. Biochem. 2002, 269, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Thevissen, K.; Osborn, R.W.; Acland, D.P.; Broekaert, W.F. Specific, high affinity binding sites for an antifungal plant defensin on Neurospora crassa hyphae and microsomal membranes. J. Biol. Chem. 1997, 272, 32176–32181. [Google Scholar] [CrossRef] [Green Version]
- Vriens, K.; Cammue, B.P.A.; Thevissen, K. Antifungal plant defensins: Mechanisms of action and production. Molecules 2014, 19, 12280–12303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Medeiros, L.N.; Angeli, R.; Sarzedas, C.G.; Barreto-Bergter, E.; Valente, A.P.; Kurtenbach, E.; Almeida, F.C.L. Backbone dynamics of the antifungal Psd1 pea defensin and its correlation with membrane interaction by NMR spectroscopy. Biochim. Biophys. Acta Biomembr. 2010, 1798, 105–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobo, D.S.; Pereira, I.B.; Fragel-Madeira, L.; Medeiros, L.N.; Cabral, L.M.; Faria, J.; Bellio, M.; Campos, R.C.; Linden, R.; Kurtenbach, E. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry 2007, 46, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Aerts, A.M.; François, I.E.J.A.; Meert, E.M.K.; Li, Q.T.; Cammue, B.P.A.; Thevissen, K. The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J. Mol. Microbiol. Biotechnol. 2007, 13, 243–247. [Google Scholar] [CrossRef]
- Hayes, B.M.E.; Bleackley, M.R.; Wiltshire, J.L.; Anderson, M.A.; Traven, A.; Van Der Weerden, N.L. Identification and mechanism of action of the plant defensin nad1 as a new member of the antifungal drug arsenal against candida albicans. Antimicrob. Agents Chemother. 2013, 57, 3667–3675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Weerden, N.L.; Lay, F.T.; Anderson, M.A. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 2008, 283, 14445–14452. [Google Scholar] [CrossRef] [Green Version]
- Wang, G. Structural insight into the mechanisms of action of antimicrobial peptides and structure-based design. In Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; Cabi: Wallingford, UK, 2017; pp. 169–187. [Google Scholar]
- Rozek, A.; Friedrich, C.L.; Hancock, R.E.W. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 2000, 39, 15765–15774. [Google Scholar] [CrossRef] [PubMed]
- Schibli, D.J.; Hwang, P.M.; Vogel, H.J. Structure of the antimicrobial peptide tritrpticin bound to micelles: A distinct membrane-bound peptide fold. Biochemistry 1999, 38, 16749–16755. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schibli, D.J.; Vogel, H.J. Structural studies and model membrane interactions of two peptides derived from bovine lactoferricin. J. Pept. Sci. 2005, 11, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tailhades, J.; O’Brien-Simpson, N.M.; Separovic, F.; Otvos, L.; Hossain, M.A.; Wade, J.D. Proline-rich antimicrobial peptides: Potential therapeutics against antibiotic-resistant bacteria. Amino Acids 2014, 46, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Krizsan, A.; Volke, D.; Weinert, S.; Sträter, N.; Knappe, D.; Hoffmann, R. Insect-Derived Proline-Rich Antimicrobial Peptides Kill Bacteria by Inhibiting Bacterial Protein Translation at the 70 S Ribosome. Angew. Chemie Int. Ed. 2014, 53, 12236–12239. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Grzela, R.; Giglione, C.; Meinnel, T.; Gennaro, R.; Mergaert, P.; Scocchi, M. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 2014, 21, 1639–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seefeldt, A.C.; Nguyen, F.; Antunes, S.; Pérébaskine, N.; Graf, M.; Arenz, S.; Inampudi, K.K.; Douat, C.; Guichard, G.; Wilson, D.N.; et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 2015, 22, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.Y.; Wang, G.; Tam, Y.T.; Tam, C. Membrane-active epithelial keratin 6A fragments (KAMPs) are unique human antimicrobial peptides with a non-αβ structure. Front. Microbiol. 2016, 7, 1799. [Google Scholar] [CrossRef] [Green Version]
- Koehbach, J.; Craik, D.J. The Vast Structural Diversity of Antimicrobial Peptides. Trends Pharmacol. Sci. 2019, 40, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsen, C.; Brede, D.A.; Nes, I.F.; Diep, D.B. Circular bacteriocins: Biosynthesis and mode of action. Appl. Environ. Microbiol. 2014, 80, 6854–6862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Visscher, L.A.; Gong, X.; Duszyk, M.; Vederas, J.C. The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 2009, 284, 28674–28681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Himeno, K.; Rosengren, K.J.; Inoue, T.; Perez, R.H.; Colgrave, M.L.; Lee, H.S.; Chan, L.Y.; Henriques, S.T.; Fujita, K.; Ishibashi, N.; et al. Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium. Biochemistry 2015, 54, 4863–4876. [Google Scholar] [CrossRef] [PubMed]
- Craik, D.J.; Clark, R.J.; Daly, N.L. Potential therapeutic applications of the cyclotides and related cystine knot mini-proteins. Expert Opin. Investig. Drugs 2007, 16, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Lu, Y.A.; Yang, J.L.; Chiu, K.W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 1999, 96, 8913–8918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, N.L.; Gustafson, K.R.; Craik, D.J. The role of the cyclic peptide backbone in the anti-HIV activity of the cyclotide kalata B1. FEBS Lett. 2004, 574, 69–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craik, D.J.; Daly, N.L.; Waine, C. The cystine knot motif in toxins and implications for drug design. Toxicon 2001, 39, 43–60. [Google Scholar] [CrossRef]
- Daly, N.L.; Koltay, A.; Gustafson, K.R.; Boyd, M.R.; Casas-Finet, J.R.; Craik, D.J. Solution structure by NMR of circulin A: A macrocyclic knotted peptide having anti-HIV activity. J. Mol. Biol. 1999, 285, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Koltay, A.; Daly, N.L.; Gustafson, K.R.; Craik, D.J. Structure of circulin B and implications for antimicrobial activity of the cyclotides. Int. J. Pept. Res. Ther. 2005, 11, 99–106. [Google Scholar] [CrossRef]
- Gustafson, K.R.; Sowder, R.C.; Henderson, L.E.; Parsons, I.C.; Kashman, Y.; Cardellina, J.H.; McMahon, J.B.; Buckheit Jr, R.W.; Pannell, L.K.; Boyd, M.R. Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 1994, 116, 9337–9338. [Google Scholar] [CrossRef]
- Fujitani, N.; Kouno, T.; Nakahara, T.; Takaya, K.; Osaki, T.; Kawabata, S.I.; Mizuguchi, M.; Aizawa, T.; Demura, M.; Nishimura, S.I.; et al. The solution structure of horseshoe crab antimicrobial peptide tachystatin B with an inhibitory cystine-knot motif. J. Pept. Sci. 2007, 13, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Kawulka, K.E.; Sprules, T.; Diaper, C.M.; Whittal, R.M.; McKay, R.T.; Mercier, P.; Zuber, P.; Vederas, J.C. Structure of Subtilosin A, A Cyclic Antimicrobial Peptide from Bacillus subtilis with Unusual Sulfur to α-Carbon Cross-Links: Formation and Reduction of α-Thio-α-Amino Acid Derivatives. Biochemistry 2004, 43, 3385–3395. [Google Scholar] [CrossRef] [PubMed]
- Trabi, M.; Schirra, H.J.; Craik, D.J. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from rhesus macaque leukocytes. Biochemistry 2001, 40, 4211–4221. [Google Scholar] [CrossRef] [PubMed]
- Bastian, A.; Schäfer, H. Human α-defensin 1 (HNP-1) inhibits adenoviral infection in vitro. Regul. Pept. 2001, 101, 157–161. [Google Scholar] [CrossRef]
- Edward Robinson, W.; McDougall, B.; Tran, D.; Selsted, M.E. Anti-HIV-1 activity of indolicidin, an antimicrobial peptide from neutrophils. J. Leukoc. Biol. 1998, 63, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Tamamura, H.; Ishihara, T.; Otaka, A.; Murakami, T.; Ibuka, T.; Waki, M.; Matsumoto, A.; Yamamoto, N.; Fujii, N. Analysis of the interaction of an anti-HIV peptide, T22 ([Tyr5,12, Lys7]-polyphemusin II), with gp120 and CD4 by surface plasmon resonance. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1996, 1298, 37–44. [Google Scholar] [CrossRef]
- Belaid, A.; Aouni, M.; Khelifa, R.; Trabelsi, A.; Jemmali, M.; Hani, K. In vitro antiviral activity of dermaseptins against herpes simplex virus type 1. J. Med. Virol. 2002, 66, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Motsuchi, W.; Tanaka, S.; Dosakol, S.I. Inhibition with lactoferrin of in vitro infection with human herpes VIRUS. Japanese J. Med. Sci. Biol. 1994, 47, 73–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchetti, M.; Longhi, C.; Conte, M.P.; Pisani, S.; Valenti, P.; Seganti, L. Lactoferrin inhibits herpes simplex virus type 1 adsorption to Vero cells. Antiviral Res. 1996, 29, 221–231. [Google Scholar] [CrossRef]
- Andersen, J.H.; Jenssen, H.; Sandvik, K.; Gutteberg, T.J. Anti-HSV activity of lactoferrin and lactoferricin is dependent on the presence of heparan sulphate at the cell surface. J. Med. Virol. 2004, 74, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Cheshenko, N.; Lehrer, R.I.; Herold, B.C. NP-1, a rabbit α-defensin, prevents the entry and intercellular spread of herpes simplex virus type 2. Antimicrob. Agents Chemother. 2003, 47, 494–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Gong, W.; Huang, C.C.; Herr, W.; Cheng, X. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 1999, 13, 1692–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wachinger, M.; Kleinschmidt, A.; Winder, D.; Von Pechmann, N.; Ludvigsen, A.; Neumann, M.; Holle, R.; Salmons, B.; Erfle, V.; Brack-Werner, R. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J. Gen. Virol. 1998, 79, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Albiol Matanic, V.C.; Castilla, V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int. J. Antimicrob. Agents 2004, 23, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Hayashiya, K. Red Fluorescent Protein in the Digestive Juice of the Silkworm Larvae Fed on Host-Plant Mulberry Leaves. Entomol. Exp. Appl. 1978, 24, 428–436. [Google Scholar] [CrossRef]
- Funakoshi, M.; Aizawa, K. Antiviral substance in the silkworm gut juice against a nuclear polyhedrosis virus of the silkworm, Bombyx mori. J. Invertebr. Pathol. 1989, 53, 135–136. [Google Scholar] [CrossRef]
- Sunagar, S.G.; Savanurmath, C.J.; Hinchigeri, S.B. The profiles of red fluorescent proteins with antinucleopolyhedrovirus activity in races of the silkworm Bombyx mori. J. Insect Physiol. 2011, 57, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, A.; Hirayama, E.; Kim, J. Antiviral substance from silkworm faeces: Characterization of its antiviral activity. Microbiol. Immunol. 2000, 44, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rozek, A.; Hancock, R.E.W. Interaction of Cationic Antimicrobial Peptides with Model Membranes. J. Biol. Chem. 2001, 276, 35714–35722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolym. Pept. Sci. Sect. 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.; Liu, H.; Hui Lee, W.; Zhang, Y. An anionic antimicrobial peptide from toad Bombina maxima. Biochem. Biophys. Res. Commun. 2002, 295, 796–799. [Google Scholar] [CrossRef]
- Brogden, K.A.; Ackermann, M.; Huttner, K.M. Small, anionic, and charge-neutralizing propeptide fragments of zymogens are antimicrobial. Antimicrob. Agents Chemother. 1997, 41, 1615–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gales, A.C.; Jones, R.N.; Sader, H.S. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: Results from the SENTRY antimicrobial surveillance program (2006-09). J. Antimicrob. Chemother. 2011, 66, 2070–2074. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.W. DAPTOMYCIN, its membrane-active mechanism vs. that of other antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862. [Google Scholar] [CrossRef]
- Kahne, D.; Leimkuhler, C.; Lu, W.; Walsh, C. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, P.E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 1989, 8, 943–950. [Google Scholar] [CrossRef]
- Cooper, M.A.; Williams, D.H. Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem. Biol. 1999, 6, 891–899. [Google Scholar] [CrossRef]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Cruciani, R.A.; Barker, J.L.; Durell, S.R.; Raghunathan, G.; Robert Guy, H.; Zasloff, M.; Stanley, E.F. Magainin 2, a natural antibiotic from frog skin, forms ion channels in lipid bilayer membranes. Eur. J. Pharmacol. Mol. Pharmacol. 1992, 226, 287–296. [Google Scholar] [CrossRef]
- Shai, Y.; Oren, Z. From “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides 2001, 22, 1629–1641. [Google Scholar] [CrossRef]
- Sengupta, D.; Leontiadou, H.; Mark, A.E.; Marrink, S.J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. Biophys. Acta Biomembr. 2008, 1778, 2308–2317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wadhwani, P.; Epand, R.F.; Heidenreich, N.; Bürck, J.; Ulrich, A.S.; Epand, R.M. Membrane-active peptides and the clustering of anionic lipids. Biophys. J. 2012, 103, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lohner, K.; Prossnigg, F. Biological activity and structural aspects of PGLa interaction with membrane mimetic systems. Biochim. Biophys. Acta Biomembr. 2009. [CrossRef] [PubMed] [Green Version]
- Rapaport, D.; Shai, Y. Interaction of fluorescently labeled pardaxin and its analogues with lipid bilayers. J. Biol. Chem. 1991, 266, 23769–23775. [Google Scholar] [CrossRef]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef] [Green Version]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta Biomembr. 1999, 1462, 55–70. [Google Scholar] [CrossRef] [Green Version]
- Gazit, E.; Shai, Y. Structural and Functional Characterization of the α5 Segment of Bacillus thuringiensis δ-Endotoxin. Biochemistry 1993, 32, 3429–3436. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Paramonov, A.S.; Lyukmanova, E.N.; Gizatullina, A.K.; Zhuravleva, A.V.; Tagaev, A.A.; Yakimenko, Z.A.; Telezhinskaya, I.N.; Kirpichnikov, M.P.; Ovchinnikova, T.V.; et al. Peptaibol antiamoebin I: Spatial structure, backbone dynamics, interaction with bicelles and lipid-protein nanodiscs, and pore formation in context of barrel-stave model. Chem. Biodivers. 2013, 10, 838–863. [Google Scholar] [CrossRef] [PubMed]
- Moll, G.N.; Konings, W.N.; Driessen, A.J.M. Bacteriocins: Mechanism of membrane insertion and pore formation. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 1999, 76, 185–198. [Google Scholar] [CrossRef]
- Hara, T.; Mitani, Y.; Tanaka, K.; Uematsu, N.; Takakura, A.; Tachi, T.; Kodama, H.; Kondo, M.; Mori, H.; Otaka, A.; et al. Heterodimer formation between the antimicrobial peptides magainin 2 and PGLa in lipid bilayers: A cross-linking study. Biochemistry 2001, 40, 12395–12399. [Google Scholar] [CrossRef] [PubMed]
- Campagna, S.; Saint, N.; Molle, G.; Aumelas, A. Structure and mechanism of action of the antimicrobial peptide piscidin. Biochemistry 2007, 46, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Kodama, H.; Kondo, M.; Wakamatsu, K.; Takeda, A.; Tachi, T.; Matsuzaki, K. Effects of peptide dimerization on pore formation: Antiparallel disulfide-dimerized magainin 2 analogue. Biopolymers 2001, 58, 437–446. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Balandin, S.V.; Trunov, K.I.; Paramonov, A.S.; Sukhanov, S.V.; Barsukov, L.I.; Arseniev, A.S.; Ovchinnikova, T.V. Molecular mechanism of action of β-Hairpin antimicrobial peptide arenicin: Oligomeric structure in dodecylphosphocholine micelles and pore formation in planar lipid bilayers. Biochemistry 2011, 50, 6255–6265. [Google Scholar] [CrossRef]
- Leontiadou, H.; Mark, A.E.; Marrink, S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006, 128, 12156–12161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneyama, F.; Imura, Y.; Ohno, K.; Zendo, T.; Nakayama, J.; Matsuzaki, K.; Sonomoto, K. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob. Agents Chemother. 2009, 53, 3211–3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allende, D.; Simon, S.A.; McIntosh, T.J. Melittin-induced bilayer leakage depends on lipid material properties: Evidence for toroidal pores. Biophys. J. 2005, 88, 1828–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladokhin, A.S.; White, S.H. “Detergent-like” permeabilization of anionic lipid vesicles by melittin. Biochim. Biophys. Acta Biomembr. 2001, 1514, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Saint, N.; Cadiou, H.; Bessin, Y.; Molle, G. Antibacterial peptide pleurocidin forms ion channels in planar lipid bilayers. Biochim. Biophys. Acta Biomembr. 2002, 1564, 359–364. [Google Scholar] [CrossRef] [Green Version]
- Bond, P.J.; Parton, D.L.; Clark, J.F.; Sansom, M.S.P. Coarse-grained simulations of the membrane-active antimicrobial peptide maculatin 1.1. Biophys. J. 2008, 95, 3802–3815. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Sahoo, N.; Bhunia, A. Antimicrobial Peptides and their Pore/Ion Channel Properties in Neutralization of Pathogenic Microbes. Curr. Top. Med. Chem. 2015, 16, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Sitaram, N.; Nagaraj, R. Interaction of antimicrobial peptides with biological and model membranes: Structural and charge requirements for activity. Biochim. Biophys. Acta Biomembr. 1999, 1462, 29–54. [Google Scholar] [CrossRef] [Green Version]
- Marchand, C.; Krajewski, K.; Lee, H.F.; Antony, S.; Johnson, A.A.; Amin, R.; Roller, P.; Kvaratskhelia, M.; Pommier, Y. Covalent binding of the natural antimicrobial peptide indolicidin to DNA abasic sites. Nucleic Acids Res. 2006, 34, 5157–5165. [Google Scholar] [CrossRef] [PubMed]
- Subbalakshmi, C.; Sitaram, N. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 1998, 160, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, D.I.; Le Brun, A.P.; Whitwell, T.C.; Sani, M.A.; James, M.; Separovic, F. The antimicrobial peptide aurein 1.2 disrupts model membranes via the carpet mechanism. Phys. Chem. Chem. Phys. 2012, 14, 15739–15751. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.; Bowie, J.H.; Carver, J.A. The solution structure and activity of caerin 1.1, an antimicrobial peptide from the Australian green tree frog, Litoria splendida. Eur. J. Biochem. 1997, 247, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Monaco, V.; Formaggio, F.; Crisma, M.; Toniolo, C.; Hanson, P.; Millhauser, G.L. Orientation and immersion depth of a helical lipopeptaibol in membranes using TOAC as an ESR probe. Biopolymers 1999, 50, 239–253. [Google Scholar] [CrossRef]
- Otvos, L. Antibacterial peptides and proteins with multiple cellular targets. J. Pept. Sci. 2005, 11, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Harrison, S.D. Cell-penetrating peptides in drug development: Enabling intracellular targets. Biochem. Soc. Trans. 2007, 35, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Kim, M.S.; Kim, S.C. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. Commun. 1996, 218, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.B.; Yi, K.S.; Matsuzaki, K.; Kim, M.S.; Kim, S.C. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: The proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA 2000, 97, 8245–8250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustillo, M.E.; Fischer, A.L.; Labouyer, M.A.; Klaips, J.A.; Webb, A.C.; Elmore, D.E. Modular analysis of hipposin, a histone-derived antimicrobial peptide consisting of membrane translocating and membrane permeabilizing fragments. Biochim. Biophys. Acta Biomembr. 2014, 1838, 2228–2233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta Proteins Proteomics 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Schibli, D.J.; Epand, R.F.; Vogel, H.J.; Epand, R.M. Tryptophan-rich antimicrobial peptides: Comparative properties and membrane interactions. Biochem. Cell Biol. 2002, 80, 667–677. [Google Scholar] [CrossRef]
- Sugiarto, H.; Yu, P.L. Mechanisms of action of ostrich β-defensins against Escherichia coli. FEMS Microbiol. Lett. 2007, 270, 195–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizan, J.L.; Hernandez-Chico, C.; Del Castillo, I.; Moreno, F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 1991, 10, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G.; Agerberth, B.; Boman, A. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 1993, 61, 2978–2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutkenhaus, J. Regulation of cell division in E. coli. Trends Genet. 1990, 6, 22–25. [Google Scholar] [CrossRef]
- Delgado, M.A.; Rintoul, M.R.; Farías, R.N.; Salomón, R.A. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 2001, 183, 4543–4550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chileveru, H.R.; Lim, S.A.; Chairatana, P.; Wommack, A.J.; Chiang, I.L.; Nolan, E.M. Visualizing attack of escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry 2015, 54, 1767–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Patrzykat, A.; Friedrich, C.L.; Zhang, L.; Mendoza, V.; Hancock, R.E.W. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 605–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, C.F.; Gudimella, R.; Razali, R.; Manikam, R.; Sekaran, S.D. Transcriptome analysis of Streptococcus pneumoniae treated with the designed antimicrobial peptides, DM3. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Yamazaki, K.; Kawakami, S.; Miyoshi, D.; Ooi, T.; Hashimoto, S.; Taguchi, S. In vivo target exploration of apidaecin based on Acquired Resistance induced by Gene Overexpression (ARGO assay). Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Ho, Y.H.; Shah, P.; Chen, Y.W.; Chen, C.S. Systematic analysis of intracellular-targeting antimicrobial peptides, bactenecin 7, hybrid of pleurocidin and dermaseptin, proline-arginine-rich peptide, and lactoferricin b, by using Escherichia coli proteome microarrays. Mol. Cell. Proteomics 2016, 15, 1837–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenssen, H.; Hamill, P.; Hancock, R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otvos, L.; Insug, O.; Rogers, M.E.; Consolvo, P.J.; Condie, B.A.; Lovas, S.; Bulet, P.; Blaszczyk-Thurin, M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 2000, 39, 14150–14159. [Google Scholar] [CrossRef] [PubMed]
- Chesnokova, L.S.; Slepenkov, S.V.; Witt, S.N. The insect antimicrobial peptide, L-pyrrhocoricin, binds to and stimulates the ATPase activity of both wild-type and lidless DnaK. FEBS Lett. 2004, 565, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Scocchi, M.; Lüthy, C.; Decarli, P.; Mignogna, G.; Christen, P.; Gennaro, R. The Proline-rich Antibacterial Peptide Bac7 Binds to and Inhibits in vitro the Molecular Chaperone DnaK. Int. J. Pept. Res. Ther. 2009, 15, 147–155. [Google Scholar] [CrossRef]
- Knappe, D.; Zahn, M.; Sauer, U.; Schiffer, G.; Sträter, N.; Hoffmann, R. Rational Design of Oncocin Derivatives with Superior Protease Stabilities and Antibacterial Activities Based on the High-Resolution Structure of the Oncocin-DnaK Complex. ChemBioChem 2011, 12, 874–876. [Google Scholar] [CrossRef] [PubMed]
- Knappe, D.; Piantavigna, S.; Hansen, A.; Mechler, A.; Binas, A.; Nolte, O.; Martin, L.L.; Hoffmann, R. Oncocin (VDKPPYLPRPRPPRRIYNR-NH2): A novel antibacterial peptide optimized against gram-negative human pathogens. J. Med. Chem. 2010, 53, 5240–5247. [Google Scholar] [CrossRef] [PubMed]
- Couto, M.A.; Harwig, S.S.L.; Lehrer, R.I. Selective inhibition of microbial serine proteases by eNAP-2, an antimicrobial peptide from equine neutrophils. Infect. Immun. 1993, 61, 2991–2994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogaça, A.C.; Almeida, I.C.; Eberlin, M.N.; Tanaka, A.S.; Bulet, P.; Daffre, S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides 2006, 27, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Troxler, R.F.; Offner, G.D.; Xu, T.; Vanderspek, J.C.; Oppenheim, F.G. Structural Relationship Between Human Salivary Histatins. J. Dent. Res. 1990, 69, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Mackay, B.J.; Denepitiya, L.; Iacono, V.J.; Krost, S.B.; Pollock, J.J. Growth-inhibitory and bactericidal effects of human parotid salivary histidine-rich polypeptides on Streptococcus mutans. Infect. Immun. 1984, 44, 695–701. [Google Scholar] [CrossRef] [Green Version]
- Nishikata, M.; Kanehira, T.; Oh, H.; Tani, H.; Tazaki, M.; Kuboki, Y. Salivary histatin as an inhibitor of a protease produced by the oral bacterium Bacteroides gingivalis. Biochem. Biophys. Res. Commun. 1991, 174, 625–630. [Google Scholar] [CrossRef]
- Brötz, H.; Bierbaum, G.; Reynolds, P.E.; Sahl, H.G. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur. J. Biochem. 1997, 246, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Sass, V.; Schneider, T.; Wilmes, M.; Körner, C.; Tossi, A.; Novikova, N.; Shamova, O.; Sahl, H.G. Human β-defensin 3 inhibits cell wall biosynthesis in staphylococci. Infect. Immun. 2010, 78, 2793–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Leeuw, E.; Li, C.; Zeng, P.; Li, C.; de Buin, M.D.; Lu, W.Y.; Breukink, E.; Lu, W. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010, 584, 1543–1548. [Google Scholar] [CrossRef] [Green Version]
- Song, C.; Weichbrodt, C.; Salnikov, E.S.; Dynowski, M.; Forsberg, B.O.; Bechinger, B.; Steinem, C.; De Groot, B.L.; Zachariae, U.; Zeth, K. Crystal structure and functional mechanism of a human antimicrobial membrane channel. Proc. Natl. Acad. Sci. USA 2013, 110, 4586–4591. [Google Scholar] [CrossRef] [Green Version]
- Perrin, B.S.; Tian, Y.; Fu, R.; Grant, C.V.; Chekmenev, E.Y.; Wieczorek, W.E.; Dao, A.E.; Hayden, R.M.; Burzynski, C.M.; Venable, R.M.; et al. High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion. J. Am. Chem. Soc. 2014, 136, 3491–3504. [Google Scholar] [CrossRef] [PubMed]
- Henzler Wildman, K.A.; Lee, D.K.; Ramamoorthy, A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 2003, 42, 6545–6558. [Google Scholar] [CrossRef] [PubMed]
- Fernández de Ullivarri, M.; Arbulu, S.; Garcia-Gutierrez, E.; Cotter, P.D. Antifungal Peptides as Therapeutic Agents. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.C.; Palacios, D.S.; Dailey, I.; Endo, M.M.; Uno, B.E.; Wilcock, B.C.; Burke, M.D. Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. USA 2012, 109, 2234–2239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, T.; Umegawa, Y.; Yamagami, M.; Suzuki, T.; Tsuchikawa, H.; Hanashima, S.; Matsumori, N.; Murata, M. The Perpendicular Orientation of Amphotericin B Methyl Ester in Hydrated Lipid Bilayers Supports the Barrel-Stave Model. Biochemistry 2019, 58, 2282–2291. [Google Scholar] [CrossRef] [PubMed]
- Galanth, C.; Abbassi, F.; Lequin, O.; Ayala-Sanmartin, J.; Ladram, A.; Nicolas, P.; Amiche, M. Mechanism of antibacterial action of dermaseptin B2: Interplay between helix—Hinge—Helix structure and membrane curvature strain. Biochemistry 2009, 48, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xi, X.; Ma, C.; Chen, X.; Zhou, M.; Burrows, J.F.; Chen, T.; Wang, L. A novel dermaseptin isolated from the skin secretion of phyllomedusa tarsius and its cationicity- enhanced analogue exhibiting effective antimicrobial and anti-proliferative activities. Biomolecules 2019, 9, 628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benincasa, M.; Scocchi, M.; Pacor, S.; Tossi, A.; Nobili, D.; Basaglia, G.; Busetti, M.; Gennaro, R. Fungicidal activity of five cathelicidin peptides against clinically isolated yeasts. J. Antimicrob. Chemother. 2006, 58, 950–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Cesare, G.B.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial peptides: A new frontier in antifungal therapy. Mbio 2020, 11, 1–21. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlić, A.; Bi, C.; Bluhm, W.F.; Christie, C.H.; Dutta, S.; Green, R.K.; Goodsell, D.S.; Westbrook, J.D.; Woo, J.; et al. The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015, 43, D345–D356. [Google Scholar] [CrossRef] [PubMed]
- Jhong, J.H.; Chi, Y.H.; Li, W.C.; Lin, T.H.; Huang, K.Y.; Lee, T.Y. DbAMP: An integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019, 47, D285–D297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirtskhalava, M.; Gabrielian, A.; Cruz, P.; Griggs, H.L.; Squires, R.B.; Hurt, D.E.; Grigolava, M.; Chubinidze, M.; Gogoladze, G.; Vishnepolsky, B.; et al. DBAASP v.2: An enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016, 44, D1104–D1112. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.P.; Fleming, K.G. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl. Acad. Sci. USA 2011, 108, 10174–10177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wimley, W.C.; White, S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996, 3, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 1984, 81, 140–144. [Google Scholar] [CrossRef] [Green Version]
- Koehler, J.; Woetzel, N.; Staritzbichler, R.; Sanders, C.R.; Meiler, J. A unified hydrophobicity scale for multispan membrane proteins. Proteins Struct. Funct. Bioinforma. 2009, 76, 13–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessa, T.; Kim, H.; Bihlmaier, K.; Lundin, C.; Boekel, J.; Andersson, H.; Nilsson, I.M.; White, S.H.; Von Heijne, G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 2005, 433, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Wu, H.; Huang, J.; Wang, W.; Ge, K.; Li, G.; Zhong, J.; Huang, Q. LAMP2: A major update of the database linking antimicrobial peptides. Database 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Pires, A.S.; Franco, O.L. Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnol. Adv. 2017, 35, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990, 183, 63–98. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [Green Version]
- Porto, W.F.; Souza, V.A.; Nolasco, D.O.; Franco, O.L. In silico identification of novel hevein-like peptide precursors. Peptides 2012, 38, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Fan, L.; Sun, J.; Lao, X.; Zheng, H. Computational resources and tools for antimicrobial peptides. J. Pept. Sci. 2017, 23, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Wang, P.; Hu, L.; Liu, G.; Jiang, N.; Chen, X.; Xu, J.; Zheng, W.; Li, L.; Tan, M.; Chen, Z.; et al. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PloS ONE 2011, 6, e18476. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, O.L. Peptide promiscuity: An evolutionary concept for plant defense. FEBS Lett. 2011, 585, 995–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Kotsiantis, S.B. Supervised machine learning: A review of classification techniques. Informatica 2007, 31, 249–268. [Google Scholar] [CrossRef]
- Francis, L. Unsupervised learning. In Predictive Modeling Applications in Actuarial Science: Volume I: Predictive Modeling Techniques; Cambridge University Press: Cambridge, UK, 2014; pp. 280–312. ISBN 9781139342674. [Google Scholar]
- Lee, M.W.; Lee, E.Y.; Ferguson, A.L.; Wong, G.C.L. Machine learning antimicrobial peptide sequences: Some surprising variations on the theme of amphiphilic assembly. Curr. Opin. Colloid Interface Sci. 2018, 38, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Sun, Z. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 2001, 17, 721–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, X.Y.; Rosdi, B.A.; Shahrudin, S. Prediction of antimicrobial peptides based on sequence alignment and support vector machine-pairwise algorithm utilizing LZ-complexity. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forrests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Khatun, S.; Hasan, M.; Kurata, H. Efficient computational model for identification of antitubercular peptides by integrating amino acid patterns and properties. FEBS Lett. 2019, 593, 3029–3039. [Google Scholar] [CrossRef]
- Poorinmohammad, N.; Hamedi, J.; Moghaddam, M.H.A.M. Sequence-based analysis and prediction of lantibiotics: A machine learning approach. Comput. Biol. Chem. 2018, 77, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xu, J.; Yin, Y.; Quan, X.; Zhang, H. Antimicrobial peptide identification using multi-scale convolutional network. BMC Bioinform. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Garlick, S.; Zloh, M. Deep learning for novel antimicrobial peptide design. Biomolecules 2021, 11, 471. [Google Scholar] [CrossRef]
- Sharma, R.; Shrivastava, S.; Kumar Singh, S.; Kumar, A.; Saxena, S.; Kumar Singh, R. AniAMPpred: Artificial intelligence guided discovery of novel antimicrobial peptides in animal kingdom. Brief. Bioinform. 2021. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, P.; Wang, Z.; Yu, X. Predicting antimicrobial peptides by using increment of diversity with quadratic discriminant analysis method. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2019, 16, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.Y.; Lin, T.P.; Shih, L.Y.; Wang, C.K. Analysis and prediction of the critical regions of antimicrobial peptides based on conditional random fields. PLoS ONE 2015, 10, e0119490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhao, T.; Wei, D.; Strandberg, E.; Ulrich, A.S.; Ulmschneider, J.P. How reliable are molecular dynamics simulations of membrane active antimicrobial peptides? Biochim. Biophys. Acta Biomembr. 2014, 1838, 2280–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulmschneider, J.P.; Ulmschneider, M.B. Molecular Dynamics Simulations Are Redefining Our View of Peptides Interacting with Biological Membranes. Acc. Chem. Res. 2018, 51, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.W.; Hsu, N.Y.; Wang, C.H.; Lu, C.Y.; Chang, Y.; Tsai, H.H.G.; Ruaan, R.C. Coupling Molecular Dynamics Simulations with Experiments for the Rational Design of Indolicidin-Analogous Antimicrobial Peptides. J. Mol. Biol. 2009, 392, 837–854. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, H.J.; Gangopadhyay, A.; Datta, A. Prediction and characterisation of lantibiotic structures with molecular modelling and molecular dynamics simulations. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Waghu, F.H.; Joseph, S.; Ghawali, S.; Martis, E.A.; Madan, T.; Venkatesh, K.V.; Idicula-Thomas, S. Designing antibacterial peptides with enhanced killing kinetics. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Talandashti, R.; Mehrnejad, F.; Rostamipour, K.; Doustdar, F.; Lavasanifar, A. Molecular Insights into Pore Formation Mechanism, Membrane Perturbation, and Water Permeation by the Antimicrobial Peptide Pleurocidin: A Combined All-Atom and Coarse-Grained Molecular Dynamics Simulation Study. J. Phys. Chem. B 2021, 125, 7163–7176. [Google Scholar] [CrossRef] [PubMed]
- Catte, A.; Wilson, M.R.; Walker, M.; Oganesyan, V.S. Antimicrobial action of the cationic peptide, chrysophsin-3: A coarse-grained molecular dynamics study. Soft Matter 2018, 14, 2796–2807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, Y.; Xiang, N.; Zhu, X.; Narsimhan, G. Potential of mean force for insertion of antimicrobial peptide melittin into a pore in mixed DOPC/DOPG lipid bilayer by molecular dynamics simulation. J. Chem. Phys. 2017, 146. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Bao, J.; Lao, X.; Zheng, H. Novel 3D Structure Based Model for Activity Prediction and Design of Antimicrobial Peptides. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Petkov, P.; Lilkova, E.; Ilieva, N.; Litov, L. Self-association of antimicrobial peptides: A molecular dynamics simulation study on bombinin. Int. J. Mol. Sci. 2019, 20, 5450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogdanova, L.R.; Valiullina, Y.A.; Faizullin, D.A.; Kurbanov, R.K.; Ermakova, E.A. Spectroscopic, zeta potential and molecular dynamics studies of the interaction of antimicrobial peptides with model bacterial membrane. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 242. [Google Scholar] [CrossRef] [PubMed]
- Lai, P.K.; Kaznessis, Y.N. Insights into membrane translocation of protegrin antimicrobial peptides by multistep molecular dynamics simulations. In Proceedings of the AIChE Annual Meeting, Pittsburgh, PA, USA, 28 October–2 November 2018; 2018; 3, pp. 6056–6065. [Google Scholar] [CrossRef]
- Zhao, L.; Cao, Z.; Bian, Y.; Hu, G.; Wang, J.; Zhou, Y. Molecular dynamics simulations of human antimicrobial peptide LL-37 in model POPC and POPG lipid bilayers. Int. J. Mol. Sci. 2018, 19, 1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sur, S.; Romo, T.D.; Grossfield, A. Selectivity and Mechanism of Fengycin, an Antimicrobial Lipopeptide, from Molecular Dynamics. J. Phys. Chem. B 2018, 122, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Sercu, T.; Wadhawan, K.; Padhi, I.; Gehrmann, S.; Cipcigan, F.; Chenthamarakshan, V.; Strobelt, H.; dos Santos, C.; Chen, P.Y.; et al. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 2021, 5, 613–623. [Google Scholar] [CrossRef]
- Wang, C.K.L.; Kaas, Q.; Chiche, L.; Craik, D.J. CyBase: A database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008, 36. [Google Scholar] [CrossRef] [Green Version]
- Lata, S.; Sharma, B.K.; Raghava, G.P.S. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Qu, X.; He, X.; Duan, L.; Wu, G.; Bi, D.; Deng, Z.; Liu, W.; Ou, H.Y. ThioFinder: A Web-Based Tool for the Identification of Thiopeptide Gene Clusters in DNA Sequences. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Qureshi, A.; Kumar, M. AVPpred: Collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, S.; Karnik, S.; Nilawe, P.; Jayaraman, V.K.; Idicula-Thomas, S. ClassAMP: A prediction tool for classification of antimicrobial peptides. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2012, 9, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Kamech, N.; Vukičević, D.; Ladram, A.; Piesse, C.; Vasseur, J.; Bojović, V.; Simunić, J.; Juretić, D. Improving the selectivity of antimicrobial peptides from anuran skin. J. Chem. Inf. Model. 2012, 52, 3341–3351. [Google Scholar] [CrossRef] [PubMed]
- Novković, M.; Simunić, J.; Bojović, V.; Tossi, A.; Juretić, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- Piotto, S.P.; Sessa, L.; Concilio, S.; Iannelli, P. YADAMP: Yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents 2012, 39, 346–351. [Google Scholar] [CrossRef]
- Holton, T.A.; Pollastri, G.; Shields, D.C.; Mooney, C. CPPpred: Prediction of cell penetrating peptides. Bioinformatics 2013, 29, 3094–3096. [Google Scholar] [CrossRef]
- Qureshi, A.; Thakur, N.; Kumar, M. HIPdb: A Database of Experimentally Validated HIV Inhibiting Peptides. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.T.; Lee, C.C.; Yang, J.R.; Lai, J.Z.C.; Chang, K.Y.; Ray, O. A large-scale structural classification of Antimicrobial peptides. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Di Luca, M.; Maccari, G.; Maisetta, G.; Batoni, G. BaAMPs: The database of biofilm-active antimicrobial peptides. Biofouling 2015, 31, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gupta, P.; Kumar, R.; Bhardwaj, A. DPABBs: A Novel in silico Approach for Predicting and Designing Anti-biofilm Peptides. Sci. Rep. 2016, 6, 21839. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Khater, S.; Gupta, M.; Sain, N.; Mohanty, D. RiPPMiner: A bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 2017, 45, W80–W88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meher, P.K.; Sahu, T.K.; Saini, V.; Rao, A.R. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.A.; Giraldo, P.; Orduz, S. InverPep: A database of invertebrate antimicrobial peptides. J. Glob. Antimicrob. Resist. 2017, 8, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gull, S.; Shamim, N.; Minhas, F. AMAP: Hierarchical multi-label prediction of biologically active and antimicrobial peptides. Comput. Biol. Med. 2019, 107, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Beltrán Lissabet, J.F.; Belén, L.H.; Farias, J.G. AntiVPP 1.0: A portable tool for prediction of antiviral peptides. Comput. Biol. Med. 2019, 107, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. Meta-iavp: A sequence-based meta-predictor for improving the prediction of antiviral peptides using effective feature representation. Int. J. Mol. Sci. 2019, 20, 5743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.C. MACppred: A support vector machine-based meta-predictor for identification of anticancer peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Chilimoniuk, J.; Rödiger, S.; Gagat, P. Proteomic Screening for Prediction and Design of Antimicrobial Peptides with AmpGram. Int. J. Mol. Sci. 2020, 21, 4310. [Google Scholar] [CrossRef] [PubMed]
- Kavousi, K.; Bagheri, M.; Behrouzi, S.; Vafadar, S.; Atanaki, F.F.; Lotfabadi, B.T.; Ariaeenejad, S.; Shockravi, A.; Moosavi-Movahedi, A.A. IAMPE: NMR-assisted computational prediction of antimicrobial peptides. J. Chem. Inf. Model. 2020, 60, 4691–4701. [Google Scholar] [CrossRef] [PubMed]
- Burdukiewicz, M.; Sidorczuk, K.; Rafacz, D.; Pietluch, F.; Bąkała, M.; Słowik, J.; Gagat, P. Cancergram: An effective classifier for differentiating anticancer from antimicrobial peptides. Pharmaceutics 2020, 12, 1045. [Google Scholar] [CrossRef]
- Fu, H.; Cao, Z.; Li, M.; Wang, S. ACEP: Improving antimicrobial peptides recognition through automatic feature fusion and amino acid embedding. BMC Genom. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj Vaithinathan, A.; Vanitha, A. WHO global priority pathogens list on antibiotic resistance: An urgent need for action to integrate One Health data. Perspect. Public Health 2018, 138, 87–88. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R. Road to clinical efficacy: Challenges and novel strategies for antimicrobial peptide development. Future Microbiol. 2011, 6, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms: My perspective. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; Volume 1117, pp. 3–6. [Google Scholar]
- Bucki, R.; Leszczyńska, K.; Namiot, A.; Sokołowski, W. Cathelicidin LL-37: A multitask antimicrobial peptide. Arch. Immunol. Ther. Exp. 2010, 58, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Agier, J.; Efenberger, M.; Brzezińska-Blaszczyk, E. Cathelicidin impact on inflammatory cells. Cent. Eur. J. Immunol. 2015, 40, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaison, G.; Graninger, W.; Bouza, E. Teicoplanin in the treatment of serious infection. J. Chemother. 2000, 12 (Suppl. 5), 26–33. [Google Scholar] [CrossRef] [PubMed]
- Di, L. Strategic Approaches to Optimizing Peptide ADME Properties. AAPS J. 2015, 17, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Irazazabal, L.N.; Porto, W.F.; Ribeiro, S.M.; Casale, S.; Humblot, V.; Ladram, A.; Franco, O.L. Selective amino acid substitution reduces cytotoxicity of the antimicrobial peptide mastoparan. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2699–2708. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunasekera, S.; Muhammad, T.; Strömstedt, A.A.; Rosengren, K.J.; Göransson, U. Backbone Cyclization and Dimerization of LL-37-Derived Peptides Enhance Antimicrobial Activity and Proteolytic Stability. Front. Microbiol. 2020, 11, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khaledi, A.; Weimann, A.; Schniederjans, M.; Asgari, E.; Kuo, T.; Oliver, A.; Cabot, G.; Kola, A.; Gastmeier, P.; Hogardt, M.; et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol. Med. 2020, 12, e10264. [Google Scholar] [CrossRef] [PubMed]
- Mehrnejad, F.; Zarei, M. Molecular dynamics simulation study of the interaction of piscidin 1 with dppc bilayers: Structure-activity relationship. J. Biomol. Struct. Dyn. 2010, 27, 551–559. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, J.; Han, M.L.; Jiang, X.; Azad, M.A.K.; Patil, N.A.; Lin, Y.W.; Zhao, J.; Hu, Y.; Yu, H.H.; et al. Polymyxins Bind to the Cell Surface of Unculturable Acinetobacter baumannii and Cause Unique Dependent Resistance. Adv. Sci. 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Yang, K.; Yuan, B.; Han, M.; Zhu, Y.; Roberts, K.D.; Patil, N.A.; Li, J.; Gong, B.; Hancock, R.E.W.; et al. Molecular dynamics simulations informed by membrane lipidomics reveal the structure-interaction relationship of polymyxins with the lipid A-based outer membrane of Acinetobacter baumannii. J. Antimicrob. Chemother. 2020, 75, 3534–3543. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Starr, C.G.; Troendle, E.; Wiedman, G.; Wimley, W.C.; Ulmschneider, J.P.; Ulmschneider, M.B. Simulation-Guided Rational de Novo Design of a Small Pore-Forming Antimicrobial Peptide. J. Am. Chem. Soc. 2019, 141, 4839–4848. [Google Scholar] [CrossRef] [PubMed]
- Kleandrova, V.V.; Ruso, J.M.; Speck-Planche, A.; Dias Soeiro Cordeiro, M.N. Enabling the Discovery and Virtual Screening of Potent and Safe Antimicrobial Peptides. Simultaneous Prediction of Antibacterial Activity and Cytotoxicity. ACS Comb. Sci. 2016, 18, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Li, F.F.; Brimble, M.A. Using chemical synthesis to optimise antimicrobial peptides in the fight against antimicrobial resistance. In Proceedings of the Pure and Applied Chemistry, Bangkok, Thailand, 7–8 February 2019; 2019; Volume 91, pp. 181–198. [Google Scholar]
- Wu, Z.; Li, Y.; Zhang, L.; Ding, Z.; Shi, G. Microbial production of small peptide: Pathway engineering and synthetic biology. Microb. Biotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
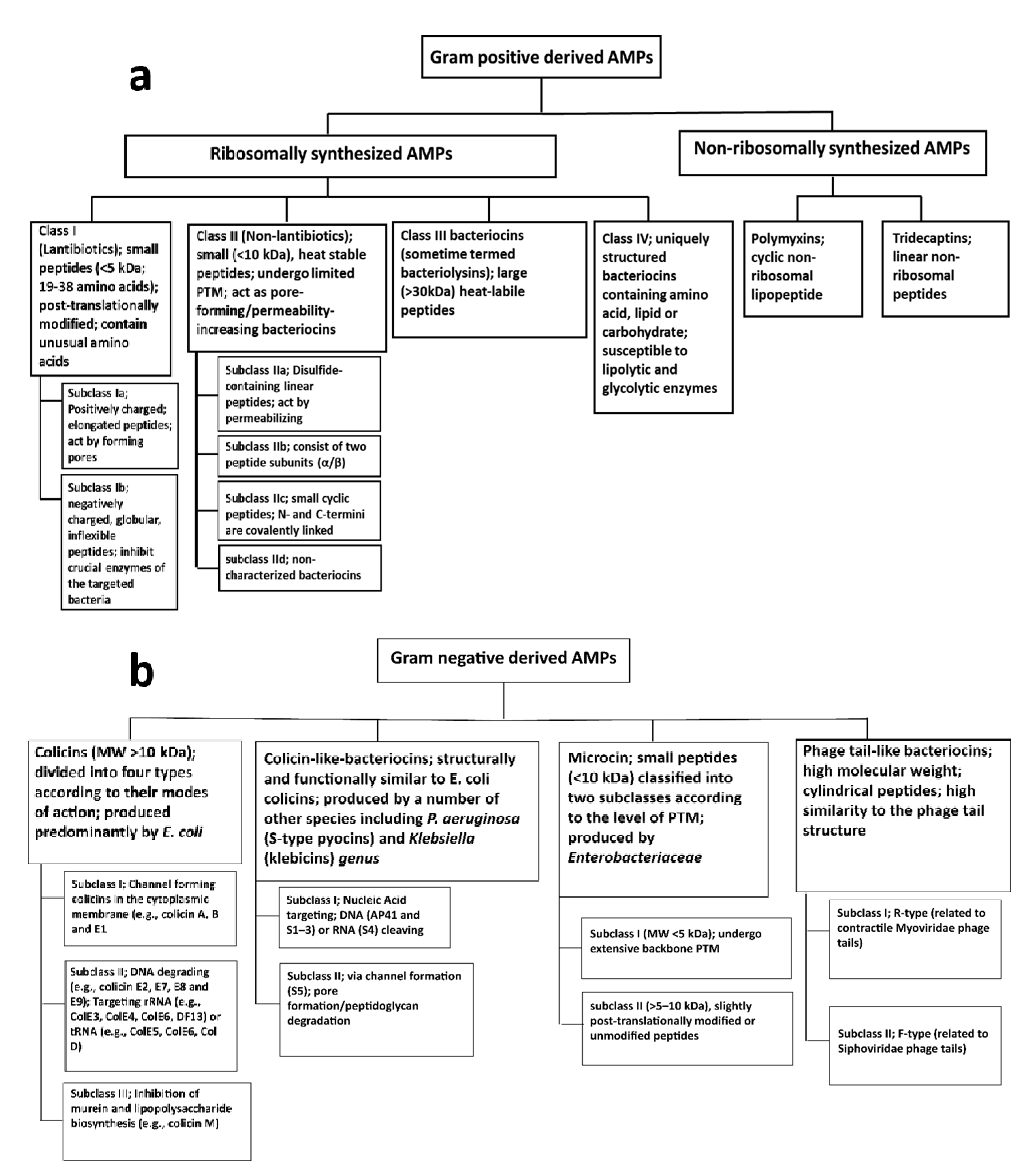




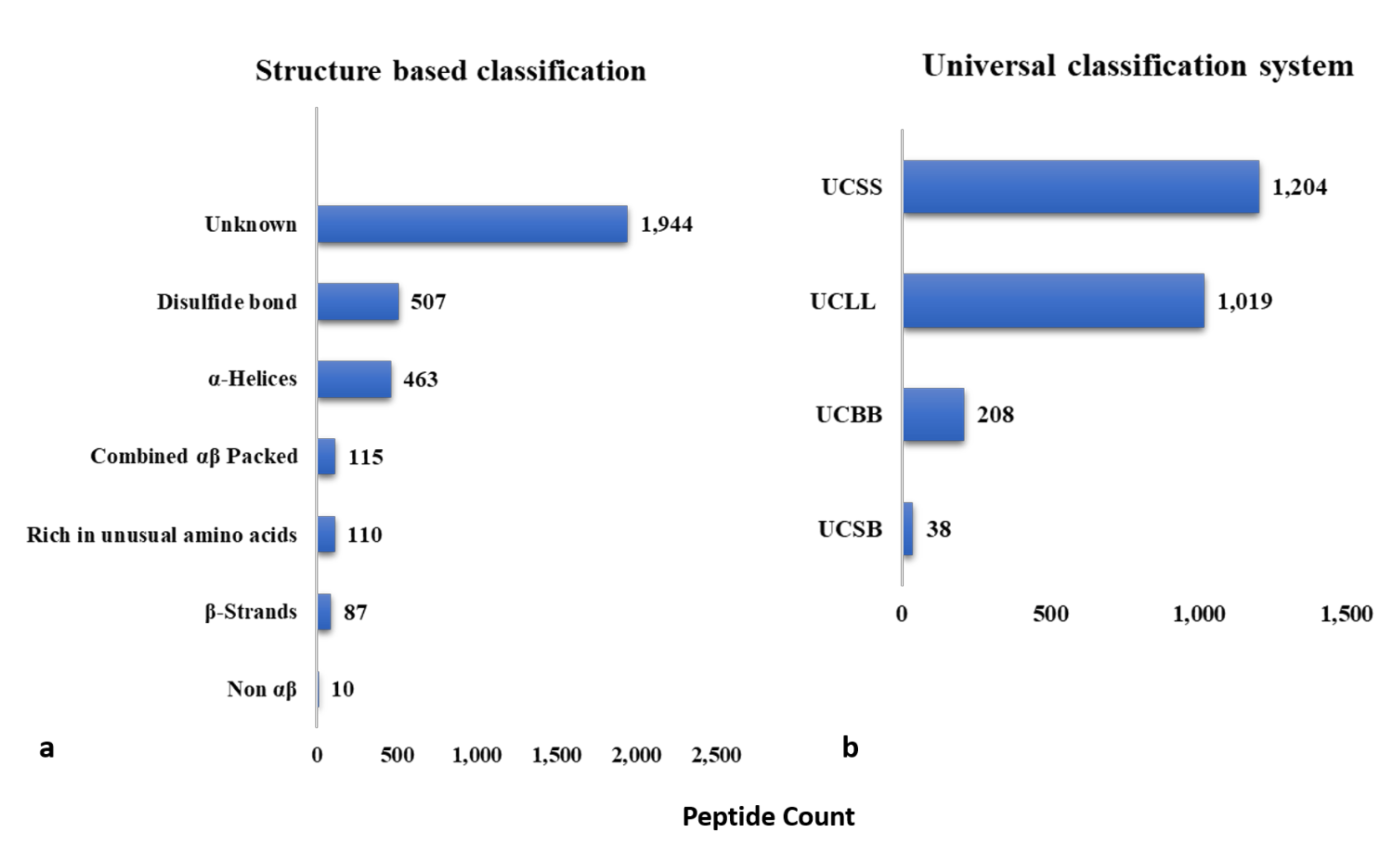
| Class | Description | Activity | References |
|---|---|---|---|
| Thionins | Cationic peptides of 45–48 amino acids containing 3–4 disulfide bridges | Antibacterial, Antifungal | [149,151] |
| Defensins | Catoinic peptides of 45 to 54 amino acids containing 4–5 disulfide bridges | Antibacterial, Antifungal | [152,153] |
| Hevein-like peptides | Basic peptides of 29–45 residues containing 3–5 disulfide bridges, rich in glycine and aromatic residues | Antifungal | [154,155] |
| Knottin-typepeptides | Peptides of ~30 residues, consisting of conserved cysteine residues and disulfide bridges | Antiviral, Antibacterial | [149,156] |
| α-Hairpinins | Rich in lysine/arginine, containing a helix-loop-helix secondary structure | Anti-HIV, Antibacterial, Antifungal | [149,157,158,159] |
| Lipid Transfer Proteins | Cationic peptides containing 70–90 residues that includes 8 cysteine residues | Antibacterial, Antifungal | [149,160,161] |
| Snakins | Catoinic small sized proteins characterized by 12 conserved cysteine residues | Antibacterial, Antifungal | [162,163] |
| Non-Cysteine Rich Peptides | Containing 0–1 cysteine residues and possessing high structural flexibility | Antibacterial, Antifungal | [149,164,165] |
| Class | Description | Activity | References |
|---|---|---|---|
| Bominin | Glycine-rich peptides with C-terminal amidated residues, isolated from skin secretions of Bombina genus | Antibacterial | [236,237] |
| Buforin | Cationic peptides rich in arginine and lysine, DNA-targeting, isolated from the stomach of Bufo gargarizans | Antibacterial, Antifungal | [238,239] |
| Cathelicidin | Amphibian cathelicidins possess homology with mammalian cathelicidins. Over 20 cathelicidins have been identified | Antibacterial | [240,241] |
| Dermaseptin | Cationic AMPs with an amphipathic structure of 24–34 residues. Dermaseptins are derived from the skin of the tree frog Phyllomedusa sauvagii and are involved in protein synthesis inhibition and induction of apoptosis | Antiviral, Antibacterial, Antifungal, Anticancer | [242,243,244] |
| Esculentin | Originally found in Rana tigrina (an edible frog). Characterized by C-terminal disulfide bond | Antibacterial, Antifungal | [245,246] |
| Fallaxin | A 25-residue AMP known as ocellatin-F1; isolated from Leptodactylus fallax | Antibacterial, Leishmanicidal | [247,248] |
| Maximin | Derived from toad related species, e.g., Chinese red belly toad (Bombina maxima) | Antibacterial | [249,250] |
| Magainin | Alpha-helical peptides isolated from the skin of X. laevis; interact with membrane phospholipids leading to disruption of the ionic gradient | Antibacterial, Antifungal, Antitumor | [251,252,253] |
| Plasticin | Dermaseptin-like peptides with 23–29 amino acids; active agasint bacteria via membrane disruption and pore formation | Antibacterial | [254,255] |
| Palustrin | Isolated from Lithobates Palustris. Characterized by the presence of disulfide bond and the rana box domain | Antibacterial | [256,257] |
| Phylloxin | Bacteriostatic AMPs; insolated from species including Phyllobates bicolor. | Antibacterial | [258] |
| Phyllospetin | C-terminal amidated peptides of 20 amino acids; isolated from South American tree frogs; inducing membrane permeabilization via a carpet-like action | Antibacterial, Antifungal, Antiparasitic, Anticancer | [259,260] |
| Psuedin | Isolated from the skin of Psuedis paradoxa; RNA targeting, ultimately stopping protein synthesis | Antibacterial, Antifungal | [261,262] |
| Ranateurin | Derived from the American bullfrog Rana catesbeiana | Antibacterial, Anticancer | [263] |
| Ranalexin | A C-terminal heptapeptide ringed 20 amino acids AMP having one disulfide bond | Antibacterial, Antifungal, Antiparasitic | [264,265] |
| Server | Algorithm | Year | Description | URL | Ref |
|---|---|---|---|---|---|
| CyBase | ellipsoid and random walk algorithm | 2008 | database of cyclic protein sequences and structures | http://www.cybase.org.au/index.php (accessed on 22 October 2021) | [567] |
| AntiBP2 | SVM | 2010 | Predict antibacterial peptides in protein sequences | www.imtech.res.in/raghava/antibp/ (accessed on 2 October 2020) | [568] |
| THIOBASE | - | 2011 | Database of thiopeptides | https://bioinfo-mml.sjtu.edu.cn/THIOBASE/index.php (accessed on 22 October 2021) | [569] |
| AvPred | SVM | 2012 | Predict antiviral peptides | http://crdd.osdd.net/servers/avppred/index.html (accessed on 2 October 2020) | [570] |
| ThioFinder | HMMs | 2012 | Identify thiopeptide antibiotic gene clusters in DNA sequences | https://bioinfo-mml.sjtu.edu.cn/ThioFinder/index.php (accessed on 22 October 2021) | [569] |
| ClassAMP | RF, SVM | 2012 | Predict AMP domains in protein sequences | http://www.bicnirrh.res.in/classamp/ (accessed on 2 October 2020) | [571] |
| Mutator 2.0 | Mutator Algorithm | 2012 | Predict the effect of single or double amino acid substitutions on the therapeutic index (TI) of helical AMPs | http://split4.pmfst.hr/mutator/ (accessed on 2 October 2020) | [572] |
| DADP | - | 2012 | Database of bioactive peptides from anuran | http://split4.pmfst.hr/dadp/ (accessed on 22 October 2021) | [573] |
| YADAMP | DSC | 2012 | AMP database of and predict AMPs | http://yadamp.unisa.it/about.aspx (accessed on 22 October 2021) | [574] |
| CPPpred | N-to-1 NN | 2013 | Predict cell penetrating peptides | http://bioware.ucd.ie/~compass/biowareweb/Server_pages/cpppred.php (accessed on 2 October 2020) | [575] |
| HIPdb | PepStr algorithm | 2013 | Database of experimentally validated anti-HIV Peptides | http://crdd.osdd.net/servers/hipdb/ (accessed on 2 October 2020) | [576] |
| ADAM | HMM, SVM | 2015 | Predict AMPs | http://bioinformatics.cs.ntou.edu.tw/ADAM/tool.html (accessed on 2 October 2020) | [577] |
| DBAASP | New algorithm DBSCAN | 2015 | Predict AMPs | https://dbaasp.org/home (accessed on 2 October 2020) | [516] |
| BaAMPs | - | 2015 | Database of anti-biofilm AMPs | http://www.baamps.it/ (accessed on 22 October 2021) | [578] |
| CAMPR3 | SVM, RF, ANN, DA | 2016 | Predict AMPs | http://www.camp3.bicnirrh.res.in/index.php (accessed on 2 October 2020) | [549] |
| APD3 | Peptide Parameter Space | 2016 | Predict AMPs | http://aps.unmc.edu/AP/main.php (accessed on 2 October 2020) | [234] |
| dPABBs | SVM, WEKA | 2016 | Predict and design anti-biofilm peptides | http://ab-openlab.csir.res.in/abp/antibiofilm/ (accessed on 2 October 2020) | [579] |
| MBPDB | - | 2017 | Database of bioactive peptides derived from milk protein | http://mbpdb.nws.oregonstate.edu/ (accessed on 22 October 2021) | [580] |
| RiPPMiner | SVM, WEKA | 2017 | Decipher RiPPs from amino acid sequence of precursor polypeptide. | http://www.nii.ac.in/rippminer.html (accessed on 22 October 2021) | [581] |
| iAMPpred | SVM | 2017 | Predict AMPs | http://cabgrid.res.in:8080/amppred/ (accessed on 22 October 2021) | [582] |
| InverPep | CALCAMP/in-house algorithm | 2017 | Database of experimentally validated AMPs from invertebrates | https://ciencias.medellin.unal.edu.co/gruposdeinvestigacion/prospeccionydisenobiomoleculas/InverPep/public/home_en (accessed on 22 October 2021) | [583] |
| BAGEL4 | HMM, Genomic context | 2018 | Predicts RiPPs and bacteriocins | http://bagel4.molgenrug.nl (accessed on 2 October 2020) | [584] |
| AMPscanner Vr.1 | RF & MARS | 2018 | Predict AMPs | https://www.dveltri.com/ascan/v1/index.html (accessed on 2 October 2020) | [585] |
| AMPscanner Vr.2 | DNN | 2018 | Predict AMPs | https://www.dveltri.com/ascan/v2/about.html (accessed on 2 October 2020) | [585] |
| dbAMP | RF/BLASTP | 2019 | Predict AMPs | http://140.138.77.240/~dbamp/index.php (accessed on 2 October 2020) | |
| ADAPT-ABLE | SR family generating, CF algorithm | 2019 | Data mining and AMP prediction | http://gec.u-picardie.fr/adaptable (accessed on 2 October 2020) | [515] |
| AMAP | SVM, XGBoost | 2019 | Predict biologically active peptides and AMPs. | http://amap.pythonanywhere.com/ (accessed on 2 October 2020) | [586] |
| AntiVPP1.0 | RF | 2019 | Predict antiviral peptides. | https://github.com/bio-coding/AntiVPP (accessed on 2 October 2020) | [587] |
| Meta-iAVP | k-NN, rpart, glm, RF, XGB, SVM | 2019 | Predict antiviral peptides. | http://codes.bio/meta-iavp/ (accessed on 2 October 2020) | [588] |
| mACPpred | SVM | 2019 | Predict anticancer peptides. | www.thegleelab.org/mACPpred (accessed on 2 October 2020) | [589] |
| Deep-AmPEP30 | CNN | 2020 | Predict AMPs ≤30 residues. | https://cbbio.cis.um.edu.mo/AxPEP (accessed on 2 October 2020) | [590] |
| AmpGram | RF | 2020 | Predict and design AMPs from proteomic data. | http://biongram.biotech.uni.wroc.pl/AmpGram/ (accessed on 2 October 2020) | [591] |
| IAMPE | NB, KNN, SVM, RF, and XGBoost | 2020 | Predict physicochemical and NMR features of peptides. | http://cbb1.ut.ac.ir/ (accessed on 2 October 2020) | [592] |
| CancerGram | n-grams and random forests | 2020 | Predict anticancer peptides. | http://biongram.biotech.uni.wroc.pl/CancerGram/ (accessed on 2 October 2020) | [593] |
| ACEP | DNN | 2020 | Predict AMPs | https://github.com/Fuhaoyi/ACEP (accessed on 2 October 2020) | [594] |
| AniAMPpred | SVM, DNN | 2021 | Identify the antimicrobial function of proteins | https://aniamppred.anvil.app/ (accessed on 22 October 2021) | [548] |
| DRAMP 3.0 | - | 2021 | Data repository of antimicrobial peptides and predict AMPs | http://dramp.cpu-bioinfor.org/ (accessed on 22 October 2021) | [595] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. https://doi.org/10.3390/ijms222111691
Bin Hafeez A, Jiang X, Bergen PJ, Zhu Y. Antimicrobial Peptides: An Update on Classifications and Databases. International Journal of Molecular Sciences. 2021; 22(21):11691. https://doi.org/10.3390/ijms222111691
Chicago/Turabian StyleBin Hafeez, Ahmer, Xukai Jiang, Phillip J. Bergen, and Yan Zhu. 2021. "Antimicrobial Peptides: An Update on Classifications and Databases" International Journal of Molecular Sciences 22, no. 21: 11691. https://doi.org/10.3390/ijms222111691
APA StyleBin Hafeez, A., Jiang, X., Bergen, P. J., & Zhu, Y. (2021). Antimicrobial Peptides: An Update on Classifications and Databases. International Journal of Molecular Sciences, 22(21), 11691. https://doi.org/10.3390/ijms222111691







