Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents
Abstract
1. Introduction
2. Factors Mediating Differential p53 Responses to DNA-Damaging Therapeutic Agents
2.1. Nuclear p53 Accumulation & Kinetics
2.2. Post-Translational Modifications of p53
2.3. p53 Binding Proteins, Cofactors, and Chromatin Remodeling
2.4. Models of p53 Target Gene Selection Regulation
3. Ongoing Efforts to Characterize Differential p53 Responses to DNA-Damaging Therapeutic Agents
3.1. Efforts to Establish a Context-Independent p53 Program
3.2. Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents
3.3. Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutics across Cell Type
3.4. Tissue Specificity in the p53 Response to DNA-Damaging Therapeutic Agents
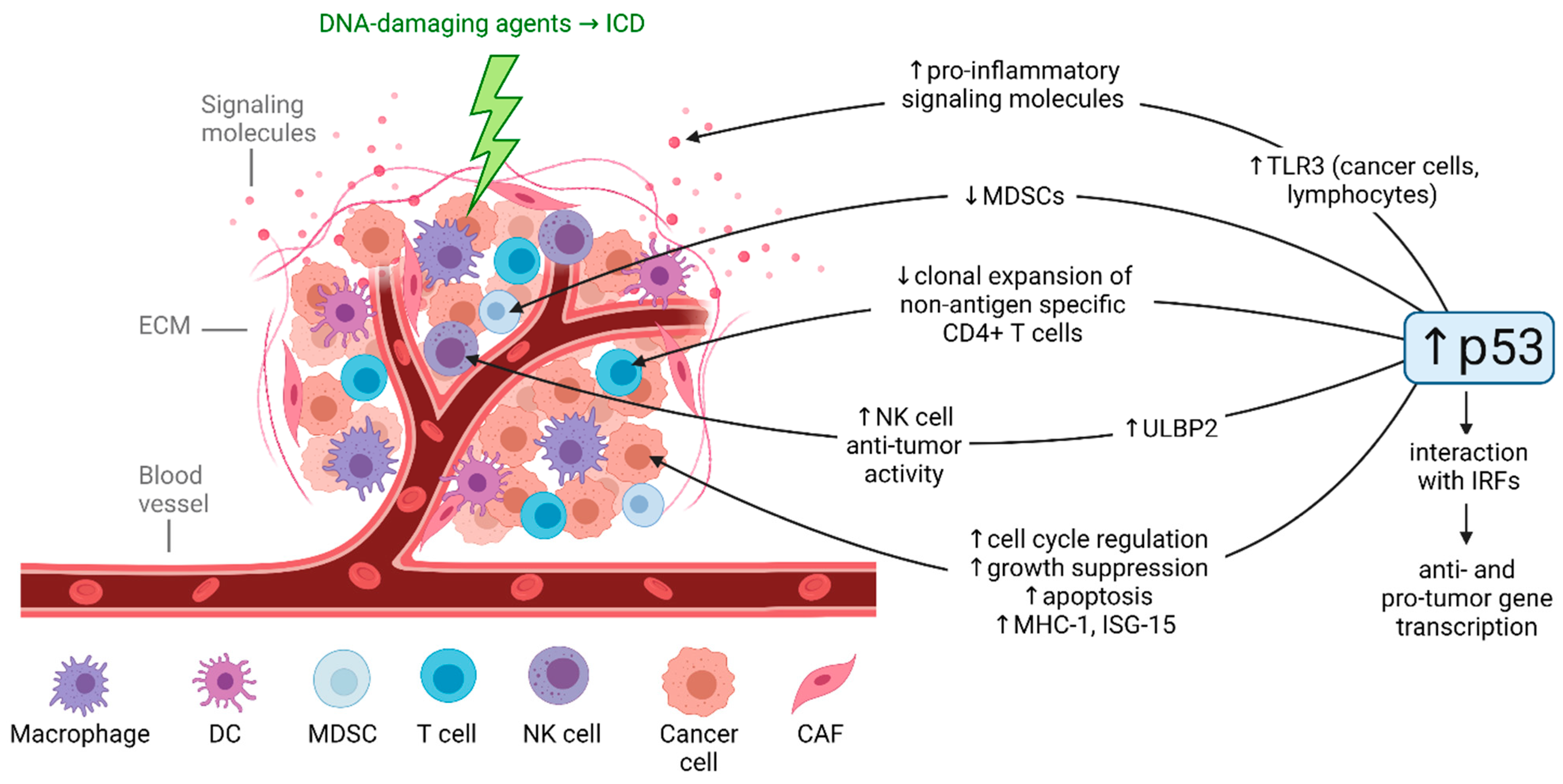
3.5. Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents in the Tumor Microenvironment
4. Conclusions and Open Questions
Funding
Acknowledgments
Conflicts of Interest
References
- Parrales, A.; Iwakuma, T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front. Oncol. 2015, 5, 288. [Google Scholar] [CrossRef] [PubMed]
- Borrero, L.J.H.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta (BBA) Bioenerg. 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Humpton, T.J.; Vousden, K.H. Regulation of Cellular Metabolism and Hypoxia by p53. Cold Spring Harb. Perspect. Med. 2016, 6, a026146. [Google Scholar] [CrossRef] [PubMed]
- Beckerman, R.; Prives, C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010, 2, a000935. [Google Scholar] [CrossRef]
- Giuriato, S.; Ryeom, S.; Fan, A.C.; Bachireddy, P.; Lynch, R.C.; Rioth, M.J.; van Riggelen, J.; Kopelman, A.M.; Passegue, E.; Tang, F.; et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc. Natl. Acad. Sci. USA 2006, 103, 16266–16271. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Ternovoi, V.V.; Curiel, D.T.; Smith, B.F.; Siegal, G.P. Adenovirus-mediated p53 tumor suppressor gene therapy of osteosarcoma. Lab. Investig. 2006, 86, 748–766. [Google Scholar] [CrossRef]
- Kaiser, A.M.; Attardi, L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018, 25, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Sammons, M.A.; Nguyen, T.-A.T.; McDade, S.S.; Fischer, M. Tumor suppressor p53: From engaging DNA to target gene regulation. Nucleic Acids Res. 2020, 48, 8848–8869. [Google Scholar] [CrossRef]
- Burns, T.F.; Bernhard, E.J.; El-Deiry, W.S. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 2001, 20, 4601–4612. [Google Scholar] [CrossRef]
- Burns, T.F.; El-Deiry, W.S. Microarray Analysis of p53 Target Gene Expression Patterns in the Spleen and Thymus in Response to Ionizing Radiation. Cancer Biol. Ther. 2003, 2, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Ornstein, J.; Lahav, G. p53 dynamics in response to DNA damage vary across cell lines and are shaped by efficiency of DNA repair and activity of the kinase ATM. Sci. Signal. 2017, 10, eaah6671. [Google Scholar] [CrossRef] [PubMed]
- Carlsen, L.; Schorl, C.; Huntington, K.; Hernandez-Borrero, L.; Jhaveri, A.; Zhang, S.; Zhou, L.; El-Deiry, W.S. Pan-drug and drug-specific mechanisms of 5-FU, irinotecan (CPT-11), oxaliplatin, and cisplatin identified by comparison of transcriptomic and cytokine responses of colorectal cancer cells. Oncotarget 2021, 12, 2006–2021. [Google Scholar] [CrossRef] [PubMed]
- Smeenk, L.; van Heeringen, S.; Koeppel, M.; Gilbert, B.; Janssen-Megens, E.; Stunnenberg, H.G.; Lohrum, M. Role of p53 Serine 46 in p53 Target Gene Regulation. PLoS ONE 2011, 6, e17574. [Google Scholar] [CrossRef]
- Botcheva, K.; McCorkle, S.R. Cell Context Dependent p53 Genome-Wide Binding Patterns and Enrichment at Repeats. PLoS ONE 2014, 9, e113492. [Google Scholar] [CrossRef]
- Kastan, M.B. Wild-Type p53: Tumors Can’t Stand It. Cell 2007, 128, 837–840. [Google Scholar] [CrossRef]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of p53 function leads to tumour regression in vivo. Nat. Cell Biol. 2007, 445, 661–665. [Google Scholar] [CrossRef]
- Shimada, H.; Matsubara, H.; Shiratori, T.; Shimizu, T.; Miyazaki, S.; Okazumi, S.; Nabeya, Y.; Shuto, K.; Hayashi, H.; Tanizawa, T.; et al. Phase I/II adenoviral p53 gene therapy for chemoradiation resistant advanced esophageal squamous cell carcinoma. Cancer Sci. 2006, 97, 554–561. [Google Scholar] [CrossRef]
- Xu, L.; Pirollo, K.F.; Tang, W.-H.; Rait, A.; Chang, E.H. Transferrin-Liposome-Mediated Systemic p53 Gene Therapy in Combination with Radiation Results in Regression of Human Head and Neck Cancer Xenografts. Hum. Gene Ther. 1999, 10, 2941–2952. [Google Scholar] [CrossRef]
- Bykov, V.N.; Issaeva, N.; Shilov, A.; Hultcrantz, M.; Pugacheva, E.; Chumakov, P.; Bergman, J.; Wiman, K.; Selivanova, G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 2002, 8, 282–288. [Google Scholar] [CrossRef]
- Burmakin, M.; Shi, Y.; Hedström, E.; Kogner, P.; Selivanova, G. Dual Targeting of Wild-Type and Mutant p53 by Small Molecule RITA Results in the Inhibition of N-Myc and Key Survival Oncogenes and Kills Neuroblastoma Cells In Vivo and In Vitro. Clin. Cancer Res. 2013, 19, 5092–5103. [Google Scholar] [CrossRef] [PubMed]
- Tchelebi, L.; Ashamalla, H.; Graves, P.R. Mutant p53 and the Response to Chemotherapy and Radiation. Subcell. Biochem. 2014, 85, 133–159. [Google Scholar] [CrossRef]
- Makhale, A.; Nanayakkara, D.; Raninga, P.; Khanna, K.; Kalimutho, M. CX-5461 Enhances the Efficacy of APR-246 via Induction of DNA Damage and Replication Stress in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2021, 22, 5782. [Google Scholar] [CrossRef]
- Maslah, N.; Salomao, N.; Drevon, L.; Verger, E.; Partouche, N.; Ly, P.; Aubin, P.; Naoui, N.; Schlageter, M.-H.; Bally, C.; et al. Synergistic effects of PRIMA-1Met (APR-246) and 5-azacitidine in TP53-mutated myelodysplastic syndromes and acute myeloid leukemia. Haematol 2019, 105, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Mohell, N.; Alfredsson, J.; Fransson, A.; Uustalu, M.; Bystrom, S.; Gullbo, J.; Hallberg, A.; Bykov, V.J.N.; Bjorklund, U.; Wiman, K. APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis. 2015, 6, e1794. [Google Scholar] [CrossRef] [PubMed]
- Saha, M.N.; Jiang, H.; Yang, Y.; Reece, N.; Chang, H. PRIMA-1Met/APR-246 Displays High Antitumor Activity in Multiple Myeloma By Induction of p73 and Noxa. Mol. Cancer Ther. 2013, 12, 2331–2341. [Google Scholar] [CrossRef] [PubMed]
- Pairawan, S.; Zhao, M.; Yuca, E.; Annis, A.; Evans, K.; Sutton, D.; Carvajal, L.; Ren, J.-G.; Santiago, S.; Guerlavais, V.; et al. First in class dual MDM2/MDMX inhibitor ALRN-6924 enhances antitumor efficacy of chemotherapy in TP53 wild-type hormone receptor-positive breast cancer models. Breast Cancer Res. 2021, 23, 29. [Google Scholar] [CrossRef]
- Jiang, L.; Zawacka-Pankau, J. The p53/MDM2/MDMX-targeted therapies-a clinical synopsis. Cell Death Dis. 2020, 11, 237. [Google Scholar] [CrossRef]
- Yogosawa, S.; Yoshida, K. Tumor suppressive role for kinases phosphorylating p53 in DNA damage-induced apoptosis. Cancer Sci. 2018, 109, 3376–3382. [Google Scholar] [CrossRef]
- Reed, S.M.; Quelle, D.E. p53 Acetylation: Regulation and Consequences. Cancers 2014, 7, 30–69. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- Bourgeois, B.; Madl, T. Regulation of cellular senescence via the FOXO4-p53 axis. FEBS Lett. 2018, 592, 2083–2097. [Google Scholar] [CrossRef] [PubMed]
- Andrysik, Z.; Kim, J.; Tan, A.C.; Espinosa, J.M. A Genetic Screen Identifies TCF3/E2A and TRIAP1 as Pathway-Specific Regulators of the Cellular Response to p53 Activation. Cell Rep. 2013, 3, 1346–1354. [Google Scholar] [CrossRef]
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-J.; Li, D.; Ou, Y.; Jiang, L.; Chen, Y.; Zhao, Y.; Gu, W. Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression. Cell Rep. 2016, 17, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.L.; Veprintsev, D.; Bycroft, M.; Fersht, A.R. Comparative Binding of p53 to its Promoter and DNA Recognition Elements. J. Mol. Biol. 2005, 348, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Morachis, J.M.; Murawsky, C.M.; Emerson, B.M. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010, 24, 135–147. [Google Scholar] [CrossRef]
- Gomes, N.P.; Espinosa, J.M. Disparate chromatin landscapes and kinetics of inactivation impact differential regulation of p53 target genes. Cell Cycle 2010, 9, 3428–3437. [Google Scholar] [CrossRef][Green Version]
- Espinosa, J.M.; Verdun, R.E.; Emerson, B.M. p53 Functions through Stress- and Promoter-Specific Recruitment of Transcription Initiation Components before and after DNA Damage. Mol. Cell 2003, 12, 1015–1027. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Speidel, D. The role of DNA damage responses in p53 biology. Arch. Toxicol. 2015, 89, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Meek, D.W.; Anderson, C.W. Posttranslational Modification of p53: Cooperative Integrators of Function. Cold Spring Harb. Perspect. Biol. 2009, 1, a000950. [Google Scholar] [CrossRef] [PubMed]
- Bruins, W.; Zwart, E.; Attardi, L.D.; Iwakuma, T.; Hoogervorst, E.M.; Beems, R.B.; Miranda, B.; van Oostrom, C.T.M.; Berg, J.V.D.; Aardweg, G.J.V.D.; et al. Increased Sensitivity to UV Radiation in Mice with a p53 Point Mutation at Ser389. Mol. Cell. Biol. 2004, 24, 8884–8894. [Google Scholar] [CrossRef]
- Huang, C.; Ma, W.-Y.; Maxiner, A.; Sun, Y.; Dong, Z. p38 Kinase Mediates UV-induced Phosphorylation of p53 Protein at Serine 389. J. Biol. Chem. 1999, 274, 12229–12235. [Google Scholar] [CrossRef]
- Hofmann, T.G.; Möller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Dröge, W.; Will, H.; Schmitz, L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2001, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Verfaillie, A.; Svetlichnyy, D.; Imrichová, H.; Davie, K.; Fiers, M.; Atak, Z.K.; Hulselmans, G.; Christiaens, V.; Aerts, S. Multiplex enhancer-reporter assays uncover unsophisticated TP53 enhancer logic. Genome Res. 2016, 26, 882–895. [Google Scholar] [CrossRef]
- Fischer, M. Census and evaluation of p53 target genes. Oncogene 2017, 36, 3943–3956. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Ohkubo, S.; Tatsuno, I.; Prives, C. hCAS/CSE1L Associates with Chromatin and Regulates Expression of Select p53 Target Genes. Cell 2007, 130, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Itahana, Y.; Zhang, J.; Göke, J.; Vardy, L.A.; Han, R.; Iwamoto, K.; Cukuroglu, E.; Robson, P.; Pouladi, M.A.; Colman, A.; et al. Histone modifications and p53 binding poise the p21 promoter for activation in human embryonic stem cells. Sci. Rep. 2016, 6, 28112. [Google Scholar] [CrossRef]
- Zhang, X.-P.; Liu, F.; Cheng, Z.; Wang, W. Cell fate decision mediated by p53 pulses. Proc. Natl. Acad. Sci. USA 2009, 106, 12245–12250. [Google Scholar] [CrossRef] [PubMed]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Grossmann, P.; Padi, M.; DeCaprio, J.A. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016, 44, 6070–6086. [Google Scholar] [CrossRef]
- Muresan, X.; Bouchal, J.; Culig, Z.; Souček, K. Toll-Like Receptor 3 in Solid Cancer and Therapy Resistance. Cancers 2020, 12, 3227. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Wang, S.-J.; Li, D.; Chu, B.; Gu, W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, 6806–6812. [Google Scholar] [CrossRef]
- Yuniati, L.; Scheijen, B.; van der Meer, L.T.; van Leeuwen, F.N. Tumor suppressors BTG1 and BTG2: Beyond growth control. J. Cell. Physiol. 2019, 234, 5379–5389. [Google Scholar] [CrossRef]
- Nikulenkov, F.; Spinnler, C.; Li, H.; Tonelli, C.; Shi, Y.; Turunen, M.; Kivioja, T.; Ignatiev, I.; Kel, A.; Taipale, J.; et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012, 19, 1992–2002. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Fei, P.; Bernhard, E.J.; El-Deiry, W.S. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002, 62, 7316–7327. [Google Scholar]
- Moyer, S.M.; Wasylishen, A.R.; Qi, Y.; Fowlkes, N.; Su, X.; Lozano, G. p53 drives a transcriptional program that elicits a non-cell-autonomous response and alters cell state in vivo. Proc. Natl. Acad. Sci. USA 2020, 117, 23663–23673. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Ornstein, J.; Iwamoto, Y.; Miller, M.A.; Prytyskach, M.A.; Ferretti, S.; Holzer, P.; Kallen, J.; Furet, P.; Jambhekar, A.; Forrester, W.C.; et al. p53 dynamics vary between tissues and are linked with radiation sensitivity. Nat. Commun. 2021, 12, 898. [Google Scholar] [CrossRef]
- Hafner, A.; Kublo, L.; Tsabar, M.; Lahav, G.; Stewart-Ornstein, J. Identification of universal and cell-type specific p53 DNA binding. BMC Mol. Cell Biol. 2020, 21, 5. [Google Scholar] [CrossRef]
- Cui, Y.; Guo, G. Immunomodulatory Function of the Tumor Suppressor p53 in Host Immune Response and the Tumor Microenvironment. Int. J. Mol. Sci. 2016, 17, 1942. [Google Scholar] [CrossRef]
- Khalaf, K.; Hana, D.; Chou, J.T.-T.; Singh, C.; Mackiewicz, A.; Kaczmarek, M. Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance. Front. Immunol. 2021, 12, 656364. [Google Scholar] [CrossRef]
- Guo, G.; Cui, Y. New perspective on targeting the tumor suppressor p53 pathway in the tumor microenvironment to enhance the efficacy of immunotherapy. J. Immunother. Cancer 2015, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, B.L.; Anderson, R. Realizing the Clinical Potential of Immunogenic Cell Death in Cancer Chemotherapy and Radiotherapy. Int. J. Mol. Sci. 2019, 20, 959. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, Z.; Safarzadeh, E.; Safaei, S.; Baradaran, A.; Mohammadi, A.; Hajiasgharzadeh, K.; Derakhshani, A.; Argentiero, A.; Silvestris, N.; Baradaran, B. Current Approaches for Combination Therapy of Cancer: The Role of Immunogenic Cell Death. Cancers 2020, 12, 1047. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Fletcher, R.; Yu, J.; Zhang, L. Immunogenic effects of chemotherapy-induced tumor cell death. Genes Dis. 2018, 5, 194–203. [Google Scholar] [CrossRef]
- Fang, D.D.; Tang, Q.; Kong, Y.; Wang, Q.; Gu, J.; Fang, X.; Zou, P.; Rong, T.; Wang, J.; Yang, D.; et al. MDM2 inhibitor APG-115 synergizes with PD-1 blockade through enhancing antitumor immunity in the tumor microenvironment. J. Immunother. Cancer 2019, 7, 7. [Google Scholar] [CrossRef]
- Guo, G.; Yu, M.; Xiao, W.; Celis, E.; Cui, Y. Local Activation of p53 in the Tumor Microenvironment Overcomes Immune Suppression and Enhances Antitumor Immunity. Cancer Res. 2017, 77, 2292–2305. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Goyama, S.; Liu, X.; Tamura, M.; Asada, S.; Tanaka, Y.; Fukuyama, T.; Wunderlich, M.; O’Brien, E.; Mizukawa, B.; et al. Antitumor immunity augments the therapeutic effects of p53 activation on acute myeloid leukemia. Nat. Commun. 2019, 10, 4869. [Google Scholar] [CrossRef]
- Bi, X.; Hameed, M.; Mirani, N.; Pimenta, E.M.; Anari, J.; Barnes, B.J. Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis. Breast Cancer Res. 2011, 13, R111. [Google Scholar] [CrossRef]
- Fresquet, V.; Robles, E.F.; Parker, A.; Martinez-Useros, J.; Mena, M.; Malumbres, R.; Agirre, X.; Catarino, S.; Arteta, D.; Osaba, L.; et al. High-throughput sequencing analysis of the chromosome 7q32 deletion reveals IRF5 as a potential tumour suppressor in splenic marginal-zone lymphoma. Br. J. Haematol. 2012, 158, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Wang, Y.; Yang, J.; Stark, G.R. IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression. Proc. Natl. Acad. Sci. USA 2018, 115, 3906–3911. [Google Scholar] [CrossRef]
- Menendez, D.; Lowe, J.M.; Snipe, J.; Resnick, M.A. Ligand dependent restoration of human TLR3 signaling and death in p53 mutant cells. Oncotarget 2016, 7, 61630–61642. [Google Scholar] [CrossRef] [PubMed]
- Shatz, M.; Menendez, D.; Resnick, M. The Human TLR Innate Immune Gene Family Is Differentially Influenced by DNA Stress and p53 Status in Cancer Cells. Cancer Res. 2012, 72, 3948–3957. [Google Scholar] [CrossRef]
- Taura, M.; Eguma, A.; Suico, M.A.; Shuto, T.; Koga, T.; Komatsu, K.; Komune, T.; Sato, T.; Saya, H.; Li, J.-D.; et al. p53 Regulates Toll-Like Receptor 3 Expression and Function in Human Epithelial Cell Lines. Mol. Cell. Biol. 2008, 28, 6557–6567. [Google Scholar] [CrossRef]
- Anwar, M.A.; Shah, M.; Kim, J.; Choi, S. Recent clinical trends in Toll-like receptor targeting therapeutics. Med. Res. Rev. 2019, 39, 1053–1090. [Google Scholar] [CrossRef]
- Le Naour, J.; Galluzzi, L.; Zitvogel, L.; Kroemer, G.; Vacchelli, E. Trial watch: TLR3 agonists in cancer therapy. OncoImmunology 2020, 9, 1771143. [Google Scholar] [CrossRef] [PubMed]
- Cen, X.; Liu, S.; Cheng, K. The Role of Toll-Like Receptor in Inflammation and Tumor Immunity. Front. Pharmacol. 2018, 9, 878. [Google Scholar] [CrossRef]
- Wang, B.; Niu, D.; Lai, L.; Ren, E.C. p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat. Commun. 2013, 4, 2359. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Yang, S.W.; Park, J.M.; Ka, S.H.; Kim, J.H.; Kong, Y.Y.; Jeon, Y.J.; Seol, J.H.; Chung, C.H. Positive feedback regulation of p53 transactivity by DNA damage-induced ISG15 modification. Nat. Commun. 2016, 7, 12513. [Google Scholar] [CrossRef]
- Gasser, S.; Orsulic, S.; Brown, E.J.; Raulet, D.H. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nat. Cell Biol. 2005, 436, 1186–1190. [Google Scholar] [CrossRef]
- Iannello, A.; Thompson, T.W.; Ardolino, M.; Lowe, S.W.; Raulet, D.H. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J. Exp. Med. 2013, 210, 2057–2069. [Google Scholar] [CrossRef]
- Li, H.; Lakshmikanth, T.; Garofalo, C.; Spinnler, C.; Enge, M.; Anichini, A.; Szekely, L.; Karre, K.; Carbone, E.; Selivanova, G. Pharmacological activation of p53 triggers anticancer innate immune response through induction of ULBP2. Cell Cycle 2011, 10, 3346–3358. [Google Scholar] [CrossRef]
- Textor, S.; Fiegler, N.; Arnold, A.; Porgador, A.; Hofmann, T.G.; Cerwenka, A. Human NK Cells Are Alerted to Induction of p53 in Cancer Cells by Upregulation of the NKG2D Ligands ULBP1 and ULBP2. Cancer Res. 2011, 71, 5998–6009. [Google Scholar] [CrossRef]
- Watanabe, M.; Moon, K.D.; Vacchio, M.S.; Hathcock, K.S.; Hodes, R.J. Downmodulation of Tumor Suppressor p53 by T Cell Receptor Signaling Is Critical for Antigen-Specific CD4+ T Cell Responses. Immunity 2014, 40, 681–691. [Google Scholar] [CrossRef]
- Kelly, R.M.; Goren, E.M.; Taylor, P.A.; Mueller, S.N.; Stefanski, H.E.; Osborn, M.J.; Scott, H.S.; Komarova, E.A.; Gudkov, A.V.; Holländer, G.A.; et al. Short-term inhibition of p53 combined with keratinocyte growth factor improves thymic epithelial cell recovery and enhances T-cell reconstitution after murine bone marrow transplantation. Blood 2010, 115, 1088–1097. [Google Scholar] [CrossRef]
- Donehower, L.A.; Soussi, T.; Korkut, A.; Liu, Y.; Schultz, A.; Cardenas, M.; Li, X.; Babur, O.; Hsu, T.K.; Lichtarge, O.; et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep. 2019, 28, 1370–1384.e1375. [Google Scholar] [CrossRef] [PubMed]
- Mello, S.S.; Attardi, L.D. Deciphering p53 signaling in tumor suppression. Curr. Opin. Cell Biol. 2018, 51, 65–72. [Google Scholar] [CrossRef]
- Athar, M.; Elmets, C.A.; Kopelovich, L. Pharmacological Activation of p53 in Cancer Cells. Curr. Pharm. Des. 2011, 17, 631–639. [Google Scholar] [CrossRef]
- Ho, T.; Tan, B.X.; Lane, D. How the Other Half Lives: What p53 Does When It Is Not Being a Transcription Factor. Int. J. Mol. Sci. 2019, 21, 13. [Google Scholar] [CrossRef]
- Williams, A.B.; Schumacher, B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef]
- Inoue, K.; Fry, E.A. Alterations of p63 and p73 in human cancers. Subcell. Biochem. 2014, 85, 17–40. [Google Scholar] [CrossRef]
- Nguyen, B.-C.; Lefort, K.; Mandinova, A.; Antonini, D.; Devgan, V.; Della Gatta, G.; Koster, M.I.; Zhang, Z.; Wang, J.; di Vignano, A.T.; et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006, 20, 1028–1042. [Google Scholar] [CrossRef]
- Carroll, D.K.; Carroll, J.; Leong, C.-O.; Cheng, F.; Brown, M.; Mills, A.A.; Brugge, J.S.; Ellisen, L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006, 8, 551–561. [Google Scholar] [CrossRef]
- Cam, H.; Griesmann, H.; Beitzinger, M.; Hofmann, L.; Beinoraviciute-Kellner, R.; Sauer, M.; Hüttinger-Kirchhof, N.; Oswald, C.; Friedl, P.; Gattenlöhner, S.; et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell 2006, 10, 281–293. [Google Scholar] [CrossRef]
- Deyoung, M.P.; Ellisen, L.W. p63 and p73 in human cancer: Defining the network. Oncogene 2007, 26, 5169–5183. [Google Scholar] [CrossRef] [PubMed]
- Fricker, M.; Papadia, S.; Hardingham, G.E.; Tolkovsky, A.M. Implication of TAp73 in the p53-independent pathway of Puma induction and Puma-dependent apoptosis in primary cortical neurons. J. Neurochem. 2010, 114, 772–783. [Google Scholar] [CrossRef] [PubMed]
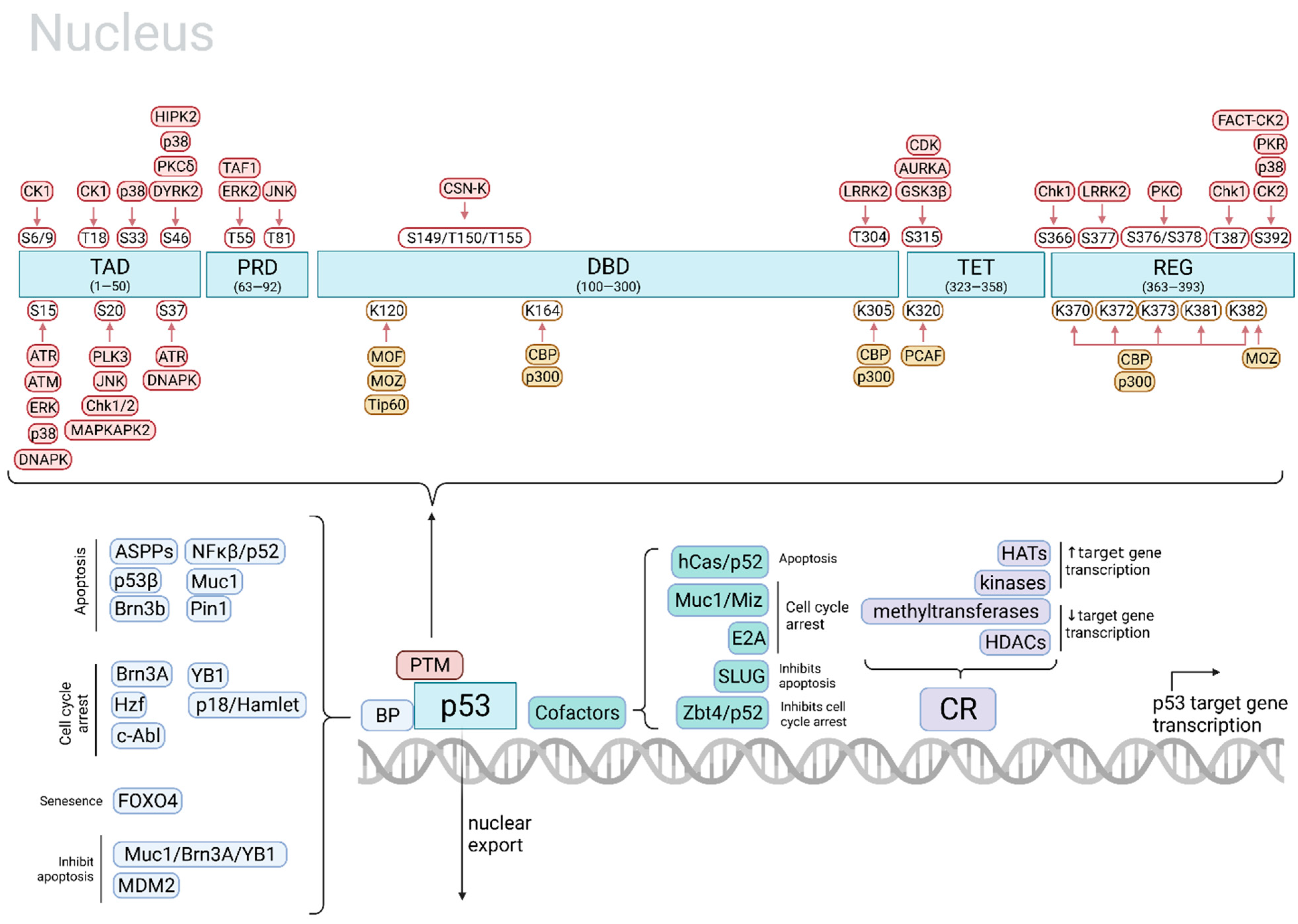
| Main Cell Fate | p53 Site and PTM | p53 Modifier | Stimulus | Main Target Gene(s) | Ref. |
|---|---|---|---|---|---|
| Apoptosis | phospho-S15 | ERKs | UV light | [31] | |
| phospho-S15 | P38 | UV light | [31] | ||
| phospho-S15, -S37 | ATR | γ-radiation, UV light | [31] | ||
| phospho-S20 | JNK | UV light | [31] | ||
| phospho-S20 | MAPKAP2 | UV light | [31] | ||
| phospho-S46 | HIPK2 | UV light | e.g., ↑ AIP1 | [31,32,35] | |
| phospho-S15 | ATM | DNA damage | [31] | ||
| acetyl-K120, -C-terminal (concurrent phospho-S46 needed) | Tip60, MOF, p300/CBP, PCAF | DNA damage, other genotoxic stresses | ↑ Bax, Fas, Noxa and Puma | [30] | |
| acetyl-K120 | hMOF, Tip60 | e.g., ↑ Puma | [32,35] | ||
| Stabilization; apoptosis | phospho-S33, -S46 | p38 | UV light | [31] | |
| Apoptosis, cell cycle arrest | phospho-S46 | HIPK2 | UV light | [31] | |
| Cell cycle arrest, apoptosis | acetyl-K164 | p300, CBP | Likely important for the activation of the majority of p53 target genes | [32,35] | |
| Cell cycle arrest, DNA repair | acetyl-C-terminal (concurrent phospho-N-terminal needed) | p300/CBP, PCAF; binding by Tip60 w/o acetylation | DNA damage, other genotoxic stresses | ↑ p21, GADD45 ↓ Noxa, Pidd | [30] |
| Cell cycle arrest, promotes cell survival | acetyl-K320 | PCAF | ↑ p21 | [32,35] | |
| Senescence | acetyl-K120, K320, K382 (concurrent phospho-S15, -S20 needed) | MOZ, PCAF, p300 | DNA damage, oncogene activation | ↑ p21 | [30] |
| Ferroptosis | acetyl-K101 | CBP | [35,36] | ||
| ↑ p53 transcription | phospho-S315 | CDK (CDC2/CDK2) | UV light | [31] | |
| ↑ p53 activity | phospho-S392 | FACT-CK2 | UV light | [31] | |
| phospho-T55 | ERK2 | Doxorubicin | [31] | ||
| Stabilization | phospho-T81 | JNK | DNA damage | [31] | |
| phospho-S6, -S9, -T18 (concurrent phospho-S15 needed) | CK1 | Topoisomerase-directed drugs and DNA damage | ↓ MDM2 | [31] | |
| phospho-S20 | Chk1/2 | Ionizing radiation | [31] | ||
| phospho-S15, -S37 | DNAPK | DNA damage | [31] | ||
| ↑ DNA-binding activity of p53 | phospho-S392 | P38 | UV light, DNA damage | [31] | |
| Ubiquitination and degradation; ↑DNA-binding affinity | phospho-S376, -S378 | PKC | Unstressed state; constitutively phosphorylated and dephosphorylated with IR light | [31] | |
| Degradation | phospho-T150, -T155, -S149 | CSN-associated kinase complex | Unstressed state | [31] | |
| Degradation or stabilization of p53 | phospho-T55 | TAF1 | Constitutively phosphorylated | [31] | |
| ↓ p53-mediated apoptosis | phospho-S315, S376 | GSK3β | Endoplasmic reticulum stress | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlsen, L.; El-Deiry, W.S. Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents. Int. J. Mol. Sci. 2021, 22, 11828. https://doi.org/10.3390/ijms222111828
Carlsen L, El-Deiry WS. Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents. International Journal of Molecular Sciences. 2021; 22(21):11828. https://doi.org/10.3390/ijms222111828
Chicago/Turabian StyleCarlsen, Lindsey, and Wafik S. El-Deiry. 2021. "Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents" International Journal of Molecular Sciences 22, no. 21: 11828. https://doi.org/10.3390/ijms222111828
APA StyleCarlsen, L., & El-Deiry, W. S. (2021). Differential p53-Mediated Cellular Responses to DNA-Damaging Therapeutic Agents. International Journal of Molecular Sciences, 22(21), 11828. https://doi.org/10.3390/ijms222111828







