Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L.
Abstract
:1. Introduction
2. Results
2.1. Identification of ARF in Sweet Cherry
2.2. Phylogenetic Analysis of PavARF
2.3. Gene Structure and Motif Analysis
2.4. Analysis of Hormone-Related Cis-Elements
2.5. Genomic Distribution and Gene Duplication
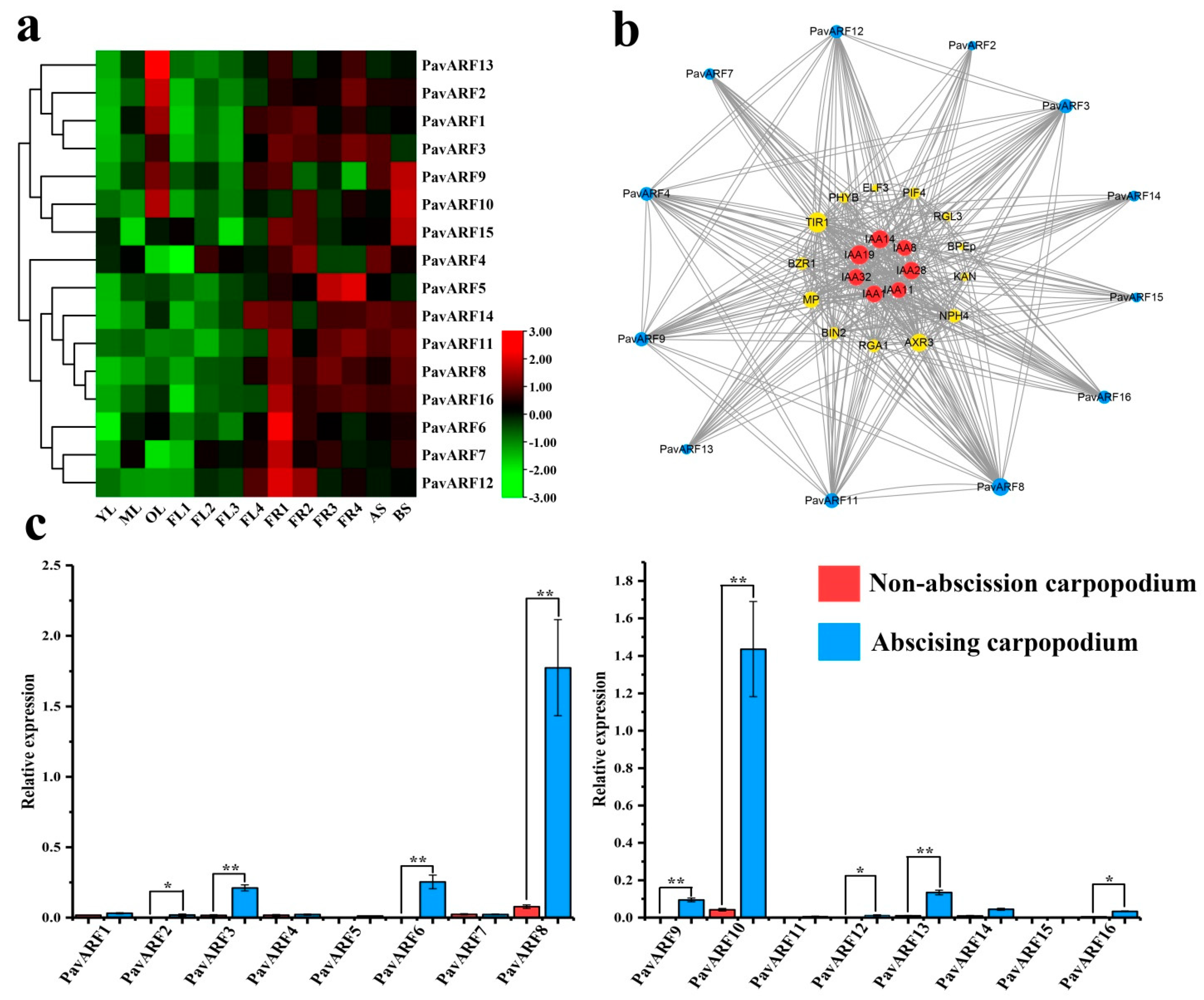
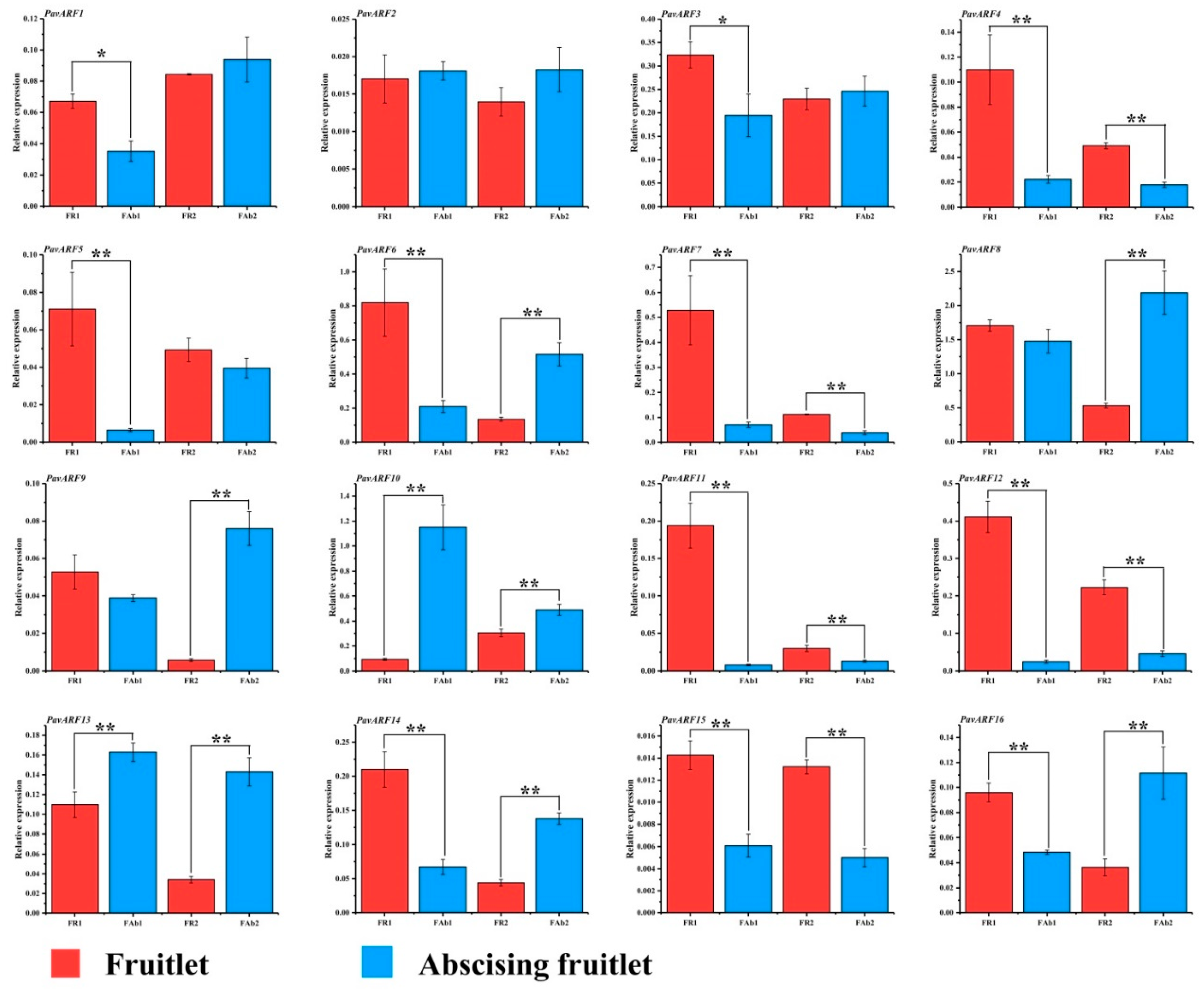
2.6. Expression Analysis of PavARF Genes in Stage-Specific and Tissue/Organ-Specific
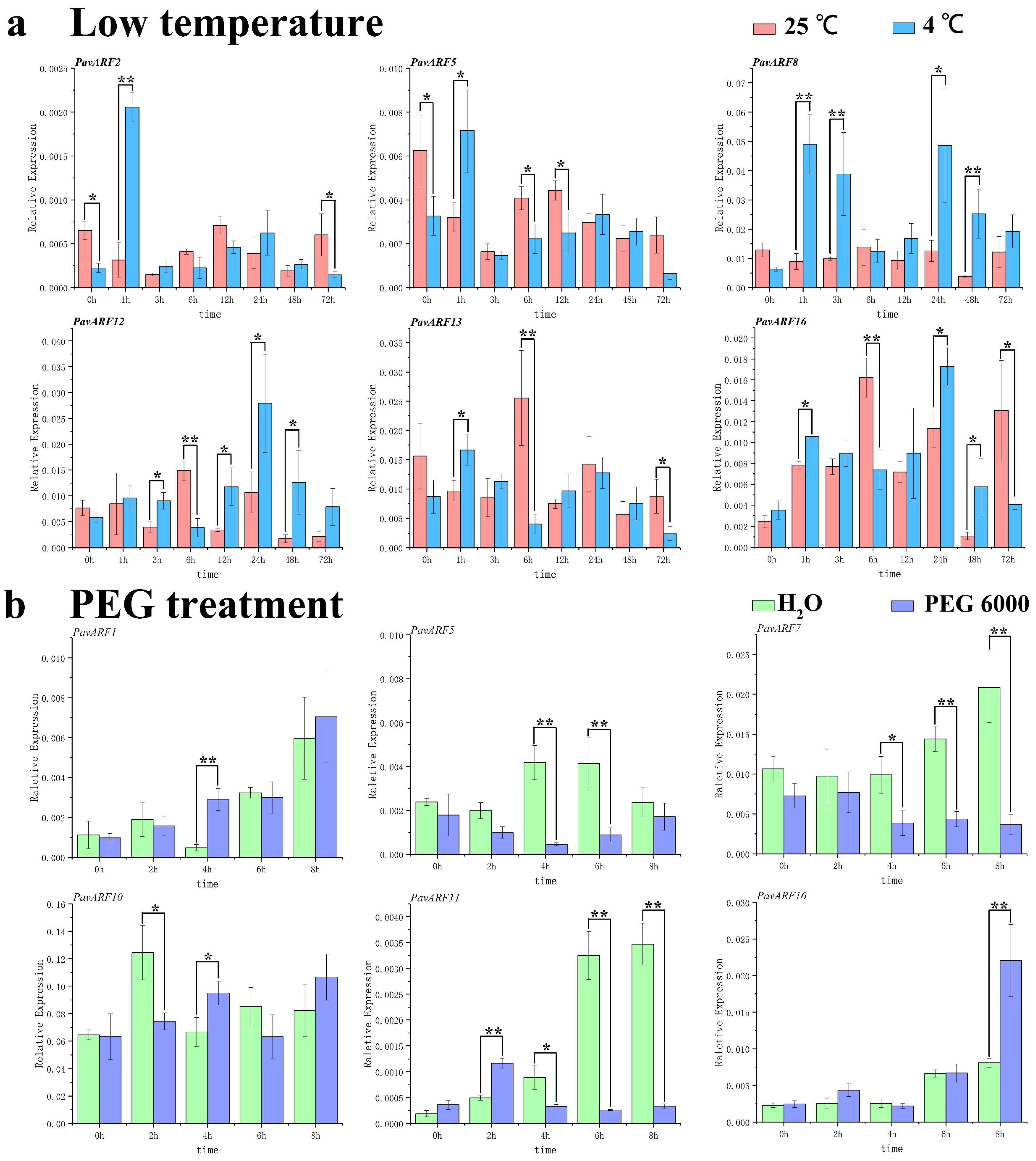
2.7. Expression of Low-Temperature and Drought Stress
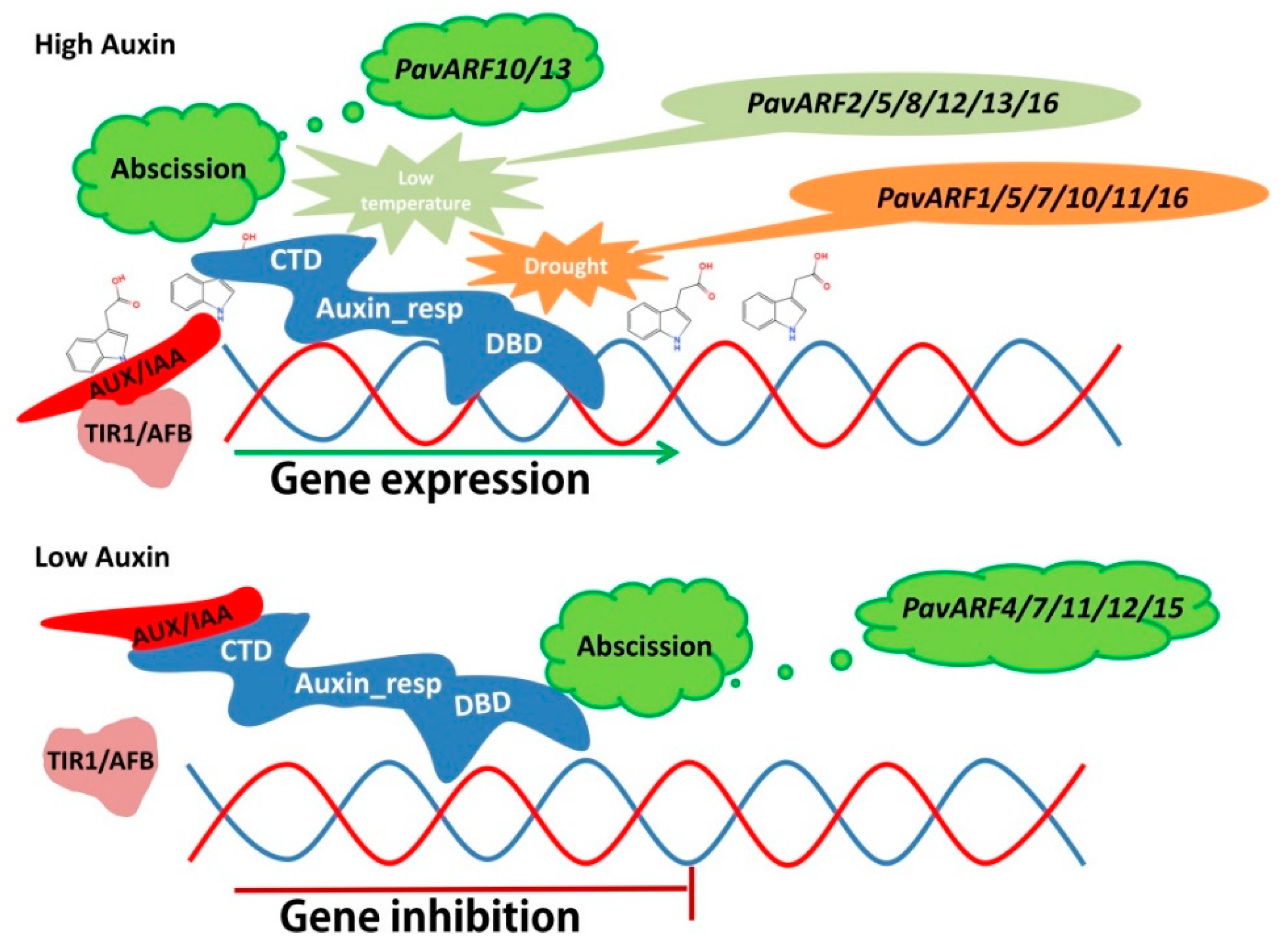
3. Discussion
4. Materials and Methods
4.1. Identification of ARF Genes in Cherry
4.2. Multiple Sequence Alignment and Phylogenetic Analyses
4.3. Sequence and Cis-Acting Element Analysis
4.4. Chromosomal Distribution and Gene Duplication of ARF Genes
4.5. Plant Materials
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Casanova-Sáez, R.; Voß, U. Auxin Metabolism Controls Developmental Decisions in Land Plants. Trends Plant Sci. 2019, 24, 741–754. [Google Scholar] [CrossRef]
- Peer, W.A. From Perception to Attenuation: Auxin Signalling and Responses. Curr. Opin. Plant Biol. 2013, 16, 561–568. [Google Scholar] [CrossRef]
- Hagen, G. Auxin Signal Transduction. Essays Biochem. 2015, 58, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of Auxin Signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [Green Version]
- Guilfoyle, T.J.; Hagen, G. Auxin Response Factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. Auxin Response Factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [Green Version]
- Korasick, D.A.; Jez, J.M.; Strader, L.C. Refining the Nuclear Auxin Response Pathway through Structural Biology. Curr. Opin. Plant Biol. 2015, 27, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Boer, D.R.; Freire-Rios, A.; Van Den Berg, W.A.M.; Saaki, T.; Manfield, I.W.; Kepinski, S.; López-Vidrieo, I.; Franco-Zorrilla, J.M.; De Vries, S.C.; Solano, R.; et al. Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: Output Control in Auxin Biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Remington, D.L.; Vision, T.J.; Guilfoyle, T.J.; Reed, J.W. Contrasting Modes of Diversification in the Aux/IAA and ARF Gene Families. Plant Physiol. 2004, 135, 1738–1752. [Google Scholar] [CrossRef] [Green Version]
- Orosa-Puente, B.; Leftley, N.; von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root Branching toward Water Involves Posttranslational Modification of Transcription Factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) Are Potential Mediators of Auxin Action in Tomato Response to Biotic and Abiotic Stress (Solanum lycopersicum). PLoS ONE 2018, 13, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Baranwal, V.K.; Negi, N.; Khurana, P. Auxin Response Factor Genes Repertoire in Mulberry: Identification, and Structural, Functional and Evolutionary Analyses. Genes 2017, 8, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Wang, W.; Wu, Y.; Xie, H.; Zhao, L.; Zeng, Q.; Zhan, Y. Expression and Distribution of the Auxin Response Factors in Sorghum Bicolor during Development and Temperature Stress. Int. J. Mol. Sci. 2019, 20, 4816. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Liu, H.; Xiong, L. Endogenous Auxin and Jasmonic Acid Levels Are Differentially Modulated by Abiotic Stresses in Rice. Front. Plant Sci. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, D.; Hu, X.; Guan, D.; Wang, W.; Yang, H.; Liu, Y. Genome-Wide Identification of the ARF (Auxin Response Factor) Gene Family in Peach and Their Expression Analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-Wide Identification and Expression Profiling of Auxin Response Factor (ARF) Gene Family in Maize. BMC Genom. 2011, 12, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-Wide Analysis of the Auxin Response Factors (ARF) Gene Family in Rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Wen, J.; Guo, P.; Ke, Y.; Liu, M.; Li, P.; Wu, Y.; Ran, F.; Wang, M.; Li, J.; Du, H. The Auxin Response Factor Gene Family in Allopolyploid Brassica Napus. PLoS ONE 2019, 14, 1–24. [Google Scholar] [CrossRef]
- Singh, V.K.; Jain, M. Genome-Wide Survey and Comprehensive Expression Profiling of Aux/IAA Gene Family in Chickpea and Soybean. Front. Plant Sci. 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wang, D.; Liu, S.; Fang, Z.; Su, S.; Guo, C.; Zhao, C.; Tang, Y. Comprehensive Atlas of Wheat (Triticum aestivum L.) Auxin Response Factor Expression During Male Reproductive Development and Abiotic Stress. Front. Plant Sci. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A Sweetpotato Auxin Response Factor Gene (IbARF5) Is Involved in Carotenoid Biosynthesis and Salt and Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, Z.; Chen, C.; Li, C.; Xia, R.; Li, J. Genome-Wide Characterization of the Auxin Response Factor (ARF) Gene Family of Litchi (Litchi Chinensis Sonn.): Phylogenetic Analysis, miRNA Regulation and Expression Changes during Fruit Abscission. PeerJ 2019, 7, e6677. [Google Scholar] [CrossRef] [Green Version]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The Genome Sequence of Sweet Cherry (Prunus avium) for Use in Genomics-Assisted Breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Baruah, P.M.; Krishnatreya, D.B.; Bordoloi, K.S.; Gill, S.S.; Agarwala, N. Genome Wide Identification and Characterization of Abiotic Stress Responsive LncRNAs in Capsicum annuum. Plant Physiol. Biochem. 2021, 162, 221–236. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Zhou, W.; Xie, L.; Wang, L.; Zhang, Y.; Zhang, Q. Genome-Wide Identification and Expression Analysis of the AT-Hook Motif Nuclear Localized Gene Family in Soybean. BMC Genom. 2021, 22, 1–26. [Google Scholar]
- Tong, T.; Fang, Y.X.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-Wide Identification and Expression Pattern Analysis of the KCS Gene Family in Barley. Plant Growth Regul. 2021, 93, 89–103. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-Scale Genome Assembly of Sweet Cherry (Prunus avium L.) Cv. Tieton Obtained Using Long-Read and Hi-C Sequencing. Hortic. Res. 2020, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.-C.; Sun, M.-H.; Xu, R.-R.; Shu, H.-R.; Wang, J.-W.; Zhang, S.-Z. Genomewide Identification and Expression Analysis of the ARF Gene Family in Apple. J. Genet. 2014, 93, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Krogan, N.T.; Marcos, D.; Weiner, A.I.; Berleth, T. The Auxin Response Factor MONOPTEROS Controls Meristem Function and Organogenesis in Both the Shoot and Root through the Direct Regulation of PIN Genes. New Phytol. 2016, 212, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, S.; Sun, C.; Xu, Y.; Chen, Y.; Yu, C.; Qian, Q.; Jiang, D.A.; Qi, Y. Auxin Response Factor (OsARF12), a Novel Regulator for Phosphate Homeostasis in Rice (Oryza Sativa). New Phytol. 2014, 201, 91–103. [Google Scholar] [CrossRef]
- Herud, O.; Weijers, D.; Lau, S.; Jürgens, G. Auxin Responsiveness of the Monopteros-Bodenlos Module in Primary Root Initiation Critically Depends on the Nuclear Import Kinetics of the Aux/IAA Inhibitor Bodenlos. Plant J. 2016, 85, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional Genomic Analysis of the Auxin Response Factor Gene Family Members in Arabidopsis thaliana: Unique and Overlapping Functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaul, S.; Koo, H.L.; Jenkins, J.; Rizzo, M.; Rooney, T.; Tallon, L.J.; Feldblyum, T.; Nierman, W.; Benito, M.I.; Lin, X.; et al. Analysis of the Genome Sequence of the Flowering Plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza Sativa Nipponbare Reference Genome Using next Generation Sequence and Optical Map Data. Rice 2013, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The Tomato Genome Sequence Provides Insights into Fleshy Fruit Evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Erratum: Genome Sequence of the Palaeopolyploid Soybean (Nature (2010) 463 (178-183)). Nature 2010, 465, 120. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The Draft Genome of Sweet Orange (Citrus sinensis). Nature Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Van De Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-Quality de Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nature Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The High-Quality Draft Genome of Peach (Prunus persica) Identifies Unique Patterns of Genetic Diversity, Domestication and Genome Evolution. Nature Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, F.; Cheng, L.; Kong, F.; Peng, Z.; Liu, S.; Yu, X.; Lu, G. Identification, Isolation and Expression Analysis of Auxin Response Factor (ARF) Genes in Solanum lycopersicum. Plant Cell Rep. 2011, 30, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Van Ha, C.; Le, D.T.; Nishiyama, R.; Watanabe, Y.; Sulieman, S.; Tran, U.T.; Mochida, K.; Van Dong, N.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; et al. The Auxin Response Factor Transcription Factor Family in Soybean: Genome-Wide Identification and Expression Analyses during Development and Water Stress. DNA Res. 2013, 20, 511–524. [Google Scholar] [CrossRef]
- Lin, S.B.; Ouyang, W.Z.; Hou, X.J.; Xie, L.L.; Hu, C.G.; Zhang, J.Z. Genome-Wide Identification, Isolation and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Sweet Orange (Citrus sinensis). Front. Plant Sci. 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Hao, L.; Zhao, P.; Xu, Y.; Zhong, N.; Zhang, H.; Liu, N. Genome-Wide Identification, Expression Profiling and Evolutionary Analysis of Auxin Response Factor Gene Family in Potato (Solanum tuberosum Group Phureja). Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Benjamins, R.; Scheres, B. Auxin: The Looping Star in Plant Development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef]
- Liu, N.; Dong, L.; Deng, X.; Liu, D.; Liu, Y.; Li, M.; Hu, Y.; Yan, Y. Genome-Wide Identification, Molecular Evolution, and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and Diversification of Genes Regulating Auxin Biosynthesis, Transport, and Signaling. Mol. Plant 2019, 12, 298–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, H.; Ishizaki, K.; Kouno, M.; Shirakawa, M.; Bowman, J.L.; Nishihama, R.; Kohchi, T. Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in Marchantia Polymorpha. PLoS Genet. 2015, 11, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, Y.; Li, S.S.; Han, G.Z. Insights into the Origin and Evolution of the Plant Hormone Signaling Machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Nishihama, R.; Weijers, D.; Kohchi, T. Evolution of Nuclear Auxin Signaling: Lessons from Genetic Studies with Basal Land Plants. J. Exp. Bot. 2018, 69, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Mutte, S.K.; Kato, H.; Rothfels, C.; Melkonian, M.; Wong, G.K.S.; Weijers, D. Origin and Evolution of the Nuclear Auxin Response System. bioRxiv 2017, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Ma, Z.; Wang, A.; Zheng, T.; Huang, L.; Sun, W.; Zhang, Y.; Jin, W.; Zhan, J.; Cai, Y.; et al. Genome-Wide Investigation of the Auxin Response Factor Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Bendahmane, M.; Varaud, E.; Brioudes, F.; Leroux, J.; Brown, S.; et al. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell. 2015, 23, 973–983. [Google Scholar]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.; Clarke, T.H.; Reed, J.W.; Van Der Knaap, E. Down-Regulation of Auxin Response Factors 6 and 8 by microRNA167 Leads to Floral Development Defects and Female Sterility in Tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef] [Green Version]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. Auxin Response Factor1 and Auxin Response FACTOR2 Regulate Senescence and Floral Organ Abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [Green Version]
- Damodharan, S.; Zhao, D.; Arazi, T. A Common MiRNA160-Based Mechanism Regulates Ovary Patterning, Floral Organ Abscission and Lamina Outgrowth in Tomato. Plant J. Cell Mol. Biol. 2016, 86, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, Y.; Huang, X.; Li, J.; Wang, H.; Li, J. An Improved Fruit Transcriptome and the Identification of the Candidate Genes Involved in Fruit Abscission Induced by Carbohydrate Stress in Litchi. Front. Plant Sci. 2015, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Xie, R.; Pang, S.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. The ARF, AUX/IAA and GH3 Gene Families in Citrus: Genome-Wide Identification and Expression Analysis during Fruitlet Drop from Abscission Zone A. Mol. Genet. Genom. 2015, 290, 2089–2105. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinform. 2017, 60, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Cash, P. 2-D Proteome Analysis Protocols. Cell Biol. Int. 1999, 23, 385. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H. Bin Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Engeset, R.V.; Udnæs, H.C.; Guneriussen, T.; Koren, H.; Malnes, E.; Solberg, R.; Alfnes, E. Improving Runoff Simulations Using Satellite-Observed Time-Series of Snow Covered Area. Nordic Hydrol. 2003, 34, 281–294. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | CDS | Protein | Mw (KDa) | pI | GRAVY | Subcellular Localization | Domain |
|---|---|---|---|---|---|---|---|---|
| PavARF1 | Pav_sc0001248.1_g270.1.mk | 3264 | 1187 | 119627.80 | 5.86 | −0.608 | Nucleus | B3,Auxin_resp,CTD |
| PavARF2 | Pav_sc0000030.1_g880.1.mk | 2547 | 848 | 93909.05 | 6.96 | −0.449 | Nucleus | B3,Auxin_resp,CTD |
| PavARF3 | Pav_sc0001196.1_g1840.1.mk | 1155 | 384 | 42848.91 | 5.20 | −0.662 | Nucleus | Auxin_resp,CTD |
| PavARF4 | Pav_sc0000586.1_g230.1.mk | 2097 | 698 | 77581.78 | 6.44 | −0.430 | Nucleus | B3,Auxin_resp,CTD |
| PavARF5 | Pav_sc0000084.1_g430.1.mk | 2151 | 716 | 79165.21 | 6.59 | −0.408 | Nucleus | B3,Auxin_resp |
| PavARF6 | Pav_sc0001345.1_g010.1.mk | 2256 | 751 | 82327.61 | 5.90 | −0.335 | Nucleus | B3,Auxin_resp,CTD |
| PavARF7 | Pav_sc0001900.1_g150.1.mk | 1920 | 639 | 69349.71 | 5.92 | −0.530 | Nucleus | B3,Auxin_resp |
| PavARF8 | Pav_sc0003135.1_g100.1.mk | 2670 | 889 | 98481.69 | 6.08 | −0.431 | Nucleus | B3,Auxin_resp,CTD |
| PavARF9 | Pav_sc0002250.1_g170.1.mk | 2046 | 681 | 75315.71 | 6.20 | −0.476 | Nucleus | B3,Auxin_resp,CTD |
| PavARF10 | Pav_sc0000042.1_g450.1.mk | 2316 | 771 | 86014.22 | 5.85 | −0.694 | Nucleus | B3,Auxin_resp,CTD |
| PavARF11 | Pav_sc0000094.1_g590.1.mk | 2061 | 686 | 75602.80 | 5.35 | −0.499 | Nucleus | B3,Auxin_resp,CTD |
| PavARF12 | Pav_sc0002446.1_g140.1.mk | 2160 | 719 | 79093.28 | 7.88 | −0.410 | Nucleus | B3,Auxin_resp |
| PavARF13 | Pav_sc0000711.1_g070.1.mk | 2172 | 723 | 79392.80 | 6.09 | −0.413 | Nucleus | B3,Auxin_resp |
| PavARF14 | Pav_sc0000129.1_g1480.1.mk | 2130 | 709 | 78539.30 | 5.34 | −0.444 | Nucleus | B3,Auxin_resp,CTD |
| PavARF15 | Pav_sc0000848.1_g300.1.mk | 2652 | 883 | 97861.21 | 5.92 | −0.444 | Nucleus | B3,Auxin_resp,CTD |
| PavARF16 | Pav_sc0001314.1_g050.1.mk | 2751 | 916 | 101887.61 | 5.76 | −0.410 | Nucleus | B3,Auxin_resp,CTD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Q.; Qiu, Z.; Wen, Z.; Zhang, H.; Li, Z.; Hong, Y.; Qiao, G.; Wen, X. Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L. Int. J. Mol. Sci. 2021, 22, 11968. https://doi.org/10.3390/ijms222111968
Hou Q, Qiu Z, Wen Z, Zhang H, Li Z, Hong Y, Qiao G, Wen X. Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L. International Journal of Molecular Sciences. 2021; 22(21):11968. https://doi.org/10.3390/ijms222111968
Chicago/Turabian StyleHou, Qiandong, Zhilang Qiu, Zhuang Wen, Huimin Zhang, Zhengchun Li, Yi Hong, Guang Qiao, and Xiaopeng Wen. 2021. "Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L." International Journal of Molecular Sciences 22, no. 21: 11968. https://doi.org/10.3390/ijms222111968
APA StyleHou, Q., Qiu, Z., Wen, Z., Zhang, H., Li, Z., Hong, Y., Qiao, G., & Wen, X. (2021). Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L. International Journal of Molecular Sciences, 22(21), 11968. https://doi.org/10.3390/ijms222111968






