(Un)expected Similarity of the Temporary Adhesive Systems of Marine, Brackish, and Freshwater Flatworms
Abstract
1. Introduction
2. Results
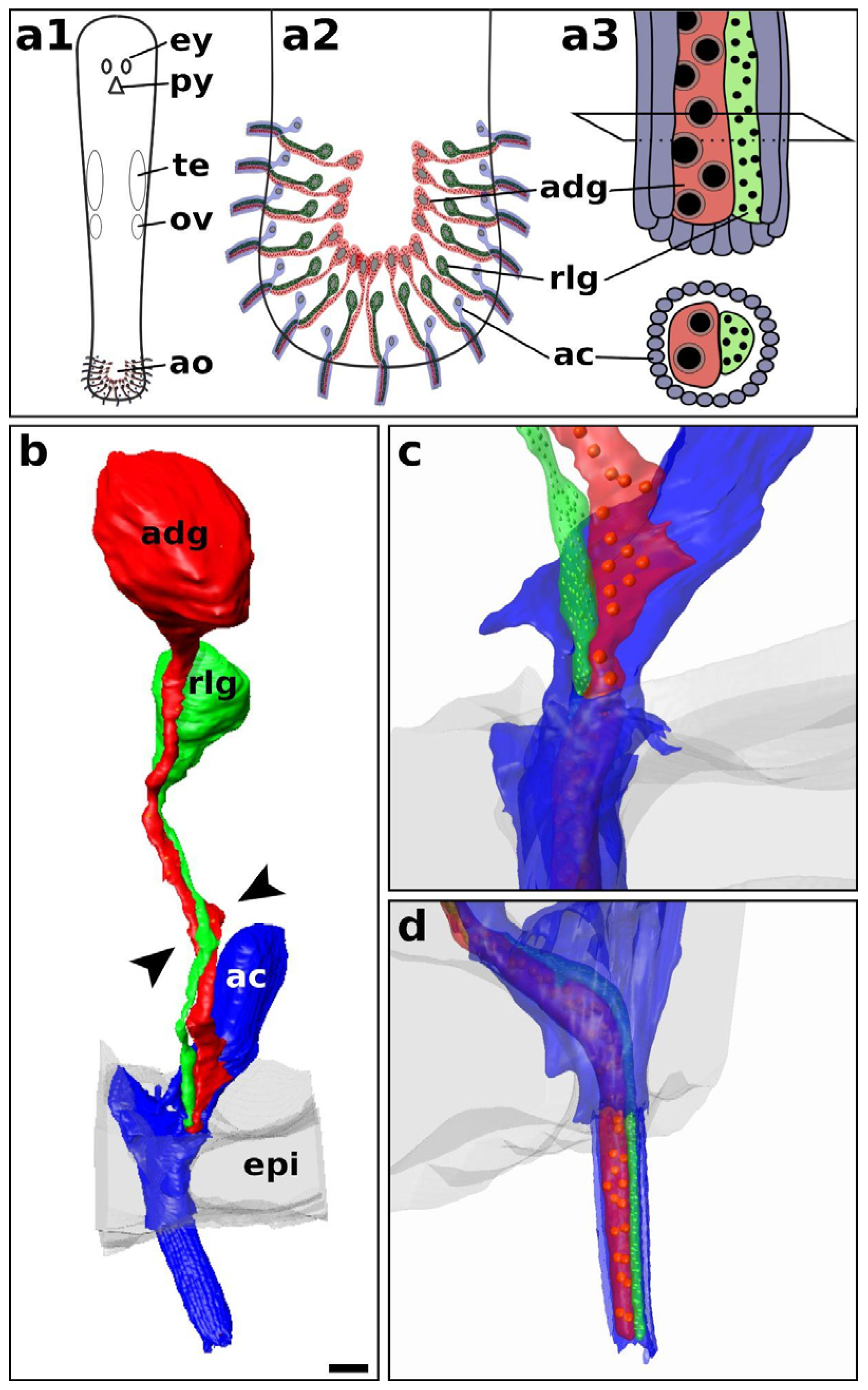
2.1. General Overview of Adhesion Organs in Macrostomum
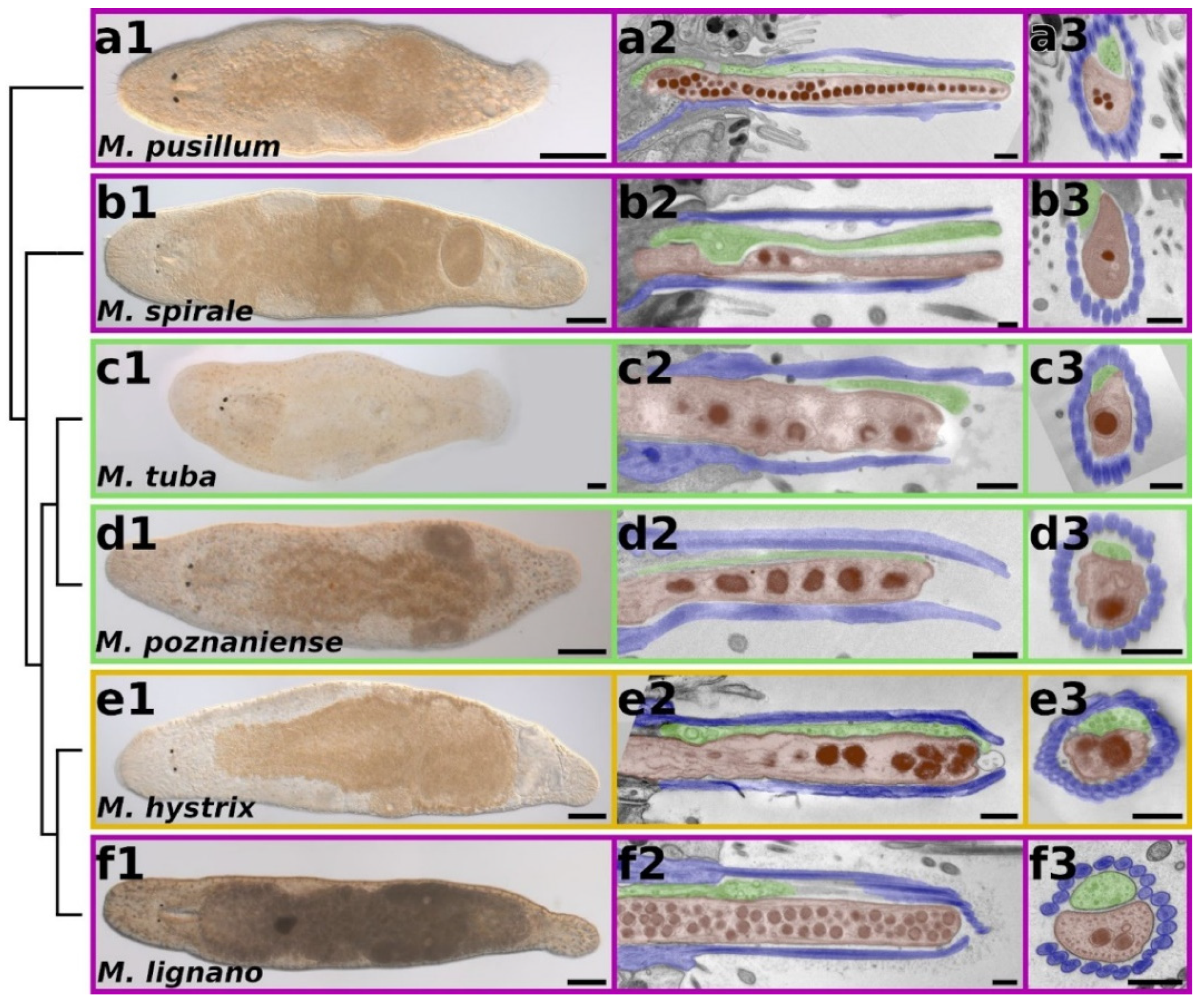
2.2. Morphology of Adhesion Organs in Different Macrostomum Species
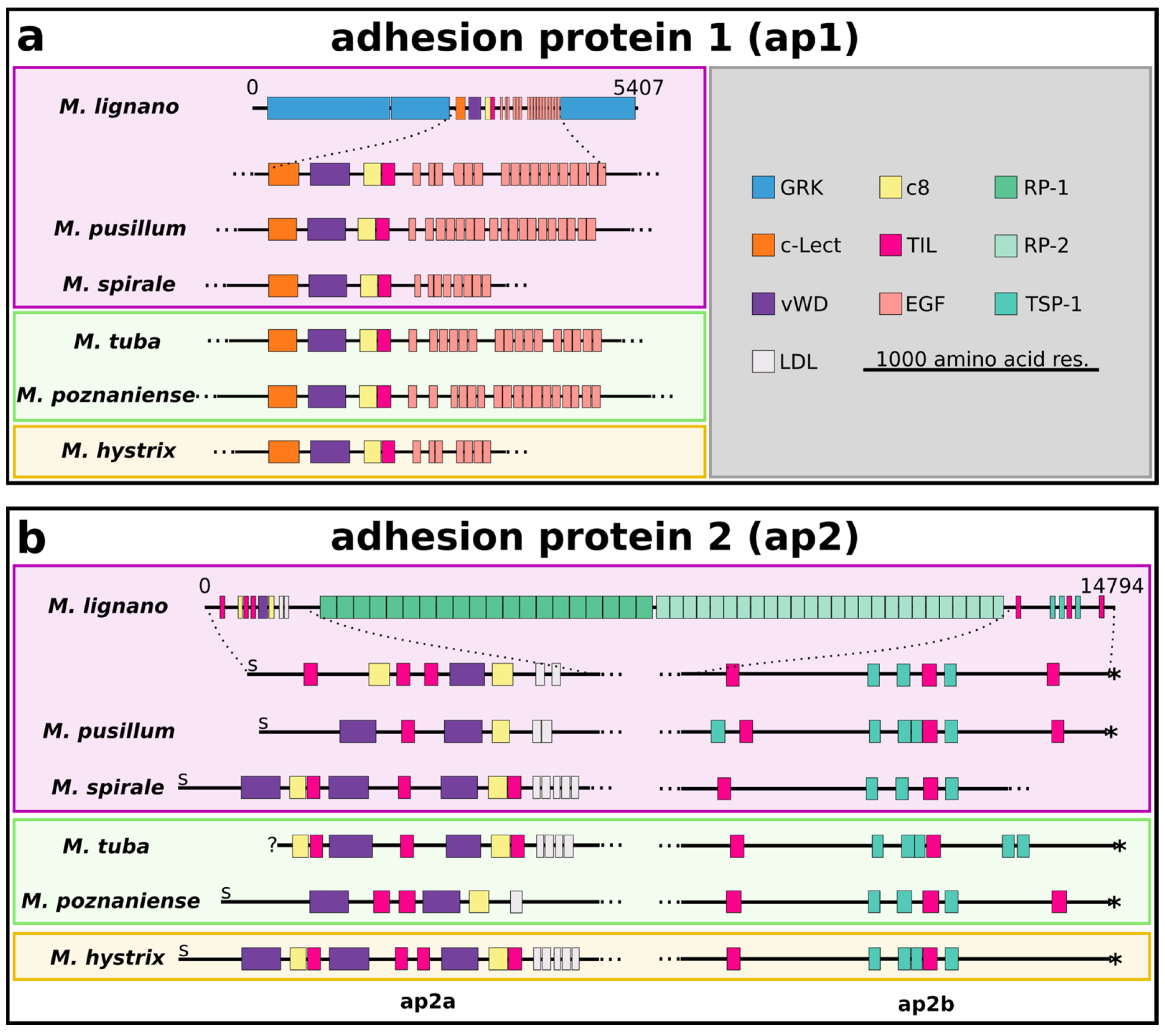
2.3. Conservation of Protein Domains in Adhesives
2.4. Adhesion Protein 1
2.5. Adhesion Protein 2
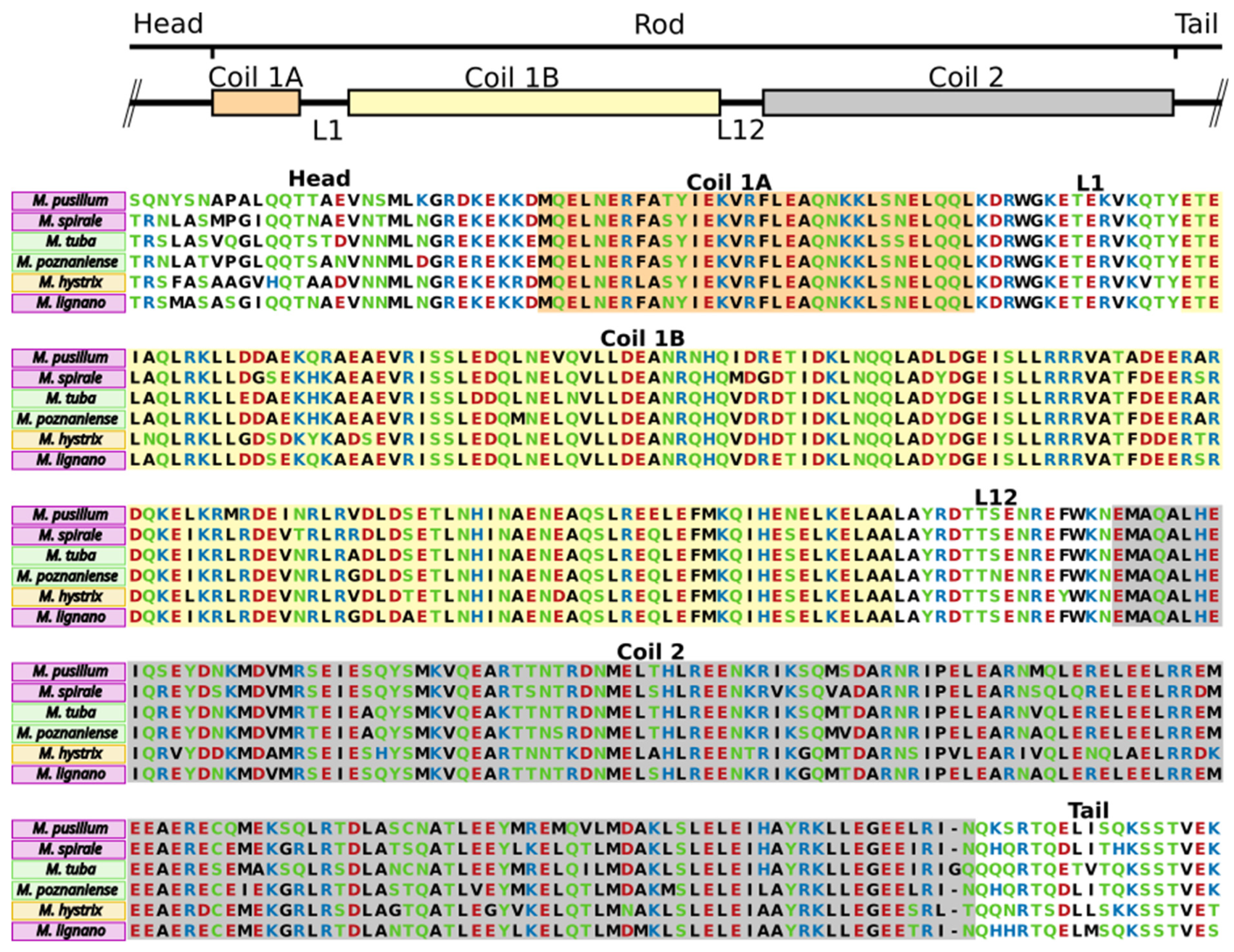
2.6. Anchor Cell-Specific Intermediate Filaments in Macrostomum Species
2.7. Adhesion Assay in Different Salt Concentrations
2.8. In Situ Hybridisation of ap1 and ap2
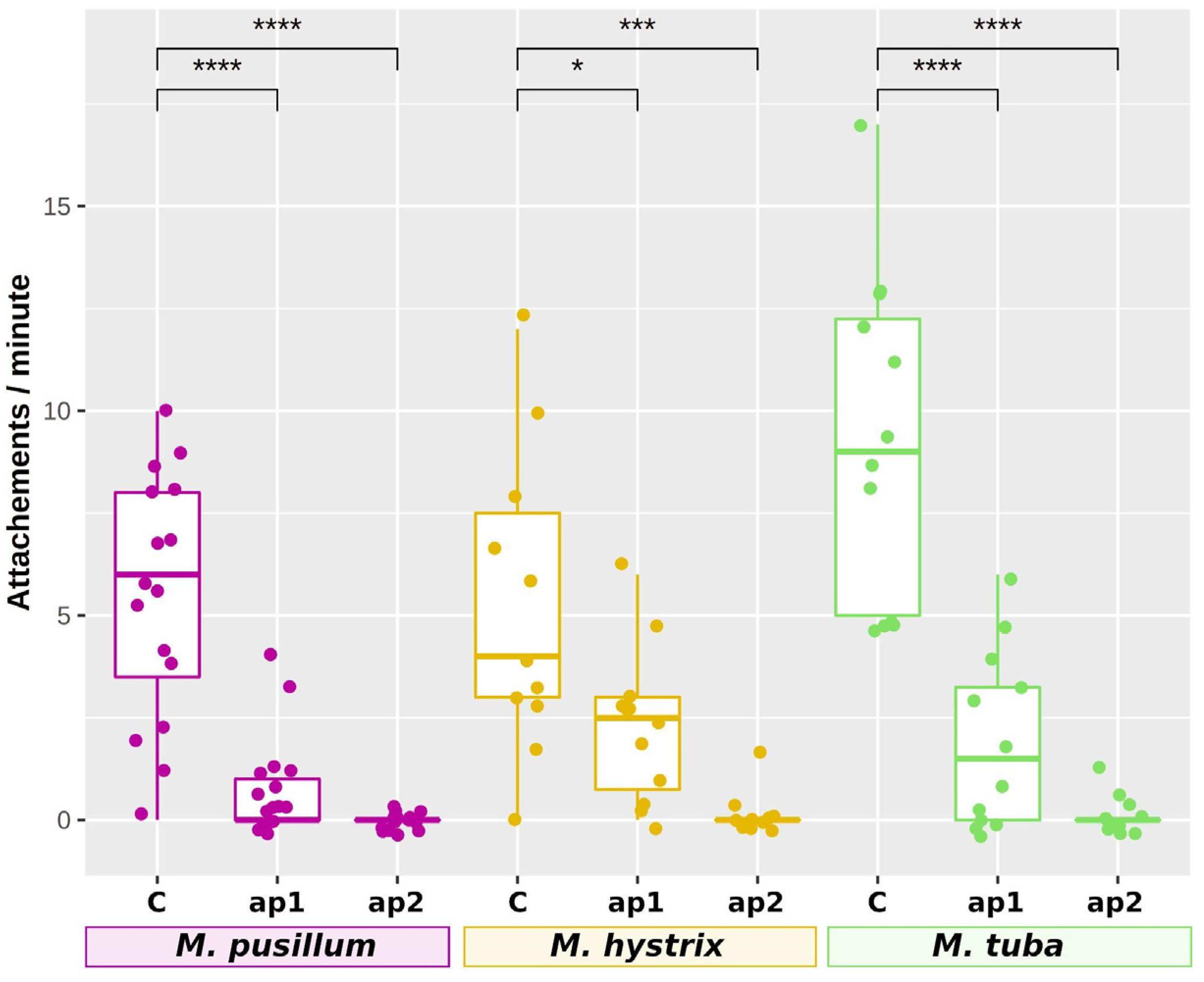
2.9. RNAi Knockdown of Adhesion Protein 1 and 2 Genes
3. Discussion
3.1. Macrostomum Adhesion Organ Morphology Is Not Influenced by the Habitat
3.2. The Two Adhesive Proteins ap1 and ap2 Are Present in All Investigated Species
3.3. Conserved Domains in Adhesive Proteins
3.4. Large Adhesive Genes Cannot Be Sequenced with Short-Read Technology
3.5. Post-Translational Modifications and Their Role in Bioadhesion
3.6. The Two-Component Flatworm Adhesive could Lead to the Development of Novel Biomimetic Glues
4. Materials and Methods
4.1. Description of Sampling Sites of the Fresh, Brackish, and Seawater Species Cultures/Sampling
4.2. Documentation of Live Squeeze—Preparations
4.3. Anaesthetization and Fixation
4.4. RNA Extraction, cDNA Synthesis, Template Synthesis by PCR, and Probe Synthesis
4.5. RNA Extraction and Sequencing of Macrostomum Tuba
4.6. Transcriptome Assembly and Annotation of Macrostomum Tuba
4.7. Transcriptome Data of M. pusillum, M. spirale, M. hystrix, and M. poznaniense
4.8. BLAST Searches and Conserved Domain Identification
4.9. In Situ Hybridisation
4.10. Double-Stranded RNA Synthesis and RNAi Adhesion Assay
4.11. Attachment Assay in Different Salt Concentrations
4.12. Lectin Staining and gSTED High-Resolution Microscopy
4.13. Transmission Electron Microscopy
4.14. Element Analysis with EELS and ESI
4.15. Serial Block-Face Scanning Electron Microscopy
4.16. Reconstruction of the Adhesive System
4.17. Figure and Movie Preparation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bianco-Peled, H.; Davidovich-Pinhas, M. Bioadhesion and Biomimetics: From Nature to Applications; Pan Stanford Publishing: Singapore, 2015; ISBN 978-981-4463-98-0. [Google Scholar]
- Byern, J.; von Grunwald, I. (Eds.) Biological Adhesive Systems: From Nature to Technical and Medical Application; Springer: Wien, Austria, 2010; ISBN 978-3-7091-0141-4. [Google Scholar]
- Smith, A.M. (Ed.) Biological Adhesives, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-46081-9. [Google Scholar]
- Gorb, E.; Gorb, S. Contact separation force of the fruit burrs in four plant species adapted to dispersal by mechanical interlocking. Plant Physiol. Biochem. 2002, 40, 373–381. [Google Scholar] [CrossRef]
- Berne, C.; Ellison, C.K.; Ducret, A.; Brun, Y.V. Bacterial adhesion at the single-cell level. Nat. Rev. Microbiol. 2018, 16, 616–627. [Google Scholar] [CrossRef]
- Autumn, K.; Peattie, A.M. Mechanisms of adhesion in geckos. Integr. Comp. Biol. 2002, 42, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Stellwagen, S.D.; Renberg, R.L. Toward spider glue: Long read scaffolding for extreme length and repetitious silk family genes AgSp1 and AgSp2 with insights into functional adaptation. G3 Genes Genomes Genet. 2019, 9, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Waite, J.H. Evidence for a repeating 3,4-Dihydroxyphenylalanine- and Hydroxyproline-containing decapeptide in the adhesive protein of the mussel, Mytilus edulis L. J. Biol. Chem. 1983, 258, 2911–2915. [Google Scholar] [CrossRef]
- Waite, J.H. Translational bioadhesion research: Embracing biology without tokenism. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190207. [Google Scholar] [CrossRef]
- Waite, J.H.; Tanzer, M.L. Polyphenolic substance of Mytilus edulis: Novel Adhesive Containing L-Dopa and Hydroxyproline. Science 1981, 212, 1038–1040. [Google Scholar] [CrossRef] [PubMed]
- Flammang, P. Adhesion in echinoderms. In Echinoderm Studies; CRC Press: Boca Raton, FL, USA, 1996; Volume 5, pp. 1–60. [Google Scholar]
- Kang, V.; Lengerer, B.; Wattiez, R.; Flammang, P. Molecular insights into the powerful mucus-based adhesion of limpets (Patella vulgata L.). Open Biol. 2019, 10, 200019. [Google Scholar] [CrossRef]
- Wunderer, J.; Lengerer, B.; Pjeta, R.; Bertemes, P.; Kremser, L.; Lindner, H.; Ederth, T.; Hess, M.W.; Stock, D.; Salvenmoser, W.; et al. A mechanism for temporary bioadhesion. Proc. Natl. Acad. Sci. USA 2019, 116, 4297–4306. [Google Scholar] [CrossRef]
- Lengerer, B.; Ladurner, P. Properties of Temporary adhesion systems of marine and freshwater organisms. J. Exp. Biol. 2018, 221, jeb182717. [Google Scholar] [CrossRef]
- Egger, B.; Lapraz, F.; Tomiczek, B.; Müller, S.; Dessimoz, C.; Girstmair, J.; Škunca, N.; Rawlinson, K.A.; Cameron, C.B.; Beli, E.; et al. A transcriptomic-phylogenomic analysis of the evolutionary relationships of flatworms. Curr. Biol. 2015, 25, 1347–1353. [Google Scholar] [CrossRef]
- Brand, J.N.; Viktorin, G.; Axel, W.; Wiberg, R.; Beisel, C.; Schärer, L. Large-scale phylogenomics of the genus Macrostomum (Platyhelminthes) reveals cryptic diversity and novel sexual traits. Mol. Phylogenet. Evol. 2021, 166, 107296. [Google Scholar] [CrossRef]
- Lin, Y.; Zhou, W.; Xiao, P.; Zheng, Y.; Lu, J.; Li, J.; Wang, A. Two new species of freshwater Macrostomum (Rhabditophora: Macrostomorpha) found in China. Zootaxa 2017, 4329, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Schärer, L.; Brand, J.N.; Singh, P.; Zadesenets, K.S.; Stelzer, C.-P.; Viktorin, G. A phylogenetically informed search for an alternative macrostomum model species, with notes on taxonomy, mating behavior, karyology, and genome size. J. Zool. Syst. Evol. Res. 2020, 58, 41–65. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, L.; Wang, A.-T.; Zhang, Y. Three new species of freshwater Macrostomum (Platyhelminthes, Macrostomida) from Southern China. Zootaxa 2015, 4012, 120–134. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tyler, S.; Hooge, M.; Bush, L. (2006–2016) Turbellarian Taxonomic Database. Version 1.7. Available online: http://turbellaria.umaine.edu/ (accessed on 18 October 2021).
- Xin, F.; Zhang, S.-Y.; Shi, Y.-S.; Wang, L.; Zhang, Y.U.; Wang, A.-T. Macrostomum shenda and M. spiriger, two new brackish-water species of Macrostomum (Platyhelminthes: Macrostomorpha) from China. Zootaxa 2019, 4603, 105–124. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, Y.; Zeng, Z.; Xin, F.; Deng, L.; Wang, A. two new brackish-water species of Macrostomum (Platyhelminthes: Macrostomorpha) from China and their phylogenetic positions. Zool. Sci. 2021, 38, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Wudarski, J.; Egger, B.; Ramm, S.A.; Schärer, L.; Ladurner, P.; Zadesenets, K.S.; Rubtsov, N.B.; Mouton, S.; Berezikov, E. The free-living flatworm Macrostomum lignano. EvoDevo 2020, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Lengerer, B.; Hennebert, E.; Flammang, P.; Salvenmoser, W.; Ladurner, P. Adhesive organ regeneration in Macrostomum lignano. BMC Dev. Biol. 2016, 16, 20. [Google Scholar] [CrossRef]
- Davey, P.A.; Power, A.M.; Santos, R.; Bertemes, P.; Ladurner, P.; Palmowski, P.; Clarke, J.; Flammang, P.; Lengerer, B.; Hennebert, E.; et al. Omics-based molecular analyses of adhesion by aquatic invertebrates. Biol. Rev. 2021, 96, 1051–1075. [Google Scholar] [CrossRef] [PubMed]
- Lengerer, B.; Pjeta, R.; Wunderer, J.; Rodrigues, M.; Arbore, R.; Schärer, L.; Berezikov, E.; Hess, M.W.; Pfaller, K.; Egger, B.; et al. Biological adhesion of the flatworm Macrostomum lignano relies on a duo-gland system and is mediated by a cell type-specific intermediate filament protein. Front. Zool. 2014, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Lengerer, B.; Wunderer, J.; Pjeta, R.; Carta, G.; Kao, D.; Aboobaker, A.; Beisel, C.; Berezikov, E.; Salvenmoser, W.; Ladurner, P. Organ specific gene expression in the regenerating tail of Macrostomum lignano. Dev. Biol. 2018, 433, 448–460. [Google Scholar] [CrossRef]
- Ax, P. Monographie der Otoplanidae (Turbellaria). Morphologie und Systematik. Abh. Math. Nat. Kl. 1956, 13, 3–298. [Google Scholar]
- Ørsted, A.S. Annulatorum Danicorum Conspectus; Sumtibus Librariæ Wahlianæ: Hafniæ, Denmark, 1843. [Google Scholar]
- Kolasa, J. Two new species of Macrostomum (Turbellaria), a redescription of an established species and new records from Poland. Ital. J. Zool. 1973, 40, 181–200. [Google Scholar]
- von Graff, L. Monographie der Turbellarien; W. Engelmann: Leipzig, Germany, 1882; Volume 1. [Google Scholar]
- Tyler, S. Comparative ultrastructure of adhesive systems in Turbellaria. Zoomorphologie 1976, 84, 1–76. [Google Scholar] [CrossRef]
- Rieger, R.M. The relationship of character variability and morphological complexity in copulatory structures of turbellaria- Macrostomida and -Haplopharyngida. Mikrofauna Meeresbod. 1977, 61, 197–216. [Google Scholar]
- Adami, M.; Damborenea, C.; Ronderos, J.R. A new limnic species of Macrostomum (Platyhelminthes: Macrostomida) from Argentina and its muscle arrangement labeled with phalloidin. Zool. Anz. 2012, 251, 197–205. [Google Scholar] [CrossRef]
- Silveira, M.; Aragao, P.H.A. Organized filaments in the adhesive system of Macrostomum tuba Graff, 1882 (Platyhelminthes, Macrostomida). Braz. J. Morphol. Sci. 2006, 23, 471–477. [Google Scholar]
- Fang, C.-Y.; Wang, L.; Zhang, Y.; Wang, A.-T. Two new species of brackish-water Macrostomum (Platyhelminthes, Macrostomida) from Southern China. Zootaxa 2016, 4170, 298–310. [Google Scholar] [CrossRef]
- Hennebert, E.; Leroy, B.; Wattiez, R.; Ladurner, P. An integrated transcriptomic and proteomic analysis of sea star epidermal secretions identifies proteins involved in defense and adhesion. J. Proteom. 2015, 128, 83–91. [Google Scholar] [CrossRef]
- Lengerer, B.; Algrain, M.; Lefevre, M.; Delroisse, J.; Hennebert, E.; Flammang, P. Interspecies comparison of sea star adhesive proteins. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190195. [Google Scholar] [CrossRef]
- Pjeta, R.; Lindner, H.; Kremser, L.; Salvenmoser, W.; Sobral, D.; Ladurner, P.; Santos, R. Integrative transcriptome and proteome analysis of the tube foot and adhesive secretions of the sea urchin Paracentrotus lividus. Int. J. Mol. Sci. 2020, 21, 946. [Google Scholar] [CrossRef]
- Pjeta, R.; Wunderer, J.; Bertemes, P.; Hofer, T.; Salvenmoser, W.; Lengerer, B.; Coassin, S.; Erhart, G.; Beisel, C.; Sobral, D.; et al. Temporary adhesion of the proseriate flatworm Minona ileanae. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190194. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-F.; Eng, E.T.; Zhu, J.; Lu, C.; Walz, T.; Springer, T.A. Sequence and structure relationships within von Willebrand factor. Blood 2012, 120, 449–458. [Google Scholar] [CrossRef]
- Dong, X.; Leksa, N.C.; Chhabra, E.S.; Arndt, J.W.; Lu, Q.; Knockenhauer, K.E.; Peters, R.T.; Springer, T.A. The von Willebrand factor D′D3 assembly and structural principles for factor VIII binding and concatemer biogenesis. Blood 2019, 133, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Yee, A.; Kretz, C.A. Von Willebrand factor: Form for function. Semin. Thromb. Hemost. 2014, 40, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Hansson, G.C.; Samuelsson, T. Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 16209–16214. [Google Scholar] [CrossRef] [PubMed]
- Javitt, G.; Khmelnitsky, L.; Albert, L.; Bigman, L.S.; Elad, N.; Morgenstern, D.; Ilani, T.; Levy, Y.; Diskin, R.; Fass, D. Assembly mechanism of mucin and von Willebrand factor polymers. Cell 2020, 183, 717–729.e16. [Google Scholar] [CrossRef]
- Hennebert, E.; Wattiez, R.; Demeuldre, M.; Ladurner, P.; Hwang, D.S.; Waite, J.H.; Flammang, P. Sea star tenacity mediated by a protein that fragments, then aggregates. Proc. Natl. Acad. Sci. USA 2014, 111, 6317–6322. [Google Scholar] [CrossRef]
- Lefevre, M.; Flammang, P.; Aranko, A.S.; Linder, M.B.; Scheibel, T.; Humenik, M.; Leclercq, M.; Surin, M.; Tafforeau, L.; Wattiez, R.; et al. Sea star-inspired recombinant adhesive proteins self-assemble and adsorb on surfaces in aqueous environments to form cytocompatible coatings. Acta Biomater. 2020, 112, 62–74. [Google Scholar] [CrossRef]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, M.; Ederth, T.; Masai, T.; Wattiez, R.; Leclère, P.; Flammang, P.; Hennebert, E. Disentangling the roles of functional domains in the aggregation and adsorption of the multimodular sea star adhesive protein Sfp1. Mar. Biotechnol. 2021, 23, 724–735. [Google Scholar] [CrossRef]
- Bania, J.; Stachowiak, D.; Polanowski, A. Primary structure and properties of the cathepsin G/Chymotrypsin inhibitor from the larval hemolymph of Apis mellifera. Eur. J. Biochem. 1999, 262, 680–687. [Google Scholar] [CrossRef]
- Shakeel, M.; Zafar, J. Molecular identification, characterization, and expression analysis of a novel trypsin inhibitor-like cysteine-rich peptide from the cotton bollworm, Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae). Egypt. J. Biol. Pest Control 2020, 30, 10. [Google Scholar] [CrossRef]
- Adams, J.C.; Lawler, J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef] [PubMed]
- Resovi, A.; Pinessi, D.; Chiorino, G.; Taraboletti, G. Current understanding of the thrombospondin-1 interactome. Matrix Biol. 2014, 37, 83–91. [Google Scholar] [CrossRef]
- Waite, J.H. Mussel adhesion—Essential footwork. J. Exp. Biol. 2017, 220, 517. [Google Scholar] [CrossRef]
- Flammang, P.; Lambert, A.; Bailly, P.; Hennebert, E. Polyphosphoprotein-containing marine adhesives. J. Adhes. 2009, 85, 447–464. [Google Scholar] [CrossRef]
- Stewart, R.J.; Weaver, J.C.; Morse, D.E.; Waite, J.H. The tube cement of Phragmatopoma californica: A solid foam. J. Exp. Biol. 2004, 207, 4727–4734. [Google Scholar] [CrossRef]
- Hennebert, E.; Gregorowicz, E.; Flammang, P. Involvement of sulfated biopolymers in adhesive secretions produced by marine invertebrates. Biol. Open 2018, 7, bio037358. [Google Scholar] [CrossRef]
- Hennebert, E.; Wattiez, R.; Flammang, P. Characterisation of the carbohydrate fraction of the temporary adhesive secreted by the tube feet of the sea star Asterias rubens. Mar. Biotechnol. 2011, 13, 484–495. [Google Scholar] [CrossRef]
- Lengerer, B.; Bonneel, M.; Lefevre, M.; Hennebert, E.; Leclère, P.; Gosselin, E.; Ladurner, P.; Flammang, P. The structural and chemical basis of temporary adhesion in the sea star Asterina gibbosa. Beilstein J. Nanotechnol. 2018, 9, 2071–2086. [Google Scholar] [CrossRef]
- Simão, M.; Moço, M.; Marques, L.; Santos, R. Characterization of the glycans involved in sea urchin Paracentrotus lividus reversible adhesion. Mar. Biol. 2020, 167, 125. [Google Scholar] [CrossRef]
- Pagett, H.E.; Abrahams, J.L.; Bones, J.; O’Donoghue, N.; Marles-Wright, J.; Lewis, R.J.; Harris, J.R.; Caldwell, G.S.; Rudd, P.M.; Clare, A.S. Structural characterisation of the N-glycan moiety of the barnacle settlement-inducing protein complex (SIPC). J. Exp. Biol. 2012, 215, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, K.; Nishida, A.; Yamamoto, H.; Waite, J.H. A glycosylated byssal precursor protein from the green mussel Perna viridis with modified Dopa side-chains. Biofouling 2004, 20, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Wunderer, J.; Salvenmoser, W.; Ederth, T.; Rothbächer, U. Identifying adhesive components in a model tunicate. Philos. Trans. R. Soc. B 2019, 374, 20190197. [Google Scholar] [CrossRef]
- Smith, A.M.; Quick, T.J.; St Peter, R.L. Differences in the composition of adhesive and non-adhesive mucus from the limpet Lottia limatula. Biol. Bull. 1999, 196, 34–44. [Google Scholar] [CrossRef]
- Singla, S.; Amarpuri, G.; Dhopatkar, N.; Blackledge, T.A.; Dhinojwala, A. Hygroscopic compounds in spider aggregate glue remove interfacial water to maintain adhesion in humid conditions. Nat. Commun. 2018, 9, 1890. [Google Scholar] [CrossRef]
- Zayas, R.M.; Cebrià, F.; Guo, T.; Feng, J.; Newmark, P.A. The use of lectins as markers for differentiated secretory cells in Planarians. Dev. Dyn. 2010, 239, 2888–2897. [Google Scholar] [CrossRef]
- Presti, M.L.; Rizzo, G.; Farinola, G.M.; Omenetto, F.G. Bioinspired biomaterial composite for all-water-based high-performance adhesives. Adv. Sci. 2021, 8, 16. [Google Scholar] [CrossRef]
- Hong, Y.; Zhou, F.; Hua, Y.; Zhang, X.; Ni, C.; Pan, D.; Zhang, Y.; Jiang, D.; Yang, L.; Lin, Q.; et al. A strongly adhesive hemostatic hydrogel for the repair of arterial and heart bleeds. Nat. Commun. 2019, 10, 2060. [Google Scholar] [CrossRef]
- Almeida, M.; Reis, R.L.; Silva, T.H. Marine invertebrates are a source of bioadhesives with biomimetic interest. Mater. Sci. Eng. 2020, 108, 110467. [Google Scholar] [CrossRef]
- Merryman, M.S.; Alvarado, A.S.; Jenkin, J.C. Culturing planarians in the laboratory. In Planarian Regeneration: Methods and Protocols; Rink, J.C., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 241–258. ISBN 978-1-4939-7802-1. [Google Scholar]
- Brand, J.N.; Wiberg, R.A.W.; Pjeta, R.; Bertemes, P.; Beisel, C.; Ladurner, P.; Schärer, L. RNA-Seq of Three free-living flatworm species suggests rapid evolution of reproduction-related genes. BMC Genom. 2020, 21, 462. [Google Scholar] [CrossRef]
- Hustedt, F. Die Bacillariaceen-Vegetation des Lunzer Seengebietes (Nieder-Österreich). Int. Rev. Gesamten Hydrobiol. Hydrogr. 1922, 10, 40–69. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Florea, L. Rcorrector: Efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 2015, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A Fast, Lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. In Gene Prediction; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1962, pp. 227–245. [Google Scholar] [CrossRef]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Priyam, A.; Woodcroft, B.J.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.; De Mulder, K.; Philipp, I.; Kuales, G.; Hrouda, M.; Eichberger, P.; Borgonie, G.; Hartenstein, V.; Ladurner, P. The exceptional stem cell system of Macrostomum lignano: Screening for gene expression and studying cell proliferation by hydroxyurea treatment and irradiation. Front. Zool. 2007, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- King, R.S.; Newmark, P.A. Whole-mount in situ hybridization of planarians. In Planarian Regeneration; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1774, pp. 379–392. [Google Scholar] [CrossRef]
- Salvenmoser, W.; Egger, B.; Achatz, J.G.; Ladurner, P.; Hess, M.W. Chapter 14—Electron microscopy of flatworms: Standard and cryo-preparation methods. In Methods in Cell Biology; Müller-Reichert, T., Ed.; Electron Microscopy of Model Systems; Academic Press: Oxford, UK, 2010; Volume 96, pp. 307–330. [Google Scholar]
- Rombout, J.H.W.M.; Lamers, C.H.J.; Hanstede, J.G. Enteroendocrine APUD cells in the digestive tract of larval Barbus conchonius (Teleostei, Cyprinidae). Development 1978, 47, 121–135. [Google Scholar] [CrossRef]
- Eisenmann, E.A.; Alfert, M. A new fixation procedure for preserving the ultrastructure of marine invertebrate tissue. J. Microsc. 1982, 125, 117. [Google Scholar] [CrossRef]
- Denk, W.; Horstmann, H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004, 2, e329. [Google Scholar] [CrossRef] [PubMed]
- Zankel, A.; Kraus, B.; Poelt, P.; Schaffer, M.; Ingolic, E. Ultramicrotomy in the ESEM, a versatile method for materials and life sciences. J. Microsc. 2009, 233, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Zankel, A.; Wagner, J.; Poelt, P. Serial sectioning methods for 3D investigations in materials science. Micron 2014, 62, 66–78. [Google Scholar] [CrossRef]
- Deerinck, T.; Bushong, E.; Thor, A.; Ellisman, M. NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy. Microscopy 2010, 1, 6–8. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]

| Salt Concentrations [ppm] | |||||||
|---|---|---|---|---|---|---|---|
| Species | 2 | 5 | 10 | 20 | 35 | 45 | 60 |
| M. pusillum | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| M. spirale | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| M. hystrix | +++ | +++ | +++ | +++ | +++ | x | x |
| M. tuba | +++ | x | x | x | x | x | x |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertemes, P.; Pjeta, R.; Wunderer, J.; Grosbusch, A.L.; Lengerer, B.; Grüner, K.; Knapp, M.; Mertens, B.; Andresen, N.; Hess, M.W.; et al. (Un)expected Similarity of the Temporary Adhesive Systems of Marine, Brackish, and Freshwater Flatworms. Int. J. Mol. Sci. 2021, 22, 12228. https://doi.org/10.3390/ijms222212228
Bertemes P, Pjeta R, Wunderer J, Grosbusch AL, Lengerer B, Grüner K, Knapp M, Mertens B, Andresen N, Hess MW, et al. (Un)expected Similarity of the Temporary Adhesive Systems of Marine, Brackish, and Freshwater Flatworms. International Journal of Molecular Sciences. 2021; 22(22):12228. https://doi.org/10.3390/ijms222212228
Chicago/Turabian StyleBertemes, Philip, Robert Pjeta, Julia Wunderer, Alexandra L. Grosbusch, Birgit Lengerer, Kevin Grüner, Magdalena Knapp, Birte Mertens, Nikolas Andresen, Michael W. Hess, and et al. 2021. "(Un)expected Similarity of the Temporary Adhesive Systems of Marine, Brackish, and Freshwater Flatworms" International Journal of Molecular Sciences 22, no. 22: 12228. https://doi.org/10.3390/ijms222212228
APA StyleBertemes, P., Pjeta, R., Wunderer, J., Grosbusch, A. L., Lengerer, B., Grüner, K., Knapp, M., Mertens, B., Andresen, N., Hess, M. W., Tomaiuolo, S., Zankel, A., Holzer, P., Salvenmoser, W., Egger, B., & Ladurner, P. (2021). (Un)expected Similarity of the Temporary Adhesive Systems of Marine, Brackish, and Freshwater Flatworms. International Journal of Molecular Sciences, 22(22), 12228. https://doi.org/10.3390/ijms222212228






