Multi-Omics Analysis Reveals Anti-Staphylococcus aureus Activity of Actinomycin D Originating from Streptomyces parvulus
Abstract
:1. Introduction
2. Results
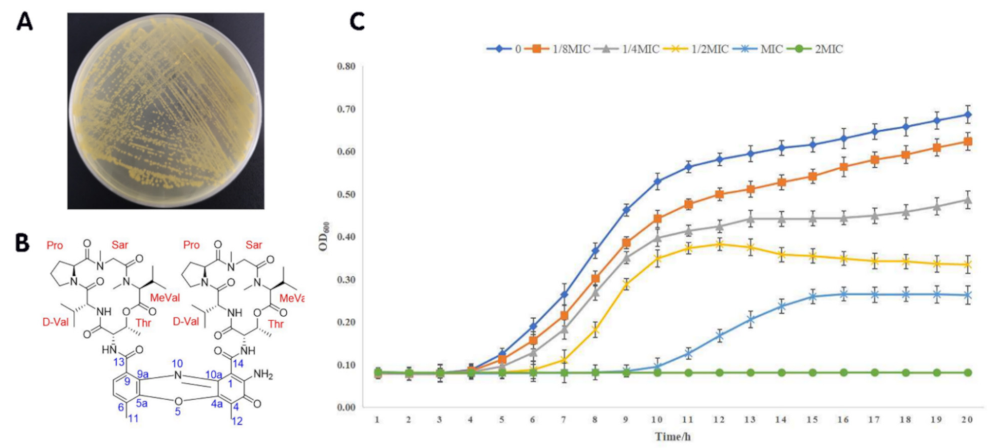
2.1. Growth of S. parvulus and Structure of Actinomycin D
2.2. Antimicrobial Activity
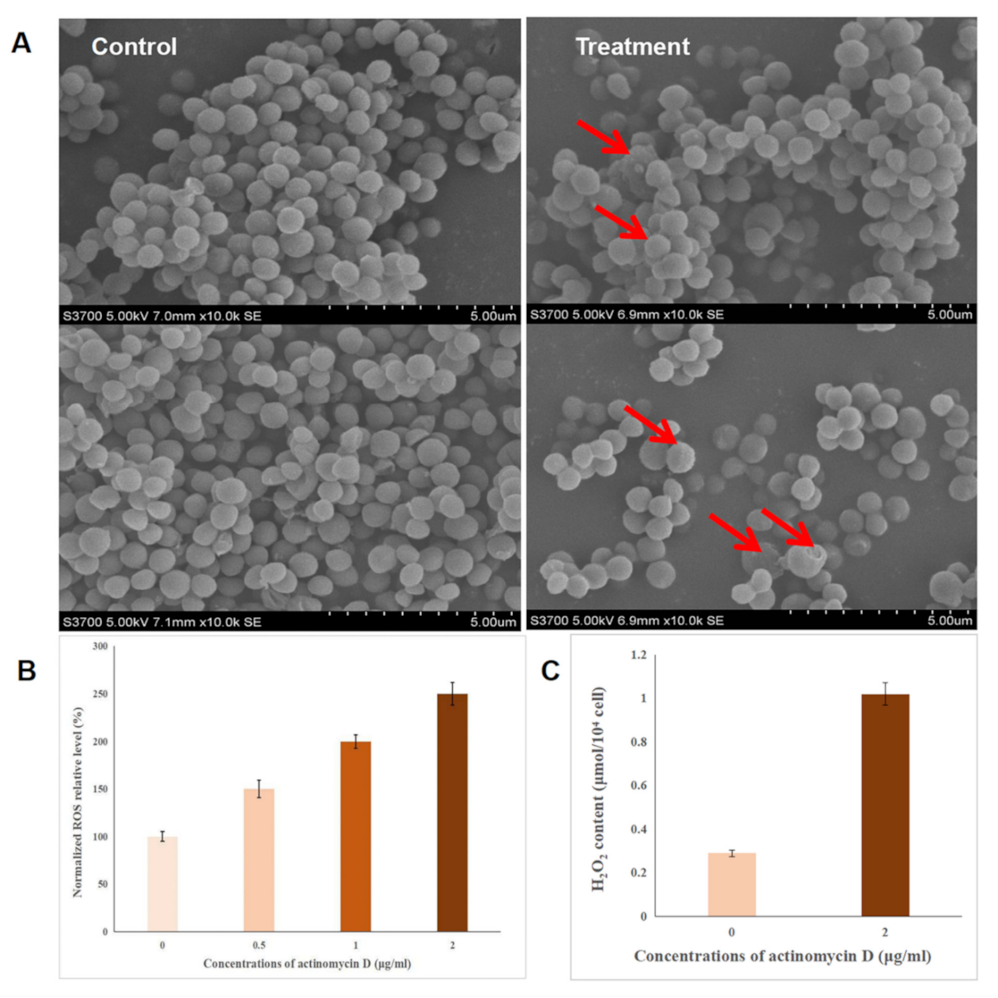
2.3. Effect of Actinomycin D on Bacterial Morphology
2.4. Intracellular Reactive Oxygen Species (ROS) Detection
2.5. The Detection of H2O2
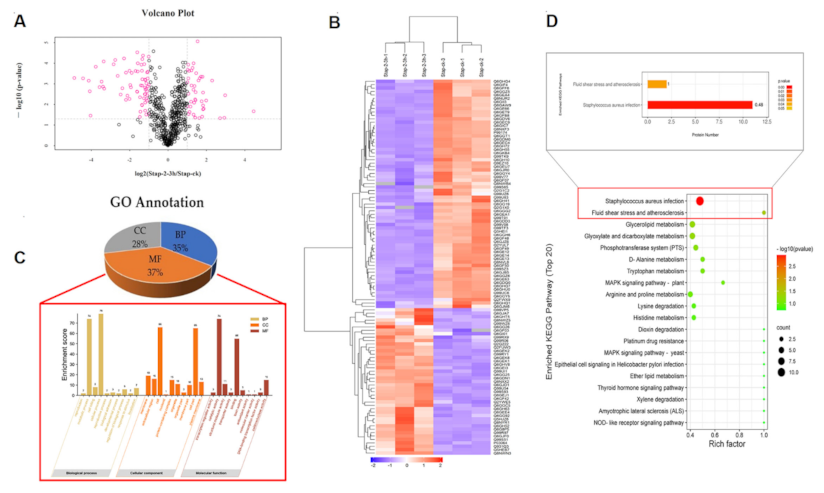
2.6. Proteomics Profile of S.aureus Treated with Actinomycin D
2.6.1. Label-Free Proteomics-Based Protein Identification
2.6.2. Gene Ontology (GO) Annotation
2.6.3. Analysis of Main Metabolic Pathways
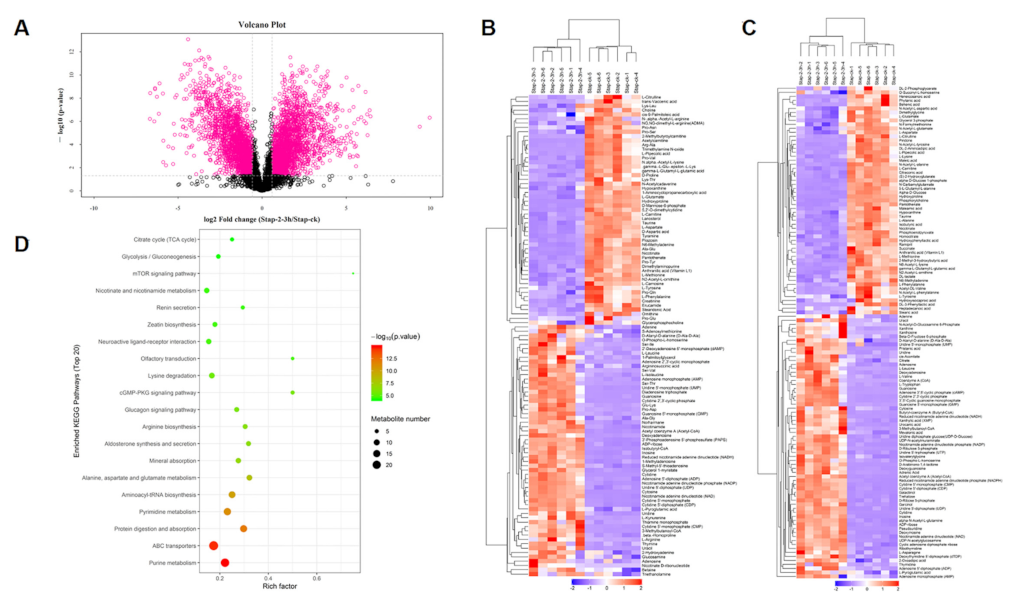
2.7. Metabolites Data of S.aureus Treated with Actinomycin D
2.7.1. Analysis of Main Metabolites
2.7.2. Analysis of the Main Metabolic Pathways
2.8. Proteomics–Metabolomics Correlation Network in S.aureus Treated with Actinomycin D
3. Discussion
4. Materials and Methods
4.1. Isolation and Identificationof Actinomycin D
4.2. Fermentation and Growth of S. aureus
4.3. MIC and MBC of Actinomycin D
4.4. Growth Curve of S. aureus Affected by Actinomycin D
4.5. Scanning Electron Microscopy (SEM) Analysis
4.6. Intracellular Detection of Reactive Oxygen Species (ROS)
4.7. Detection of H2O2
4.8. Label-Free Quantitative Proteomics
4.9. Untargeted Metabolomics Methods
4.10. Statistical Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Otto, M. Staphylococcal Infections: Mechanisms of Biofilm Maturation and Detachment as Critical Determinants of Pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef]
- Dong, L.-M.; Huang, L.-L.; Dai, H.; Xu, Q.-L.; Ouyang, J.-K.; Jia, X.-C.; Gu, W.-X.; Tan, J.-W. Anti-MRSA Sesquiterpenes from the Semi-Mangrove Plant Myoporum bontioides A. Gray. Mar. Drugs 2018, 16, 438. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Shen, H.; Wang, S.; Cao, X.; Xu, H.; Xia, Y.; Bai, T.; Liu, Y.; Peng, L.; Li, C.; et al. Spermine increases bactericidal activity of silver-nanoparticles against clinical methicillin-resistant Staphylococcus aureus. Chin. Chem. Lett. 2018, 29, 1824–1828. [Google Scholar] [CrossRef]
- Miller, L.S.; Fowler, V.G.; Shukla, S.K.; Rose, W.; Proctor, R. Development of a vaccine against Staphylococcus aureus invasive infections: Evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol. Rev. 2020, 44, 123–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassetti, M.; Carnelutti, A.; Righi, E. The role of methicillin-resistant Staphylococcus aureus in skin and soft tissue infections. Curr. Opin. Infect. Dis. 2017, 30, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Sharafutdinov, I.S.; Pavlova, A.S.; Akhatova, F.S.; Khabibrakhmanova, A.M.; Rozhina, E.V.; Romanova, Y.J.; Fakhrullin, R.; Lodochnikova, O.A.; Kurbangalieva, A.R.; Bogachev, M.I.; et al. Unraveling the Molecular Mechanism of Selective Antimicrobial Activity of 2(5H)-Furanone Derivative against Staphylococcus aureus. Int. J. Mol. Sci. 2019, 20, 694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Hal, S.J.; Jensen, S.; Vaska, V.L.; Espedido, B.A.; Paterson, D.L.; Gosbell, I.B. Predictors of Mortality in Staphylococcus aureus Bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Kim, E.; Choi, H.; Lee, J. Collismycin C from the micronesian marine bacterium Streptomyces sp. MC025 inhibits Staphylococcus aureus biofilm formation. Mar. Drugs. 2017, 15, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.; Kwon, O.-S.; Chung, B.; Lee, J.; Sun, J.; Shin, J.; Oh, K.-B. Antibacterial Activity of Chromomycins from a Marine-Derived Streptomyces microflavus. Mar. Drugs 2020, 18, 522. [Google Scholar] [CrossRef]
- De Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune evasion by Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 2. [Google Scholar] [CrossRef]
- Schrey, S.D.; Tarkka, M.T. Friends and foes: Streptomycetes as modulators of plant disease and symbiosis. Antonie van Leeuwenhoek 2008, 94, 11–19. [Google Scholar] [CrossRef]
- Ferraiuolo, S.B.; Cammarota, M.; Schiraldi, C.; Restaino, O.F. Streptomycetes as platform for biotechnological production processes of drugs. Appl. Microbiol. Biotechnol. 2021, 105, 551–568. [Google Scholar] [CrossRef]
- Lee, M.Y.; Kim, H.Y.; Lee, S.; Kim, J.-G.; Suh, J.-W.; Lee, C.H. Metabolomics-Based Chemotaxonomic Classification of Streptomyces spp. and Its Correlation with Antibacterial Activity. J. Microbiol. Biotechnol. 2015, 25, 1265–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortes, C.; Veiga, S.R.; Almacellas, E.; Hernández-Losa, J.; Ferreres, J.C.; Kozma, S.C.; Ambrosio, S.; Thomas, G.; Tauler, A. Effect of low doses of actinomycin D on neuroblastoma cell lines. Mol. Cancer. 2016, 4, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, Y.; Xie, T.; Zeng, H.; Chen, W.; Wan, C.; Zhang, L. Streptomyces -derived actinomycin D inhibits biofilm formation via downregulating ica locus and decreasing production of PIA in Staphylococcus epidermidis. J. Appl. Microbiol. 2019, 128, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.G.; Lee, K.; Kim, C.J.; Park, D.J.; Ju, Y.; Lee, J.C.; Wood, T.K.; Lee, J. Streptomyces-derived actinomycin D inhibits biofilm formation by Staphylococcus aureus and its hemolytic activity. Biofouling 2016, 32, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-S.; Tseng, W.-H.; Chuang, C.-Y.; Hou, M.-H. The structural basis of actinomycin D–binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013, 41, 4284–4294. [Google Scholar] [CrossRef]
- Sobell, H.M.; Jain, S. Stereochemistry of actinomycin binding to DNA: II. Detailed molecular model of actinomycin-DNA complex and its implications. J. Mol. Biol. 1972, 68, 21–34. [Google Scholar] [CrossRef]
- Kamitori, S.; Takusagawa, F. Crystal structure of the 2:1 complex between d(GAAGCTTC) and the anticancer drug actinomycin D. J. Mol. Biol. 1992, 225, 445–456. [Google Scholar] [CrossRef]
- Rhee, S.G. Redox signaling: Hydrogen peroxide as intracellular messenger. Exp. Mol. Med. 1999, 31, 53–59. [Google Scholar] [CrossRef] [Green Version]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Biological Production, Detection, and Fate of Hydrogen Peroxide. Antioxid. Redox Sign. 2018, 29, 541–551. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.; Holden, M. Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 2004, 12, 378–385. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [Green Version]
- Zhou, K.; Li, C.; Chen, D.; Pan, Y.; Tao, Y.; Qu, W.; Liu, Z.; Wang, X.; Xie, S. A review on nanosystems as an effective approach against infections of Staphylococcus aureus. Int. J. Nanomed. 2018, 13, 7333–7347. [Google Scholar] [CrossRef] [Green Version]
- Sharafutdinov, I.; Shigapova, Z.; Baltin, M.; Akhmetov, N.; Bogachev, M.; Kayumov, A. HtrA Protease from Bacillus subtilis Suppresses the Bacterial Fouling of the Rat Skin Injuries. BioNanoScience 2016, 6, 564–567. [Google Scholar] [CrossRef]
- Sharafutdinov, I.; Trizna, E.Y.; Baidamshina, D.R.; Ryzhikova, M.N.; Sibgatullina, R.; Khabibrakhmanova, A.M.; Latypova, L.Z.; Kurbangalieva, A.R.; Rozhina, E.V.; Klinger-Strobel, M.; et al. Antimicrobial Effects of Sulfonyl Derivative of 2(5H)-Furanone against Planktonic and Biofilm Associated Methicillin-Resistant and -Susceptible Staphylococcus aureus. Front. Microbiol. 2017, 8, 2246. [Google Scholar] [CrossRef]
- Kong, C.; Chee, C.-F.; Richter, K.; Thomas, N.; Rahman, N.A.; Nathan, S. Suppression of Staphylococcus aureus biofilm formation and virulence by a benzimidazole derivative, UM-C162. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Messner, K.; Imlay, J.A. The Identification of Primary Sites of Superoxide and Hydrogen Peroxide Formation in the Aerobic Respiratory Chain and Sulfite Reductase Complex of Escherichia coli. J. Biol. Chem. 1999, 274, 10119–10128. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-M.; Chen, H.; He, C.-L.; Wang, Q. Nitrogen Starvation Induced Oxidative Stress in an Oil-Producing Green Alga Chlorella sorokiniana C3. PLoS ONE 2013, 8, e69225. [Google Scholar] [CrossRef]
- Li, X.; Hu, H.Y.; Zhang, Y.P. Growth and lipid accumulation properties of a freshwater microalga Scenedesmus sp. under different cultivation temperature. Bioresource Technol. 2011, 102, 3098–3102. [Google Scholar]
- Yu, Y.; Li, T.; Wu, N.; Ren, L.; Jiang, L.; Ji, X.; Huang, H. Mechanism of Arachidonic Acid Accumulation during Aging in Mortierella alpina: A Large-Scale Label-Free Comparative Proteomics Study. J. Agric. Food Chem. 2016, 64, 9124–9134. [Google Scholar] [CrossRef]
- Candela, T.; Fouet, A. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 2006, 60, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Matasova, L.V.; Popova, T.N. Aconitate hydratase of mammals under oxidative stress. Biochemistry (Moscow) 2008, 73, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Hartuti, E.D.; Inaoka, D.K.; Komatsuya, K.; Miyazaki, Y.; Miller, R.J.; Xinying, W.; Sadikin, M.; Prabandari, E.E.; Waluyo, D.; Kuroda, M.; et al. Biochemical studies of membrane bound Plasmodium falciparum mitochondrial L-malate:quinone oxidoreductase, a potential drug target. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1859, 191–200. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, P.C. Functional Expression and Extension of Staphylococcal Staphyloxanthin Biosynthetic Pathway in Escherichia coli. J. Biol. Chem. 2012, 287, 21575–21583. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, N.A.; O’Bryan, C.A.; Martin, E.M.; Ricke, S.C.; Johnson, M.G.; Crandall, P.G. Further Evidence of How Unbuffered Starvation at 4 °C Influences Listeria monocytogenes EGD-e, HCC23, F2365, and Scott A. J. Food Prot. 2017, 80, 1749–1759. [Google Scholar] [CrossRef]
- Phang, J.M. Proline Metabolism in Cell Regulation and Cancer Biology: Recent Advances and Hypotheses. Antioxidants Redox Signal. 2019, 30, 635–649. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.-J.; Lin, C.-H.; Ren, E.-D.; Su, Y.; Zhu, W.-Y. Effects of Early Intervention with Maternal Fecal Bacteria and Antibiotics on Liver Metabolome and Transcription in Neonatal Pigs. Front. Physiol. 2019, 10, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubert, S.; Alban, C.; Bligny, R.; Douce, R. Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: Evidence for a role of MCCase in leucine catabolism. FEBS Lett. 1996, 383, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Pai, P.-J.; Zhang, W.; Hu, Y.; Dong, X.; Qian, P.-Y.; Chen, D.; Lam, H. Proteomic response of methicillin-resistant S. aureus to a synergistic antibacterial drug combination: A novel erythromycin derivative and oxacillin. Sci. Rep. 2016, 6, 19841. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Chen, H.; Hao, G.; Yang, B.; Feng, Y.; Wang, Y.; Feng, L.; Zhao, J.; Song, Y.; Zhang, H.; et al. Role of the Phenylalanine-Hydroxylating System in Aromatic Substance Degradation and Lipid Metabolism in the Oleaginous Fungus Mortierella alpina. Appl. Environ. Microbiol. 2013, 79, 3225–3233. [Google Scholar] [CrossRef] [Green Version]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellavalle, P.D.; Cabrera, A.; Alem, D.; Larrañaga, P.; Ferreira, F.; Rizza, M.D. Antifungal activity of medicinal plant extracts against phytopathogenic fungus Alternaria spp. Chil. J. Agric. Res. 2011, 71, 231–239. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, S.; Wahab, R.; Khan, F.; Mishra, Y.; Musarrat, J.; Al-Khedhairy, A. Reactive Oxygen Species Mediated Bacterial Biofilm Inhibition via Zinc Oxide Nanoparticles and Their Statistical Determination. PLoS ONE 2014, 9, e111289. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.; Huang, L.; Zhang, X.; Yang, J.; Chen, X.; Li, F.; Liu, J.; Huang, R. Multi-Omics Analysis Reveals Anti-Staphylococcus aureus Activity of Actinomycin D Originating from Streptomyces parvulus. Int. J. Mol. Sci. 2021, 22, 12231. https://doi.org/10.3390/ijms222212231
Lin Y, Huang L, Zhang X, Yang J, Chen X, Li F, Liu J, Huang R. Multi-Omics Analysis Reveals Anti-Staphylococcus aureus Activity of Actinomycin D Originating from Streptomyces parvulus. International Journal of Molecular Sciences. 2021; 22(22):12231. https://doi.org/10.3390/ijms222212231
Chicago/Turabian StyleLin, Yuqi, Li Huang, Xiaoyong Zhang, Jiajia Yang, Xiaodan Chen, Fengming Li, Jun Liu, and Riming Huang. 2021. "Multi-Omics Analysis Reveals Anti-Staphylococcus aureus Activity of Actinomycin D Originating from Streptomyces parvulus" International Journal of Molecular Sciences 22, no. 22: 12231. https://doi.org/10.3390/ijms222212231
APA StyleLin, Y., Huang, L., Zhang, X., Yang, J., Chen, X., Li, F., Liu, J., & Huang, R. (2021). Multi-Omics Analysis Reveals Anti-Staphylococcus aureus Activity of Actinomycin D Originating from Streptomyces parvulus. International Journal of Molecular Sciences, 22(22), 12231. https://doi.org/10.3390/ijms222212231






