Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease
Abstract
1. Introduction
2. Types of Epigenetic Mechanisms
2.1. DNA Methylation
2.2. Histone Modifications
2.3. Chromatin Remodeling Enzymes
2.4. Long Non-Coding RNAs
3. Inhibitors and Activators of Epigenetic Modifications
3.1. Pharmacological Manipulations
3.2. Genetic Manipulations of Epigenetic Modifications
4. Epigenetic Code in Synaptic Plasticity and Memory Formation
4.1. DNA Methylation, Synaptic Plasticity, and Memory Formation
4.2. Histone Modifications, Synaptic Plasticity, and Memory Formation
4.3. Chromatin Remodeling Enzymes and lncRNAs in Synaptic Plasticity and Memory Formation
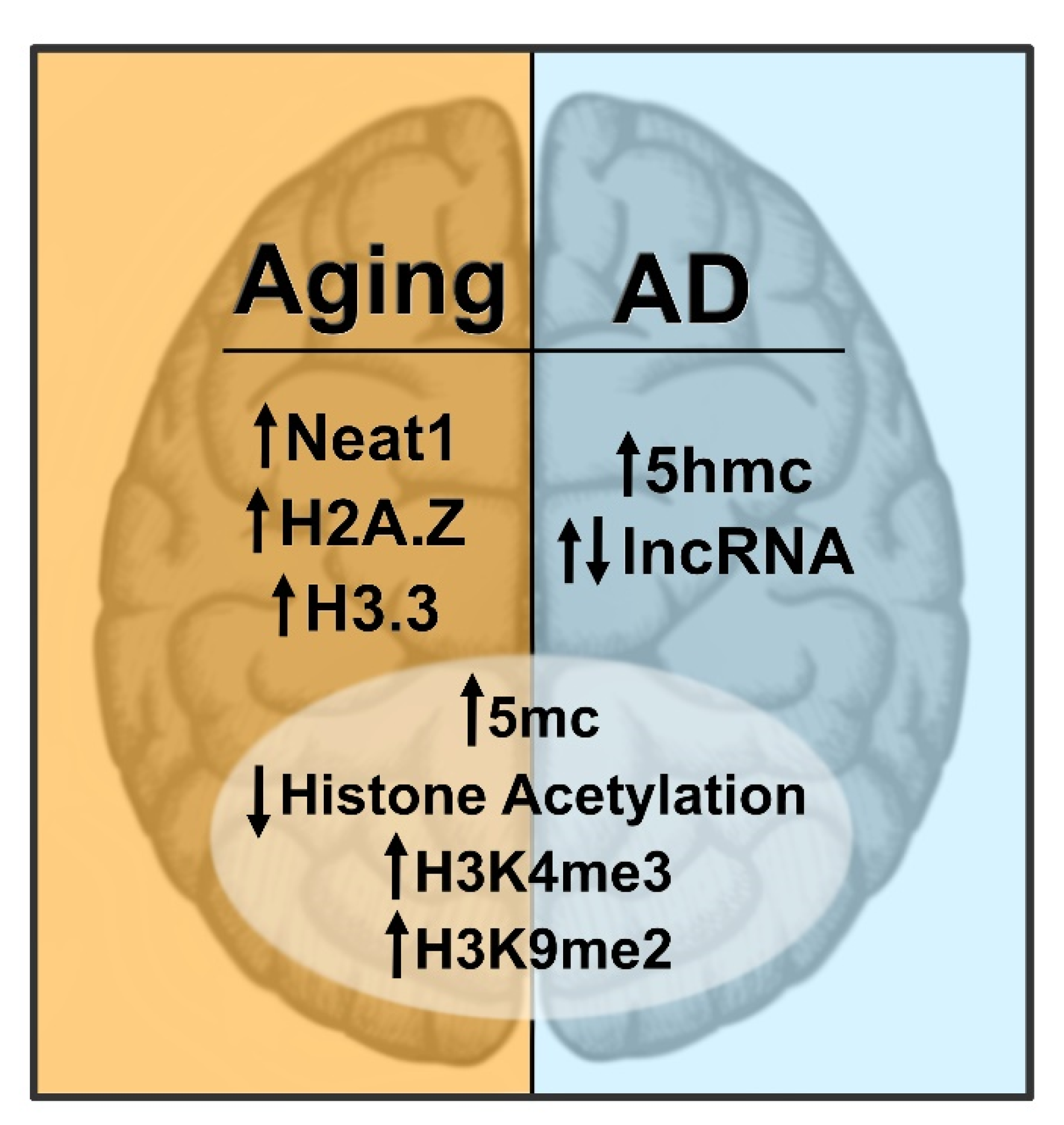
5. Epigenetics in Age-Related Memory Decline
6. Epigenetics in Alzheimer’s Disease-Related Memory Loss
7. Future Directions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waddington, C.H. The Strategy of the Genes; Routledge: England, UK, 2014. [Google Scholar]
- Deans, C.; Maggert, K.A. What do you mean, “epigenetic”? Genetics 2015, 199, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kaang, B.K. Epigenetic regulation and chromatin remodeling in learning and memory. Exp. Mol. Med. 2017, 49, e281. [Google Scholar] [CrossRef] [PubMed]
- Kwapis, J.L.; Wood, M.A. Epigenetic mechanisms in fear conditioning: Implications for treating post-traumatic stress disorder. Trends Neurosci. 2014, 37, 706–720. [Google Scholar] [CrossRef]
- Creighton, S.D.; Stefanelli, G.; Reda, A.; Zovkic, I.B. Epigenetic Mechanisms of Learning and Memory: Implications for Aging. Int. J. Mol. Sci. 2020, 21, 6918. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Aromolaran, K.A.; Zukin, R.S. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat. Rev. Neurosci. 2017, 18, 347–361. [Google Scholar] [CrossRef]
- Quina, A.; Buschbeck, M.; Di Croce, L. Chromatin structure and epigenetics. Biochem. Pharmacol. 2006, 72, 1563–1569. [Google Scholar] [CrossRef]
- Happel, N.; Doenecke, D. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene 2009, 431, 1–12. [Google Scholar] [CrossRef]
- Arney, K.L.; Fisher, A.G. Epigenetic aspects of differentiation. J. Cell Sci. 2004, 117, 4355–4363. [Google Scholar] [CrossRef][Green Version]
- Chiang, P.K.; Gordon, R.K.; Tal, J.; Zeng, G.; Doctor, B.; Pardhasaradhi, K.; McCann, P.P. S-Adenosylmetliionine and methylation. FASEB J. 1996, 10, 471–480. [Google Scholar] [CrossRef]
- Turker, M.S. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin. Cancer Biol. 1999, 9, 329–337. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Price, J.C.; Guan, S.; Burlingame, A.; Prusiner, S.B.; Ghaemmaghami, S. Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. USA 2010, 107, 14508–14513. [Google Scholar] [CrossRef]
- Nakao, M. Epigenetics: Interaction of DNA methylation and chromatin. Gene 2001, 278, 25–31. [Google Scholar] [CrossRef]
- Iguchi-Ariga, S.M.; Schaffner, W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989, 3, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Karymov, M.A.; Tomschik, M.; Leuba, S.H.; Caiafa, P.; Zlatanova, J. DNA methylation-dependent chromatin fiber compaction in vivo and in vitro: Requirement for linker histone. FASEB J. 2001, 15, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Fuks, F.; Hurd, P.J.; Wolf, D.; Nan, X.; Bird, A.P.; Kouzarides, T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003, 278, 4035–4040. [Google Scholar] [CrossRef]
- Drewell, R.A.; Goddard, C.J.; Thomas, J.O.; Surani, M.A. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res. 2002, 30, 1139–1144. [Google Scholar] [CrossRef] [PubMed]
- Chahrour, M.; Jung, S.Y.; Shaw, C.; Zhou, X.; Wong, S.T.; Qin, J.; Zoghbi, H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 2008, 320, 1224–1229. [Google Scholar] [CrossRef]
- Cohen, S.; Zhou, Z.; Greenberg, M.E. Medicine. Activating a repressor. Science 2008, 320, 1172–1173. [Google Scholar] [CrossRef]
- Chen, T.; Ueda, Y.; Xie, S.; Li, E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002, 277, 38746–38754. [Google Scholar] [CrossRef]
- Kotini, A.G.; Mpakali, A.; Agalioti, T. Dnmt3a1 upregulates transcription of distinct genes and targets chromosomal gene clusters for epigenetic silencing in mouse embryonic stem cells. Mol. Cell Biol. 2011, 31, 1577–1592. [Google Scholar] [CrossRef]
- Mellen, M.; Ayata, P.; Heintz, N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci. USA 2017, 114, E7812–E7821. [Google Scholar] [CrossRef]
- Jang, H.S.; Shin, W.J.; Lee, J.E.; Do, J.T. CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function. Genes 2017, 8, 148. [Google Scholar] [CrossRef]
- Lee, J.H.; Saito, Y.; Park, S.J.; Nakai, K. Existence and possible roles of independent non-CpG methylation in the mammalian brain. DNA Res. 2020, 27, dsaa020. [Google Scholar] [CrossRef] [PubMed]
- Kangaspeska, S.; Stride, B.; Métivier, R.; Polycarpou-Schwarz, M.; Ibberson, D.; Carmouche, R.P.; Benes, V.; Gannon, F.; Reid, G. Transient cyclical methylation of promoter DNA. Nature 2008, 452, 112–115. [Google Scholar] [CrossRef]
- Métivier, R.; Gallais, R.; Tiffoche, C.; Le Péron, C.; Jurkowska, R.Z.; Carmouche, R.P.; Ibberson, D.; Barath, P.; Demay, F.; Reid, G.; et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008, 452, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Ma, D.K.; Mo, H.; Ball, M.P.; Jang, M.H.; Bonaguidi, M.A.; Balazer, J.A.; Eaves, H.L.; Xie, B.; Ford, E.; et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011, 14, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.L.; Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef]
- Barrès, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Mühlbacher, F.; Schiessel, H.; Holm, C. Tail-induced attraction between nucleosome core particles. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2006, 74, 031919. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Mde, L.; Gutierrez, C. Novel insights into the plant histone code: Lessons from ORC1. Epigenetics 2009, 4, 205–208. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanner, K.G.; Trievel, R.C.; Kuo, M.H.; Howard, R.M.; Berger, S.L.; Allis, C.D.; Marmorstein, R.; Denu, J.M. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 1999, 274, 18157–18160. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.G.; Langer, M.R.; Denu, J.M. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 2000, 39, 11961–11969. [Google Scholar] [CrossRef]
- Tanner, K.G.; Langer, M.R.; Kim, Y.; Denu, J.M. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem. 2000, 275, 22048–22055. [Google Scholar] [CrossRef]
- Lau, O.D.; Courtney, A.D.; Vassilev, A.; Marzilli, L.A.; Cotter, R.J.; Nakatani, Y.; Cole, P.A. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. Kinetic analysis of the catalytic mechanism. J. Biol. Chem. 2000, 275, 21953–21959. [Google Scholar] [CrossRef]
- Hebbes, T.R.; Thorne, A.W.; Crane-Robinson, C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988, 7, 1395–1402. [Google Scholar] [CrossRef]
- Mujtaba, S.; Zeng, L.; Zhou, M.M. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 2007, 26, 5521–5527. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.M.M.; Wood, M.A.; McDonough, C.B.; Abel, T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn. Mem. 2007, 14, 564–572. [Google Scholar] [CrossRef][Green Version]
- Alarcón, J.M.; Malleret, G.; Touzani, K.; Vronskaya, S.; Ishii, S.; Kandel, E.R.; Barco, A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 2004, 42, 947–959. [Google Scholar] [CrossRef]
- Korzus, E.; Rosenfeld, M.G.; Mayford, M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 2004, 42, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Sun, Y.E. To learn better, keep the HAT on. Neuron 2004, 42, 879–881. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vecsey, C.G.; Hawk, J.D.; Lattal, K.M.; Stein, J.M.; Fabian, S.A.; Attner, M.A.; Cabrera, S.M.; McDonough, C.B.; Brindle, P.K.; Abel, T.; et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 2007, 27, 6128–6140. [Google Scholar] [CrossRef] [PubMed]
- Murray, K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry 1964, 3, 10–15. [Google Scholar] [CrossRef]
- Cheung, P.; Lau, P. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 2005, 19, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.H.; Schübeler, D. Methylation of histones: Playing memory with DNA. Curr. Opin. Cell Biol. 2005, 17, 230–238. [Google Scholar] [CrossRef]
- Binda, O.; LeRoy, G.; Bua, D.J.; Garcia, B.A.; Gozani, O.; Richard, S. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: Regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 2010, 5, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, E.M.; Inglis, R.J.; Matthews, H.R.; Sarner, N. Phosphorylation of very-lysine-rich histone in Physarum polycephalum. Correlation with chromosome condensation. Eur. J. Biochem. 1973, 33, 131–139. [Google Scholar] [CrossRef]
- Gurley, L.R.; Walters, R.A.; Tobey, R.A. Cell cycle-specific changes in histone phosphorylation associated with cell proliferation and chromosome condensation. J. Cell Biol. 1974, 60, 356–364. [Google Scholar] [CrossRef]
- Gurley, L.R.; D’Anna, J.A.; Barham, S.S.; Deaven, L.L.; Tobey, R.A. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 1978, 84, 1–15. [Google Scholar] [CrossRef]
- Mahadevan, L.C.; Willis, A.C.; Barratt, M.J. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 1991, 65, 775–783. [Google Scholar] [CrossRef]
- Sassone-Corsi, P.; Mizzen, C.A.; Cheung, P.; Crosio, C.; Monaco, L.; Jacquot, S.; Hanauer, A.; Allis, C.D. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 1999, 285, 886–891. [Google Scholar] [CrossRef]
- Thomson, S.; Clayton, A.L.; Hazzalin, C.A.; Rose, S.; Barratt, M.J.; Mahadevan, L.C. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999, 18, 4779–4793. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Sun, Z.W.; Li, X.; Reuben, M.; Tatchell, K.; Bishop, D.K.; Grushcow, J.M.; Brame, C.J.; Caldwell, J.A.; Hunt, D.F.; et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 2000, 102, 279–291. [Google Scholar] [CrossRef]
- Goto, H.; Yasui, Y.; Nigg, E.A.; Inagaki, M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 2002, 7, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Ajiro, K.; Yoda, K.; Utsumi, K.; Nishikawa, Y. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 1996, 271, 13197–13201. [Google Scholar] [CrossRef] [PubMed]
- Nowak, S.J.; Pai, C.-Y.; Corces, V.G. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 2003, 23, 6129–6138. [Google Scholar] [CrossRef] [PubMed]
- Musaus, M.; Navabpour, S.; Jarome, T.J. The diversity of linkage-specific polyubiquitin chains and their role in synaptic plasticity and memory formation. Neurobiol. Learn. Mem. 2020, 174, 107286. [Google Scholar] [CrossRef]
- Goldknopf, I.L.; Taylor, C.W.; Baum, R.M.; Yeoman, L.C.; Olson, M.O.; Prestayko, A.W.; Busch, H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975, 250, 7182–7187. [Google Scholar] [CrossRef]
- West, M.H.; Bonner, W.M. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980, 8, 4671–4680. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Sun, J.M.; Zhang, Y.; Davie, J.R.; Meistrich, M.L. Ubiquitination of histone H3 in elongating spermatids of rat testes. J. Biol. Chem. 1998, 273, 13165–13169. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.D.; Sauer, F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 2000, 289, 2357–2360. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Ishiguro, K.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef]
- Gearhart, M.D.; Corcoran, C.M.; Wamstad, J.A.; Bardwell, V.J. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 2006, 26, 6880–6889. [Google Scholar] [CrossRef]
- Sun, Z.W.; Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef]
- Ryu, H.Y.; Hochstrasser, M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef]
- Farrelly, L.A.; Thompson, R.E.; Zhao, S.; Lepack, A.E.; Lyu, Y.; Bhanu, N.V.; Zhang, B.; Loh, Y.E.; Ramakrishnan, A.; Vadodaria, K.C.; et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 2019, 567, 535–539. [Google Scholar] [CrossRef]
- Zhao, S.; Chuh, K.N.; Zhang, B.; Dul, B.E.; Thompson, R.E.; Farrelly, L.A.; Liu, X.; Xu, N.; Xue, Y.; Roeder, R.G.; et al. Histone H3Q5 serotonylation stabilizes H3K4 methylation and potentiates its readout. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Lepack, A.E.; Werner, C.T.; Stewart, A.F.; Fulton, S.L.; Zhong, P.; Farrelly, L.A.; Smith, A.C.W.; Ramakrishnan, A.; Lyu, Y.; Bastle, R.M.; et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 2020, 368, 197–201. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 2017, 18, 115–126. [Google Scholar] [CrossRef]
- Hanly, D.J.; Esteller, M.; Berdasco, M. Interplay between long non-coding RNAs and epigenetic machinery: Emerging targets in cancer? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170074. [Google Scholar] [CrossRef]
- Butler, A.A.; Johnston, D.R.; Kaur, S.; Lubin, F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, J.; Wang, M.; Li, X.; Gong, H.; Tang, H.; Chen, L.; Wan, L.; Liu, Q. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 2018, 9, 1726. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef] [PubMed]
- Stresemann, C.; Brueckner, B.; Musch, T.; Stopper, H.; Lyko, F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006, 66, 2794–2800. [Google Scholar] [CrossRef] [PubMed]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef]
- Zhou, L.; Cheng, X.; Connolly, B.A.; Dickman, M.J.; Hurd, P.J.; Hornby, D.P. Zebularine: A novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 2002, 321, 591–599. [Google Scholar] [CrossRef]
- Liu, K.; Wang, Y.F.; Cantemir, C.; Muller, M.T. Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell Biol. 2003, 23, 2709–2719. [Google Scholar] [CrossRef][Green Version]
- Weisenberger, D.J.; Velicescu, M.; Cheng, J.C.; Gonzales, F.A.; Liang, G.; Jones, P.A. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2004, 2, 62–72. [Google Scholar]
- Momparler, R.L. Pharmacology of 5-Aza-2’-deoxycytidine (decitabine). Semin. Hematol. 2005, 42, S9–S16. [Google Scholar] [CrossRef]
- Issa, J.P. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: Dose, duration, and patient selection. Nat. Clin. Pract. Oncol. 2005, 2 (Suppl. 1), S24–S29. [Google Scholar] [CrossRef]
- Gore, S.D.; Baylin, S.; Sugar, E.; Carraway, H.; Miller, C.B.; Carducci, M.; Grever, M.; Galm, O.; Dauses, T.; Karp, J.E.; et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006, 66, 6361–6369. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, K.; Bai, S. DNA methyltransferases as targets for cancer therapy. Drugs Today 2007, 43, 395–422. [Google Scholar] [CrossRef] [PubMed]
- Tsujioka, T.; Yokoi, A.; Uesugi, M.; Kishimoto, M.; Tochigi, A.; Suemori, S.; Tohyama, Y.; Tohyama, K. Effects of DNA methyltransferase inhibitors (DNMTIs) on MDS-derived cell lines. Exp. Hematol. 2013, 41, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. The CBP co-activator is a histone acetyltransferase. Nature 1996, 384, 641–643. [Google Scholar] [CrossRef]
- Ogryzko, V.V.; Schiltz, R.L.; Russanova, V.; Howard, B.H.; Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996, 87, 953–959. [Google Scholar] [CrossRef]
- Yang, X.J.; Ogryzko, V.V.; Nishikawa, J.; Howard, B.H.; Nakatani, Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 1996, 382, 319–324. [Google Scholar] [CrossRef]
- Mizzen, C.A.; Allis, C.D. Linking histone acetylation to transcriptional regulation. Cell Mol. Life Sci. 1998, 54, 6–20. [Google Scholar] [CrossRef]
- Struhl, K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998, 12, 599–606. [Google Scholar] [CrossRef]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef]
- Imhof, A.; Yang, X.J.; Ogryzko, V.V.; Nakatani, Y.; Wolffe, A.P.; Ge, H. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 1997, 7, 689–692. [Google Scholar] [CrossRef]
- Vetting, M.W.; LP, S.d.C.; Yu, M.; Hegde, S.S.; Magnet, S.; Roderick, S.L.; Blanchard, J.S. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005, 433, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Cullis, P.M.; Wolfenden, R.; Cousens, L.S.; Alberts, B.M. Inhibition of histone acetylation by N-[2-(S-coenzyme A)acetyl] spermidine amide, a multisubstrate analog. J. Biol. Chem. 1982, 257, 12165–12169. [Google Scholar] [CrossRef]
- Erwin, B.G.; Persson, L.; Pegg, A.E. Differential inhibition of histone and polyamine acetylases by multisubstrate analogues. Biochemistry 1984, 23, 4250–4255. [Google Scholar] [CrossRef]
- Balasubramanyam, K.; Swaminathan, V.; Ranganathan, A.; Kundu, T.K. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 2003, 278, 19134–19140. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanyam, K.; Varier, R.A.; Altaf, M.; Swaminathan, V.; Siddappa, N.B.; Ranga, U.; Kundu, T.K. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004, 279, 51163–51171. [Google Scholar] [CrossRef] [PubMed]
- Bowers, E.M.; Yan, G.; Mukherjee, C.; Orry, A.; Wang, L.; Holbert, M.A.; Crump, N.T.; Hazzalin, C.A.; Liszczak, G.; Yuan, H.; et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem. Biol. 2010, 17, 471–482. [Google Scholar] [CrossRef]
- Marks, P.A.; Richon, V.M.; Rifkind, R.A. Histone deacetylase inhibitors: Inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 2000, 92, 1210–1216. [Google Scholar] [CrossRef] [PubMed]
- Thiagalingam, S.; Cheng, K.H.; Lee, H.J.; Mineva, N.; Thiagalingam, A.; Ponte, J.F. Histone deacetylases: Unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 2003, 983, 84–100. [Google Scholar] [CrossRef] [PubMed]
- Dokmanovic, M.; Clarke, C.; Marks, P.A. Histone deacetylase inhibitors: Overview and perspectives. Mol. Cancer Res. 2007, 5, 981–989. [Google Scholar] [CrossRef]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Ruiz-Llorente, L.; Sánchez-Martínez, R.; García, L.; Zambrano, A.; Aranda, A. Histone deacetylase inhibitors: Mechanism of action and therapeutic use in cancer. Clin. Transl. Oncol. 2008, 10, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Ishikawa, C.; Senba, M.; Kimura, M.; Okano, Y. Effects of AZD1152, a selective Aurora B kinase inhibitor, on Burkitt’s and Hodgkin’s lymphomas. Biochem. Pharm. 2011, 81, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Cui, Y.; Lubecka, K.; Stefanska, B.; Irudayaraj, J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016, 7, 46545–46556. [Google Scholar] [CrossRef]
- Stepper, P.; Kungulovski, G.; Jurkowska, R.Z.; Chandra, T.; Krueger, F.; Reinhardt, R.; Reik, W.; Jeltsch, A.; Jurkowski, T.P. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res. 2017, 45, 1703–1713. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Krzisch, M.; Wu, X.; Graef, J.; Muffat, J.; Hnisz, D.; Li, C.H.; Yuan, B.; Xu, C.; et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018, 172, 979–992.e6. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Zhao, Y.T.; Lamonica, J.M.; Zhou, Z. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nat. Commun. 2017, 8, 15315. [Google Scholar] [CrossRef]
- Savell, K.E.; Bach, S.V.; Zipperly, M.E.; Revanna, J.S.; Goska, N.A.; Tuscher, J.J.; Duke, C.G.; Sultan, F.A.; Burke, J.N.; Williams, D.; et al. A Neuron-Optimized CRISPR/dCas9 Activation System for Robust and Specific Gene Regulation. eNeuro 2019, 6. [Google Scholar] [CrossRef]
- Shechner, D.M.; Hacisuleyman, E.; Younger, S.T.; Rinn, J.L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 2015, 12, 664–670. [Google Scholar] [CrossRef]
- Griffith, J.S.; Mahler, H.R. DNA ticketing theory of memory. Nature 1969, 223, 580–582. [Google Scholar] [CrossRef]
- Crick, F. Memory and molecular turnover. Nature 1984, 312, 101. [Google Scholar] [CrossRef]
- Holliday, R. Is there an epigenetic component in long-term memory? J. Biol. 1999, 200, 339–341. [Google Scholar] [CrossRef]
- Feng, J.; Chang, H.; Li, E.; Fan, G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005, 79, 734–746. [Google Scholar] [CrossRef]
- Veldic, M.; Guidotti, A.; Maloku, E.; Davis, J.M.; Costa, E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc. Natl. Acad. Sci. USA 2005, 102, 2152–2157. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Fan, G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 2009, 89, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Hutnick, L.K.; Golshani, P.; Namihira, M.; Xue, Z.; Matynia, A.; Yang, X.W.; Silva, A.J.; Schweizer, F.E.; Fan, G. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum. Mol. Genet. 2009, 18, 2875–2888. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.J.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Veldic, M.; Caruncho, H.J.; Liu, W.S.; Davis, J.; Satta, R.; Grayson, D.R.; Guidotti, A.; Costa, E. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc. Natl. Acad. Sci. USA 2004, 101, 348–353. [Google Scholar] [CrossRef]
- Miller, C.A.; Sweatt, J.D. Covalent modification of DNA regulates memory formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef]
- Lubin, F.D.; Roth, T.L.; Sweatt, J.D. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008, 28, 10576–10586. [Google Scholar] [CrossRef]
- Levenson, J.M.; Roth, T.L.; Lubin, F.D.; Miller, C.A.; Huang, I.C.; Desai, P.; Malone, L.M.; Sweatt, J.D. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006, 281, 15763–15773. [Google Scholar] [CrossRef]
- Chen, W.G.; Chang, Q.; Lin, Y.; Meissner, A.; West, A.E.; Griffith, E.C.; Jaenisch, R.; Greenberg, M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 2003, 302, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Frankland, P.W.; Bontempi, B.; Talton, L.E.; Kaczmarek, L.; Silva, A.J. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science 2004, 304, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; Gavin, C.F.; White, J.A.; Parrish, R.R.; Honasoge, A.; Yancey, C.R.; Rivera, I.M.; Rubio, M.D.; Rumbaugh, G.; Sweatt, J.D. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010, 13, 664–666. [Google Scholar] [CrossRef]
- Barreto, G.; Schäfer, A.; Marhold, J.; Stach, D.; Swaminathan, S.K.; Handa, V.; Döderlein, G.; Maltry, N.; Wu, W.; Lyko, F.; et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 2007, 445, 671–675. [Google Scholar] [CrossRef]
- Ma, D.K.; Jang, M.H.; Guo, J.U.; Kitabatake, Y.; Chang, M.L.; Pow-Anpongkul, N.; Flavell, R.A.; Lu, B.; Ming, G.L.; Song, H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009, 323, 1074–1077. [Google Scholar] [CrossRef]
- Jarome, T.J.; Butler, A.A.; Nichols, J.N.; Pacheco, N.L.; Lubin, F.D. NF-kappaB mediates Gadd45beta expression and DNA demethylation in the hippocampus during fear memory formation. Front. Mol. Neurosci. 2015, 8, 54. [Google Scholar] [CrossRef]
- Wood, M.A.; Hawk, J.D.; Abel, T. Combinatorial chromatin modifications and memory storage: A code for memory? Learn. Mem. 2006, 13, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.D.; Sweatt, J.D. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron 2007, 55, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.M.; Wood, M.A. Beyond transcription factors: The role of chromatin modifying enzymes in regulating transcription required for memory. Learn. Mem. 2008, 15, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Gräff, J.; Mansuy, I.M. Epigenetic codes in cognition and behaviour. Behav. Brain Res. 2008, 192, 70–87. [Google Scholar] [CrossRef]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone methylation regulates memory formation. J. Neurosci. 2010, 30, 3589–3599. [Google Scholar] [CrossRef]
- Levenson, J.M.; O’Riordan, K.J.; Brown, K.D.; Trinh, M.A.; Molfese, D.L.; Sweatt, J.D. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004, 279, 40545–40559. [Google Scholar] [CrossRef]
- Levenson, J.M.; Sweatt, J.D. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005, 6, 108–118. [Google Scholar] [CrossRef]
- Reul, J.M.; Chandramohan, Y. Epigenetic mechanisms in stress-related memory formation. Psychoneuroendocrinology 2007, 32 (Suppl. 1), S21–S25. [Google Scholar] [CrossRef]
- Roth, T.L.; Sweatt, J.D. Regulation of chromatin structure in memory formation. Curr. Opin. Neurobiol. 2009, 19, 336–342. [Google Scholar] [CrossRef]
- Kandel, E.R. The molecular biology of memory storage: A dialogue between genes and synapses. Science 2001, 294, 1030–1038. [Google Scholar] [CrossRef]
- Guan, Z.; Giustetto, M.; Lomvardas, S.; Kim, J.H.; Miniaci, M.C.; Schwartz, J.H.; Thanos, D.; Kandel, E.R. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 2002, 111, 483–493. [Google Scholar] [CrossRef]
- Federman, N.; Fustiñana, M.S.; Romano, A. Histone acetylation is recruited in consolidation as a molecular feature of stronger memories. Learn. Mem. 2009, 16, 600–606. [Google Scholar] [CrossRef]
- Peixoto, L.; Abel, T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 2013, 38, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Petrij, F.; Giles, R.H.; Dauwerse, H.G.; Saris, J.J.; Hennekam, R.C.; Masuno, M.; Tommerup, N.; van Ommen, G.J.; Goodman, R.H.; Peters, D.J.; et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 1995, 376, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Maurice, T.; Duclot, F.; Meunier, J.; Naert, G.; Givalois, L.; Meffre, J.; Célérier, A.; Jacquet, C.; Copois, V.; Mechti, N.; et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology 2008, 33, 1584–1602. [Google Scholar] [CrossRef] [PubMed]
- Fontán-Lozano, A.; Romero-Granados, R.; Troncoso, J.; Múnera, A.; Delgado-García, J.M.; Carrión, A.M. Histone deacetylase inhibitors improve learning consolidation in young and in KA-induced-neurodegeneration and SAMP-8-mutant mice. Mol. Cell. Neurosci. 2008, 39, 193–201. [Google Scholar] [CrossRef]
- Bredy, T.W.; Wu, H.; Crego, C.; Zellhoefer, J.; Sun, Y.E.; Barad, M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn. Mem. 2007, 14, 268–276. [Google Scholar] [CrossRef]
- Stefanko, D.P.; Barrett, R.M.; Ly, A.R.; Reolon, G.K.; Wood, M.A. Modulation of long-term memory for object recognition via HDAC inhibition. Proc. Natl. Acad. Sci. USA 2009, 106, 9447–9452. [Google Scholar] [CrossRef]
- Koshibu, K.; Gräff, J.; Beullens, M.; Heitz, F.D.; Berchtold, D.; Russig, H.; Farinelli, M.; Bollen, M.; Mansuy, I.M. Protein phosphatase 1 regulates the histone code for long-term memory. J. Neurosci. 2009, 29, 13079–13089. [Google Scholar] [CrossRef]
- Miller, C.A.; Campbell, S.L.; Sweatt, J.D. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn. Mem. 2008, 89, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Abel, T.; Martin, K.C.; Bartsch, D.; Kandel, E.R. Memory suppressor genes: Inhibitory constraints on the storage of long-term memory. Science 1998, 279, 338–341. [Google Scholar] [CrossRef]
- Guan, J.S.; Haggarty, S.J.; Giacometti, E.; Dannenberg, J.H.; Joseph, N.; Gao, J.; Nieland, T.J.; Zhou, Y.; Wang, X.; Mazitschek, R.; et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 2009, 459, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Chwang, W.B.; Arthur, J.S.; Schumacher, A.; Sweatt, J.D. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J. Neurosci. 2007, 27, 12732–12742. [Google Scholar] [CrossRef]
- Jakovcevski, M.; Ruan, H.; Shen, E.Y.; Dincer, A.; Javidfar, B.; Ma, Q.; Peter, C.J.; Cheung, I.; Mitchell, A.C.; Jiang, Y.; et al. Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. J. Neurosci. 2015, 35, 5097–5108. [Google Scholar] [CrossRef] [PubMed]
- Webb, W.M.; Sanchez, R.G.; Perez, G.; Butler, A.A.; Hauser, R.M.; Rich, M.C.; O’Bierne, A.L.; Jarome, T.J.; Lubin, F.D. Dynamic association of epigenetic H3K4me3 and DNA 5hmC marks in the dorsal hippocampus and anterior cingulate cortex following reactivation of a fear memory. Neurobiol. Learn. Mem. 2017, 142, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Gupta-Agarwal, S.; Franklin, A.V.; Deramus, T.; Wheelock, M.; Davis, R.L.; McMahon, L.L.; Lubin, F.D. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J. Neurosci. 2012, 32, 5440–5453. [Google Scholar] [CrossRef]
- Jarome, T.J.; Perez, G.A.; Hauser, R.M.; Hatch, K.M.; Lubin, F.D. EZH2 Methyltransferase Activity Controls Pten Expression and mTOR Signaling during Fear Memory Reconsolidation. J. Neurosci. 2018, 38, 7635–7648. [Google Scholar] [CrossRef]
- Zhang, J.; Ji, F.; Liu, Y.; Lei, X.; Li, H.; Ji, G.; Yuan, Z.; Jiao, J. Ezh2 regulates adult hippocampal neurogenesis and memory. J. Neurosci. 2014, 34, 5184–5199. [Google Scholar] [CrossRef]
- Gupta-Agarwal, S.; Jarome, T.J.; Fernandez, J.; Lubin, F.D. NMDA receptor- and ERK-dependent histone methylation changes in the lateral amygdala bidirectionally regulate fear memory formation. Learn. Mem. 2014, 21, 351–362. [Google Scholar] [CrossRef]
- Jarome, T.J.; Perez, G.A.; Webb, W.M.; Hatch, K.M.; Navabpour, S.; Musaus, M.; Farrell, K.; Hauser, R.M.; McFadden, T.; Martin, K.; et al. Ubiquitination of Histone H2B by Proteasome Subunit RPT6 Controls Histone Methylation Chromatin Dynamics During Memory Formation. Biol. Psychiatry 2021, 89, 1176–1187. [Google Scholar] [CrossRef]
- Zovkic, I.B.; Paulukaitis, B.S.; Day, J.J.; Etikala, D.M.; Sweatt, J.D. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 2014, 515, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Stefanelli, G.; Azam, A.B.; Walters, B.J.; Brimble, M.A.; Gettens, C.P.; Bouchard-Cannon, P.; Cheng, H.M.; Davidoff, A.M.; Narkaj, K.; Day, J.J.; et al. Learning and Age-Related Changes in Genome-wide H2A.Z Binding in the Mouse Hippocampus. Cell Rep. 2018, 22, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Narkaj, K.; Stefanelli, G.; Wahdan, M.; Azam, A.B.; Ramzan, F.; Steininger, C.F.D., Jr.; Walters, B.J.; Zovkic, I.B. Blocking H2A.Z Incorporation via Tip60 Inhibition Promotes Systems Consolidation of Fear Memory in Mice. eNeuro 2018, 5. [Google Scholar] [CrossRef]
- Ramzan, F.; Creighton, S.D.; Hall, M.; Baumbach, J.; Wahdan, M.; Poulson, S.J.; Michailidis, V.; Stefanelli, G.; Narkaj, K.; Tao, C.S.; et al. Sex-specific effects of the histone variant H2A.Z on fear memory, stress-enhanced fear learning and hypersensitivity to pain. Sci. Rep. 2020, 10, 14331. [Google Scholar] [CrossRef]
- Madabhushi, R.; Gao, F.; Pfenning, A.R.; Pan, L.; Yamakawa, S.; Seo, J.; Rueda, R.; Phan, T.X.; Yamakawa, H.; Pao, P.C.; et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell 2015, 161, 1592–1605. [Google Scholar] [CrossRef]
- Navabpour, S.; Rogers, J.; McFadden, T.; Jarome, T.J. DNA Double-Strand Breaks Are a Critical Regulator of Fear Memory Reconsolidation. Int. J. Mol. Sci. 2020, 21, 8995. [Google Scholar] [CrossRef]
- Stott, R.T.; Kritsky, O.; Tsai, L.H. Profiling DNA break sites and transcriptional changes in response to contextual fear learning. PLoS ONE 2021, 16, e0249691. [Google Scholar] [CrossRef]
- Schoberleitner, I.; Mutti, A.; Sah, A.; Wille, A.; Gimeno-Valiente, F.; Piatti, P.; Kharitonova, M.; Torres, L.; Lopez-Rodas, G.; Liu, J.J.; et al. Role for Chromatin Remodeling Factor Chd1 in Learning and Memory. Front. Mol. Neurosci. 2019, 12, 3. [Google Scholar] [CrossRef]
- Vogel Ciernia, A.; Kramar, E.A.; Matheos, D.P.; Havekes, R.; Hemstedt, T.J.; Magnan, C.N.; Sakata, K.; Tran, A.; Azzawi, S.; Lopez, A.; et al. Mutation of neuron-specific chromatin remodeling subunit BAF53b: Rescue of plasticity and memory by manipulating actin remodeling. Learn. Mem. 2017, 24, 199–209. [Google Scholar] [CrossRef]
- Vogel-Ciernia, A.; Matheos, D.P.; Barrett, R.M.; Kramar, E.A.; Azzawi, S.; Chen, Y.; Magnan, C.N.; Zeller, M.; Sylvain, A.; Haettig, J.; et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 2013, 16, 552–561. [Google Scholar] [CrossRef]
- Tamming, R.J.; Siu, J.R.; Jiang, Y.; Prado, M.A.; Beier, F.; Berube, N.G. Mosaic expression of Atrx in the mouse central nervous system causes memory deficits. Dis. Models Mech. 2017, 10, 119–126. [Google Scholar] [CrossRef]
- Korneev, S.; Garaliene, J.; Taylor, G.; Kemenes, I.; Kemenes, G. Time dependent differential regulation of a novel long non-coding natural antisense RNA during long-term memory formation. Sci. Rep. 2021, 11, 3594. [Google Scholar] [CrossRef]
- Spadaro, P.A.; Flavell, C.R.; Widagdo, J.; Ratnu, V.S.; Troup, M.; Ragan, C.; Mattick, J.S.; Bredy, T.W. Long Noncoding RNA-Directed Epigenetic Regulation of Gene Expression Is Associated With Anxiety-like Behavior in Mice. Biol. Psychiatry 2015, 78, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Ianov, L.; Riva, A.; Kumar, A.; Foster, T.C. DNA Methylation of Synaptic Genes in the Prefrontal Cortex Is Associated with Aging and Age-Related Cognitive Impairment. Front. Aging Neurosci. 2017, 9, 249. [Google Scholar] [CrossRef]
- Penner, M.R.; Parrish, R.R.; Hoang, L.T.; Roth, T.L.; Lubin, F.D.; Barnes, C.A. Age-related changes in Egr1 transcription and DNA methylation within the hippocampus. Hippocampus 2016, 26, 1008–1020. [Google Scholar] [CrossRef]
- Penner, M.R.; Roth, T.L.; Chawla, M.K.; Hoang, L.T.; Roth, E.D.; Lubin, F.D.; Sweatt, J.D.; Worley, P.F.; Barnes, C.A. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging 2011, 32, 2198–2210. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Thakur, M.K. Reduced recognition memory is correlated with decrease in DNA methyltransferase1 and increase in histone deacetylase2 protein expression in old male mice. Biogerontology 2014, 15, 339–346. [Google Scholar] [CrossRef]
- Oliveira, A.M.; Hemstedt, T.J.; Bading, H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 2012, 15, 1111–1113. [Google Scholar] [CrossRef] [PubMed]
- Gontier, G.; Iyer, M.; Shea, J.M.; Bieri, G.; Wheatley, E.G.; Ramalho-Santos, M.; Villeda, S.A. Tet2 Rescues Age-Related Regenerative Decline and Enhances Cognitive Function in the Adult Mouse Brain. Cell Rep. 2018, 22, 1974–1981. [Google Scholar] [CrossRef]
- Peleg, S.; Sananbenesi, F.; Zovoilis, A.; Burkhardt, S.; Bahari-Javan, S.; Agis-Balboa, R.C.; Cota, P.; Wittnam, J.L.; Gogol-Doering, A.; Opitz, L.; et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 2010, 328, 753–756. [Google Scholar] [CrossRef]
- Singh, P.; Thakur, M.K. Histone Deacetylase 2 Inhibition Attenuates Downregulation of Hippocampal Plasticity Gene Expression during Aging. Mol. Neurobiol. 2018, 55, 2432–2442. [Google Scholar] [CrossRef] [PubMed]
- Reolon, G.K.; Maurmann, N.; Werenicz, A.; Garcia, V.A.; Schroder, N.; Wood, M.A.; Roesler, R. Posttraining systemic administration of the histone deacetylase inhibitor sodium butyrate ameliorates aging-related memory decline in rats. Behav. Brain Res. 2011, 221, 329–332. [Google Scholar] [CrossRef]
- Kwapis, J.L.; Alaghband, Y.; Kramar, E.A.; Lopez, A.J.; Vogel Ciernia, A.; White, A.O.; Shu, G.; Rhee, D.; Michael, C.M.; Montellier, E.; et al. Epigenetic regulation of the circadian gene Per1 contributes to age-related changes in hippocampal memory. Nat. Commun. 2018, 9, 3323. [Google Scholar] [CrossRef]
- Kwapis, J.L.; Alaghband, Y.; Lopez, A.J.; Long, J.M.; Li, X.; Shu, G.; Bodinayake, K.K.; Matheos, D.P.; Rapp, P.R.; Wood, M.A. HDAC3-Mediated Repression of the Nr4a Family Contributes to Age-Related Impairments in Long-Term Memory. J. Neurosci. 2019, 39, 4999–5009. [Google Scholar] [CrossRef]
- Snigdha, S.; Prieto, G.A.; Petrosyan, A.; Loertscher, B.M.; Dieskau, A.P.; Overman, L.E.; Cotman, C.W. H3K9me3 Inhibition Improves Memory, Promotes Spine Formation, and Increases BDNF Levels in the Aged Hippocampus. J. Neurosci. 2016, 36, 3611–3622. [Google Scholar] [CrossRef]
- Morse, S.J.; Butler, A.A.; Davis, R.L.; Soller, I.J.; Lubin, F.D. Environmental enrichment reverses histone methylation changes in the aged hippocampus and restores age-related memory deficits. Biology 2015, 4, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Maze, I.; Wenderski, W.; Noh, K.M.; Bagot, R.C.; Tzavaras, N.; Purushothaman, I.; Elsasser, S.J.; Guo, Y.; Ionete, C.; Hurd, Y.L.; et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 2015, 87, 77–94. [Google Scholar] [CrossRef]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef]
- Jahn, H. Memory loss in Alzheimer’s disease. Dialogues Clin. Neurosci. 2013, 15, 445–454. [Google Scholar]
- Kilgore, M.; Miller, C.A.; Fass, D.M.; Hennig, K.M.; Haggarty, S.J.; Sweatt, J.D.; Rumbaugh, G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology 2010, 35, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Francis, Y.I.; Fa, M.; Ashraf, H.; Zhang, H.; Staniszewski, A.; Latchman, D.S.; Arancio, O. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer’s disease. J. Alzheimer’s Dis. 2009, 18, 131–139. [Google Scholar] [CrossRef]
- Sung, Y.M.; Lee, T.; Yoon, H.; DiBattista, A.M.; Song, J.M.; Sohn, Y.; Moffat, E.I.; Turner, R.S.; Jung, M.; Kim, J.; et al. Mercaptoacetamide-based class II HDAC inhibitor lowers Abeta levels and improves learning and memory in a mouse model of Alzheimer’s disease. Exp. Neurol. 2013, 239, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, A.; Wang, Z.J.; Cao, Q.; Wang, W.; Lin, L.; Ma, K.; Zhang, F.; Wei, J.; Matas, E.; et al. Inhibition of EHMT1/2 rescues synaptic and cognitive functions for Alzheimer’s disease. Brain 2019, 142, 787–807. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Wang, W.; Williams, J.B.; Yang, F.; Wang, Z.J.; Yan, Z. Targeting histone K4 trimethylation for treatment of cognitive and synaptic deficits in mouse models of Alzheimer’s disease. Sci. Adv. 2020, 6, eabc8096. [Google Scholar] [CrossRef]
- Chouliaras, L.; Mastroeni, D.; Delvaux, E.; Grover, A.; Kenis, G.; Hof, P.R.; Steinbusch, H.W.; Coleman, P.D.; Rutten, B.P.; van den Hove, D.L. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging 2013, 34, 2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Chouliaras, L.; Lardenoije, R.; Kenis, G.; Mastroeni, D.; Hof, P.R.; van Os, J.; Steinbusch, H.W.M.; van Leeuwen, F.W.; Rutten, B.P.F.; van den Hove, D.L.A. Age-related Disturbances in DNA (hydroxy)methylation in APP/PS1 Mice. Transl. Neurosci. 2018, 9, 190–202. [Google Scholar] [CrossRef]
- Sandoval-Hernandez, A.G.; Hernandez, H.G.; Restrepo, A.; Munoz, J.I.; Bayon, G.F.; Fernandez, A.F.; Fraga, M.F.; Cardona-Gomez, G.P.; Arboleda, H.; Arboleda, G.H. Liver X Receptor Agonist Modifies the DNA Methylation Profile of Synapse and Neurogenesis-Related Genes in the Triple Transgenic Mouse Model of Alzheimer’s Disease. J. Mol. Neurosci. 2016, 58, 243–253. [Google Scholar] [CrossRef]
- Monti, N.; Cavallaro, R.A.; Stoccoro, A.; Nicolia, V.; Scarpa, S.; Kovacs, G.G.; Fiorenza, M.T.; Lucarelli, M.; Aronica, E.; Ferrer, I. CpG and non-CpG Presenilin1 methylation pattern in course of neurodevelopment and neurodegeneration is associated with gene expression in human and murine brain. Epigenetics 2020, 15, 781–799. [Google Scholar] [CrossRef]
- Bakulski, K.M.; Dolinoy, D.C.; Sartor, M.A.; Paulson, H.L.; Konen, J.R.; Lieberman, A.P.; Albin, R.L.; Hu, H.; Rozek, L.S. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J. Alzheimer’s Dis. 2012, 29, 571–588. [Google Scholar] [CrossRef]
- Wang, S.-C.; Oelze, B.; Schumacher, A. Age-Specific Epigenetic Drift in Late-Onset Alzheimer’s Disease. PLoS ONE 2008, 3, e2698. [Google Scholar] [CrossRef] [PubMed]
- Iwata, A.; Nagata, K.; Hatsuta, H.; Takuma, H.; Bundo, M.; Iwamoto, K.; Tamaoka, A.; Murayama, S.; Saido, T.; Tsuji, S. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum. Mol. Genet. 2014, 23, 648–656. [Google Scholar] [CrossRef] [PubMed]
- West, R.L.; Lee, J.M.; Maroun, L.E. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J. Mol. Neurosci. 1995, 6, 141–146. [Google Scholar] [CrossRef]
- Barrachina, M.; Ferrer, I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J. Neuropathol. Exp. Neurol. 2009, 68, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Foraker, J.; Millard, S.P.; Leong, L.; Thomson, Z.; Chen, S.; Keene, C.D.; Bekris, L.M.; Yu, C.-E. The APOE gene is differentially methylated in Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 48, 745–755. [Google Scholar] [CrossRef]
- Blanco-Luquin, I.; Altuna, M.; Sanchez-Ruiz de Gordoa, J.; Urdánoz-Casado, A.; Roldán, M.; Cámara, M.; Zelaya, V.; Erro, M.E.; Echavarri, C.; Mendioroz, M. PLD3 epigenetic changes in the hippocampus of Alzheimer’s disease. Clin. Epigenet. 2018, 10, 1–10. [Google Scholar] [CrossRef]
- Xiao, X.; Liu, X.; Jiao, B. Epigenetics: Recent Advances and Its Role in the Treatment of Alzheimer’s Disease. Front. Neurol. 2020, 11, 538301. [Google Scholar] [CrossRef]
- Tang, L.; Liu, L.; Li, G.; Jiang, P.; Wang, Y.; Li, J. Expression Profiles of Long Noncoding RNAs in Intranasal LPS-Mediated Alzheimer’s Disease Model in Mice. Biomed. Res. Int. 2019, 2019, 9642589. [Google Scholar] [CrossRef]
- Song, Z.; Li, F.; He, C.; Yu, J.; Li, P.; Li, Z.; Yang, M.; Cheng, S. In-depth transcriptomic analyses of LncRNA and mRNA expression in the hippocampus of APP/PS1 mice by Danggui-Shaoyao-San. Aging 2020, 12, 23945–23959. [Google Scholar] [CrossRef]
- Jarome, T.J.; Lubin, F.D. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol. Learn. Mem. 2014, 115, 116–127. [Google Scholar] [CrossRef]
- Hemstedt, T.J.; Lattal, K.M.; Wood, M.A. Reconsolidation and extinction: Using epigenetic signatures to challenge conventional wisdom. Neurobiol. Learn. Mem. 2017, 142, 55–65. [Google Scholar] [CrossRef] [PubMed]

| The Epigenetic Mark | Brain Region | Direction |
|---|---|---|
| 5mC | Hippocampus | Up (121) |
| 5hmC | Hippocampus and Anterior Cingulate Cortex | Up (155) |
| H2BubiK120 | Hippocampus | Up (160) |
| H2B Acetylation | Hippocampus | Up (149) |
| H2A.Z | Hippocampus and Cortex | Down (161) |
| H2A.X | Hippocampus | Up (165–167) |
| H3K4me3 | Hippocampus | Up (135, 154, 155) |
| Amygdala | No Change (159) | |
| H3K9me2 | Hippocampus, Entorhinal and Amygdala | Up (156, 159) |
| H3K27me3 | Hippocampus | Up (157, 158) |
| H3 Acetylation | PFC, Hippocampus, Amygdala | Up (136, 146, 147, 149) |
| H4 Acetylation | Hippocampus | No Change (136, 147) |
| Up (143) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maity, S.; Farrell, K.; Navabpour, S.; Narayanan, S.N.; Jarome, T.J. Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 12280. https://doi.org/10.3390/ijms222212280
Maity S, Farrell K, Navabpour S, Narayanan SN, Jarome TJ. Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease. International Journal of Molecular Sciences. 2021; 22(22):12280. https://doi.org/10.3390/ijms222212280
Chicago/Turabian StyleMaity, Sabyasachi, Kayla Farrell, Shaghayegh Navabpour, Sareesh Naduvil Narayanan, and Timothy J. Jarome. 2021. "Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease" International Journal of Molecular Sciences 22, no. 22: 12280. https://doi.org/10.3390/ijms222212280
APA StyleMaity, S., Farrell, K., Navabpour, S., Narayanan, S. N., & Jarome, T. J. (2021). Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease. International Journal of Molecular Sciences, 22(22), 12280. https://doi.org/10.3390/ijms222212280






