Identification of Raf-Like Kinases B Subfamily Genes in Gossypium Species Revealed GhRAF42 Enhanced Salt Tolerance in Cotton
Abstract
1. Introduction
2. Results
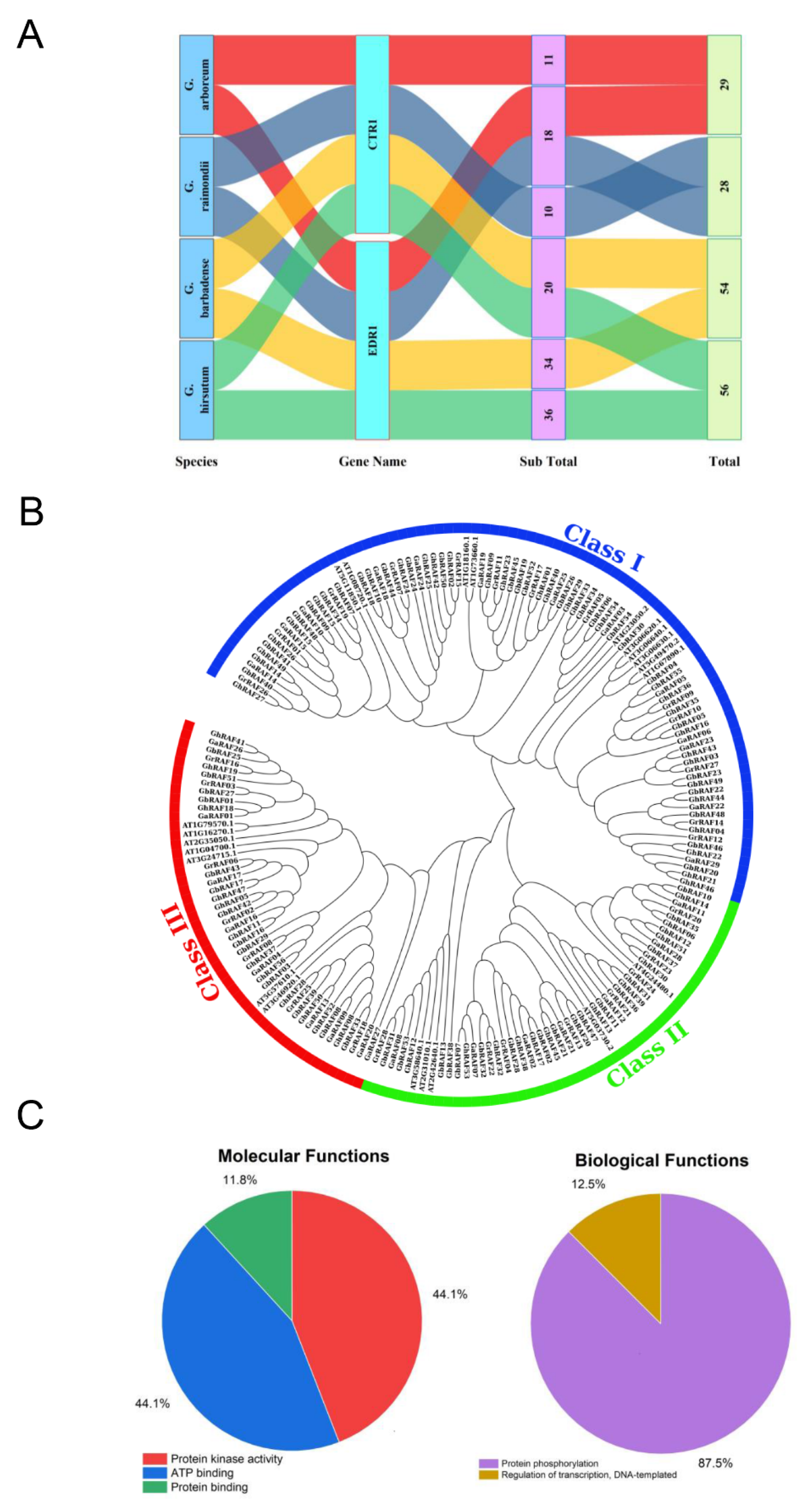
2.1. RAF Sequence Analysis and Characterization of Four Cotton Species
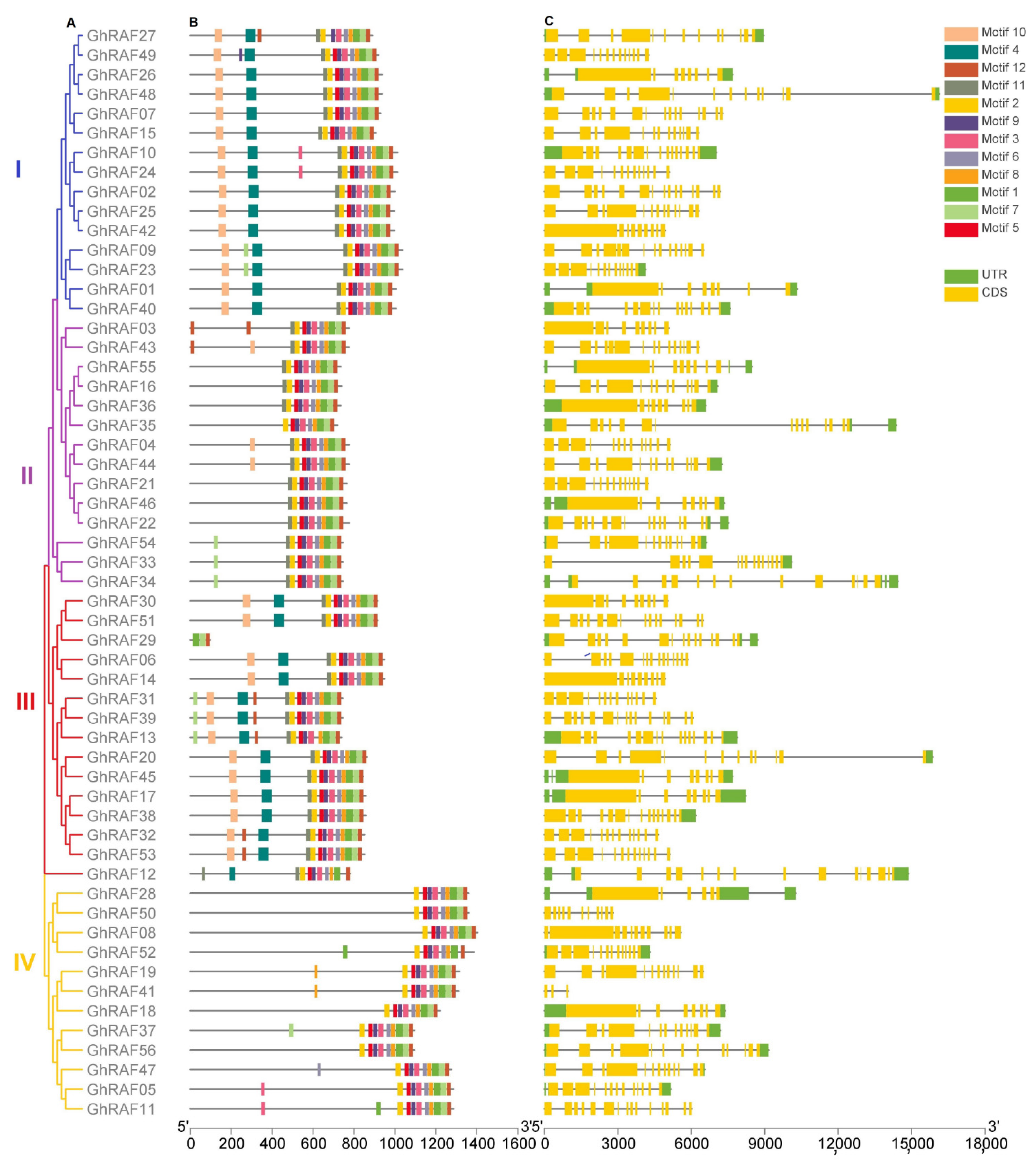
2.2. Cotton RAF Gene Family Tree Diagram, Exon-Intron Structure, Motif Assay, and Gene Ontology Analysis
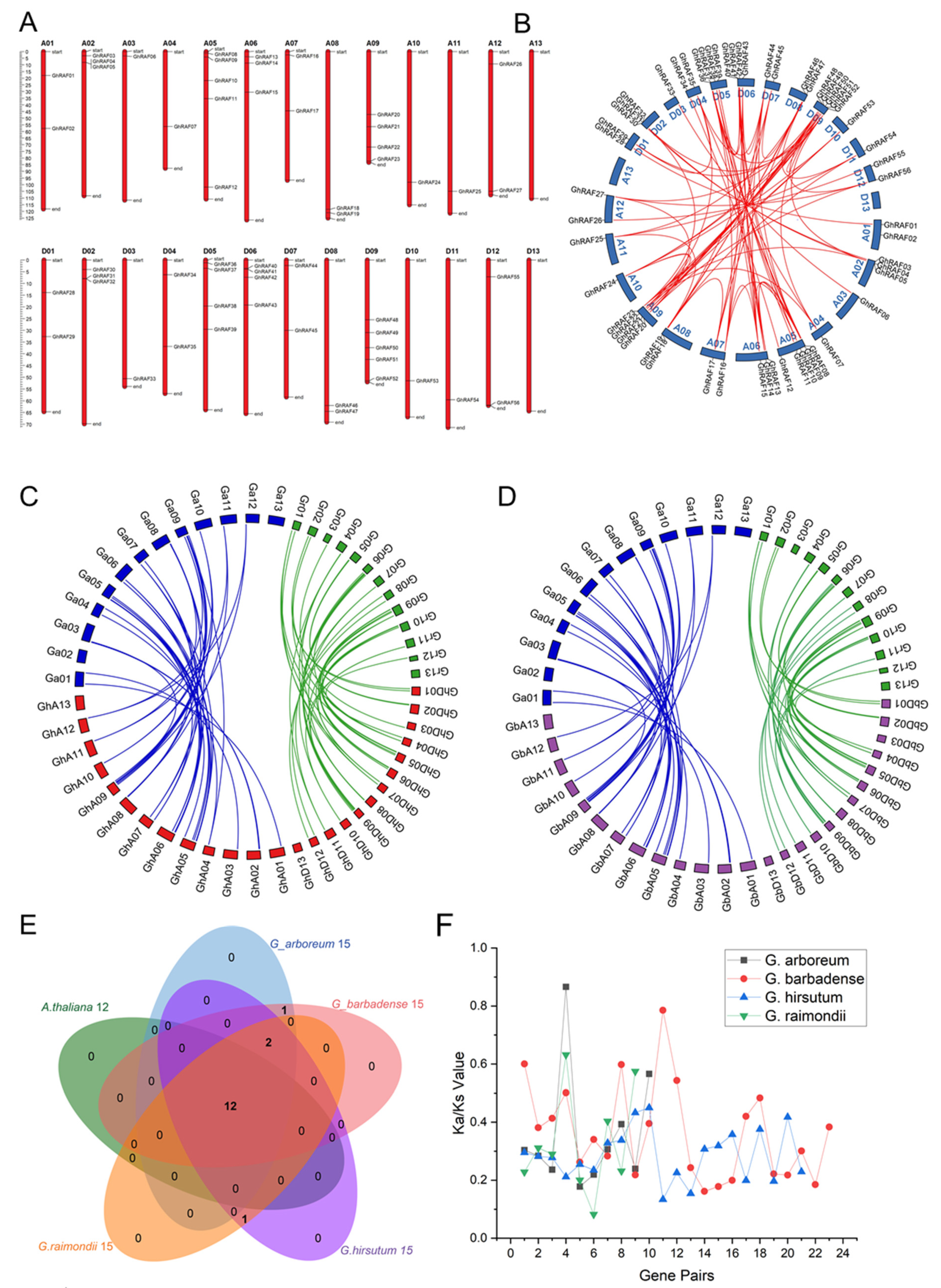
2.3. Chromosomal Gene Location and Synteny Analysis
2.4. Orthologous Gene Clusters Identification and Synonymous and Nonsynonymous Ratio
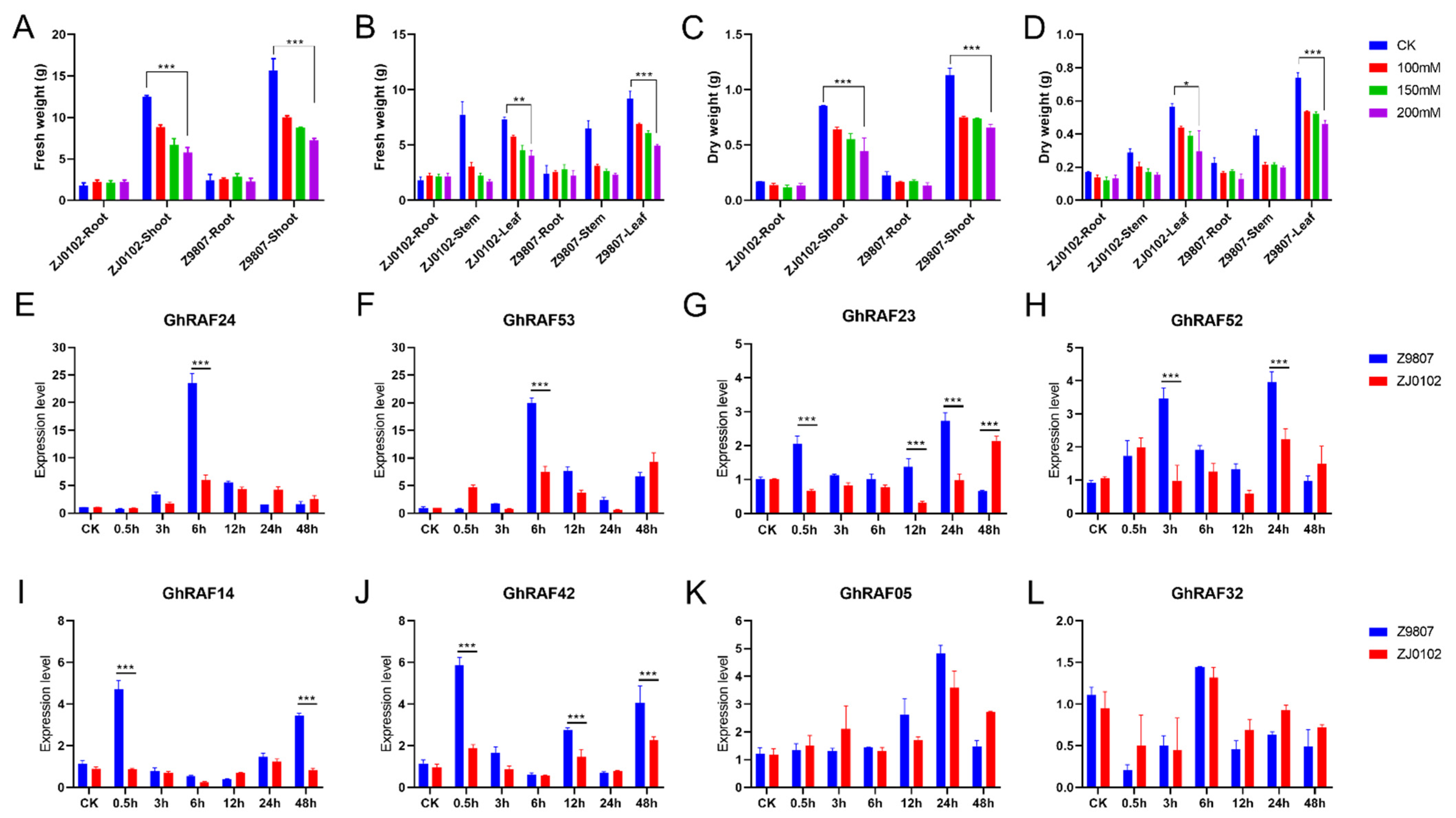
2.5. Screening of Salt Tolerance Genes Based on Transcriptome Data
2.6. GhRAF Gene Expression Characteristics Analysis
2.7. Gene Cloning and Subcellular Localization
2.8. Silencing of Genes in Upland Cotton and Its Detection
3. Discussion
4. Material and Methods
4.1. Classification and Characterization of RAF Proteins in Cotton
4.2. Multiple Sequence Alignment and Intron or Exon Structure Analysis of RAF Gene Family
4.3. Phylogenetic Analysis of RAF Genes and Gene Ontology Analysis
4.4. Chromosomal Location, Synteny Analysis, and Collinearity Analysis of RAF Genes
4.5. Identification of Orthologous RAF Genes Based on Sequence
4.6. Heat-Map Analysis
4.7. Plant Materials
4.8. RNA Extraction from Upland Cotton for Real-Time Quantitative PCR (RT-qPCR) Analysis
4.9. Subcellular Localization
4.10. Virus-Induced Gene Silencing (VIGS) in Cotton
4.11. Assessment of Enzyme Activity Content of Gene Silencing Cotton
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bin Huang, Z.; Sun, Z.J.; Lu, Z.H. Effects of Soil Amendments on Coastal Saline-Alkali Soil Improvement and the Growth of Plants. Adv. Mater. Res. 2013, 634-638, 152–159. [Google Scholar] [CrossRef]
- Hamwieh, A.; Tuyen, D.D.; Cong, H.; Benitez, E.R.; Takahashi, R.; Xu, D.H. Identification and validation of a major QTL for salt tolerance in soybean. Euphytica 2011, 179, 451–459. [Google Scholar] [CrossRef]
- Farooq, M.; Gogoi, N.; Barthakur, S.; Baroowa, B.; Bharadwaj, N.; Alghamdi, S.S.; Siddique, K.H.M. Drought Stress in Grain Legumes during Reproduction and Grain Filling. J. Agron. Crop. Sci. 2017, 203, 81–102. [Google Scholar] [CrossRef]
- Dagar, J.C.; Minhas, P.S. Global Perspectives on Agroforestry for the Management of Salt-affected Soils. In Agroforestry for the Management of Waterlogged Saline Soils and Poor-Quality Waters; Springer: New Delhi, India, 2016; pp. 5–32. [Google Scholar]
- Han, J.; Shi, J.; Zeng, L.; Xu, J.; Wu, L. Effects of nitrogen fertilization on the acidity and salinity of greenhouse soils. Environ. Sci. Pollut. Res. 2014, 22, 2976–2986. [Google Scholar] [CrossRef]
- Demiral, T.; Turkan, I. Exogenous glycinebetaine affects growth and proline accumulation and retards senescence in two rice cultivars under NaCl stress. Environ. Exp. Bot. 2006, 56, 72–79. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Flowers, T.J.; Galal, H.K.; Bromham, L. Evolution of halophytes: Multiple origins of salt tolerance in land plants. Funct. Plant Biol. 2010, 37, 604–612. [Google Scholar] [CrossRef]
- Rodriguez-Uribe, L.; Higbie, S.M.; Stewart, J.M.; Wilkins, T.; Lindemann, W.; Sengupta-Gopalan, C.; Zhang, J. Identification of salt responsive genes using comparative microarray analysis in Upland cotton (Gossypium hirsutum L.). Plant Sci. 2011, 180, 461–469. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Yuan, D.; Tang, Z.; Wang, M.; Gao, W.; Tu, L.; Jin, X.; Chen, L.; He, Y.; Zhang, L.; Zhu, L.; et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2016, 5, 17662. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef]
- Qu, J.; Ye, J.; Geng, Y.-F.; Sun, Y.-W.; Gao, S.-Q.; Zhang, B.-P.; Chen, W.; Chua, N.-H. Dissecting Functions of KATANIN and WRINKLED1 in Cotton Fiber Development by Virus-Induced Gene Silencing. Plant Physiol. 2012, 160, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-L.; Liang, S.; Wang, H.-Y.; Han, L.-B.; Wang, F.-X.; Cheng, H.-Q.; Wu, X.-M.; Qu, Z.-L.; Wu, J.-H.; Xia, G.-X. Cotton Major Latex Protein 28 Functions as a Positive Regulator of the Ethylene Responsive Factor 6 in Defense against Verticillium dahliae. Mol. Plant 2015, 8, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yu, Y.; Wang, D.; Wu, C.; Yang, G.; Huang, J.; Zheng, C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009, 183, 62–75. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yang, X.; Wang, L.; Zhu, L.; Zhou, T.; Deng, J.; Zhang, X. Molecular cloning and functional characterization of a novel cotton CBL-interacting protein kinase gene (GhCIPK6) reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2013, 435, 209–215. [Google Scholar] [CrossRef]
- Gao, S.; Yang, L.; Zeng, H.; Zhou, Z.S.; Yang, Z.M.; Fuliang, X.; Sun, D.; Xie, F.; Zhang, B. A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 2016, 6, 19736. [Google Scholar] [CrossRef]
- MAPK Group; Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef]
- Nishihama, R.; Banno, H.; Shibata, W.; Hirano, K.; Nakashima, M.; Usami, S.; Machida, Y. Plant Homologues of Components of MAPK (Mitogen-Activated Protein Kinase) Signal Pathways in Yeast and Animal Cells. Plant Cell Physiol. 1995, 36, 749–757. [Google Scholar] [CrossRef]
- Sanan-Mishra, N.; Tuteja, R.; Tuteja, N. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006, 452, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Opdenakker, K.; Remans, T.; Vangronsveld, J.; Cuypers, A. Mitogen-Activated Protein (MAP) Kinases in Plant Metal Stress: Regulation and Responses in Comparison to Other Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2012, 13, 7828–7853. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, J.; Wang, D.; Fan, W.; Wang, S.; Ye, W. The MAPKKK Gene Family in Gossypium raimondii: Genome-Wide Identification, Classification and Expression Analysis. Int. J. Mol. Sci. 2013, 14, 18740–18757. [Google Scholar] [CrossRef]
- Jonak, C. Complexity, Cross Talk and Integration of Plant MAP Kinase Signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Rao, K.P.; Richa, T.; Kumar, K.; Raghuram, B.; Sinha, A.K. In Silico Analysis Reveals 75 Members of Mitogen-Activated Protein Kinase Kinase Kinase Gene Family in Rice. DNA Res. 2010, 17, 139–153. [Google Scholar] [CrossRef]
- Shahzad, Z.; Canut, M.; Tournaire-Roux, C.; Martinière, A.; Boursiac, Y.; Loudet, O.; Maurel, C. A Potassium-Dependent Oxygen Sensing Pathway Regulates Plant Root Hydraulics. Cell 2016, 167, 87–98.e14. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, S.R.; Kamisugi, Y.; Trinh, C.H.; Schmutz, J.; Jenkins, J.W.; Grimwood, J.; Muchero, W.; A Tuskan, G.; A Rensing, S.; Lang, D.; et al. Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE (ANR), a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 2016, 28, 1310–1327. [Google Scholar] [CrossRef]
- Frye, C.A.; Tang, D.; Innes, R.W. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. 2001, 98, 373–378. [Google Scholar] [CrossRef]
- Jakubowicz, M.; Nowak, W.; Gałgański, Ł.; Babula-Skowrońska, D. Expression profiling of CTR1-like and EIN2-like genes in buds and leaves of Populus tremula, and in vitro study of the interaction between their polypeptides. Plant Physiol. Biochem. 2019, 139, 660–671. [Google Scholar] [CrossRef]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between Abscisic Acid and Ethylene Signaling Cascades. Plant Cell 2000, 12, 1103–1115. [Google Scholar] [CrossRef]
- Lee, S.-J.; Lee, M.H.; Kim, J.-I.; Kim, S.Y. Arabidopsis Putative MAP Kinase Kinase Kinases Raf10 and Raf11 are Positive Regulators of Seed Dormancy and ABA Response. Plant Cell Physiol. 2014, 56, 84–97. [Google Scholar] [CrossRef]
- Gao, L.; Xiang, C.-B. The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 2008, 67, 125–134. [Google Scholar] [CrossRef]
- Kim, J.-A.; Agrawal, G.K.; Rakwal, R.; Han, K.-S.; Kim, K.-N.; Yun, C.-H.; Heu, S.; Park, S.-Y.; Lee, Y.-H.; Jwa, N.-S. Molecular cloning and mRNA expression analysis of a novel rice (Oryzasativa L.) MAPK kinase kinase, OsEDR1, an ortholog of ArabidopsisAtEDR1, reveal its role in defense/stress signalling pathways and development. Biochem. Biophys. Res. Commun. 2003, 300, 868–876. [Google Scholar] [CrossRef]
- Lin, Z.; Alexander, L.; Hackett, R.; Grierson, D. LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J. 2008, 54, 1083–1093. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowicz-Matuk, A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012, 185-186, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Lohani, N.; Golicz, A.A.; Singh, M.B.; Bhalla, P.L. Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus. Funct. Integr. Genom. 2019, 19, 515–531. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, D.; Wang, Q.; Xu, W.; Wei, Q.; Wang, C.; Liu, C.; Zhang, C.; Yan, H.; Ling, Y.; et al. mRNA-seq Analysis of the Gossypium arboreum transcriptome Reveals Tissue Selective Signaling in Response to Water Stress during Seedling Stage. PLoS ONE 2013, 8, e54762. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, F.; Zhang, C.; Zhang, J.; Chen, Y.; Liu, G.; Zhao, Y.; Hao, F.; Zhang, J. A novel VIGS method by agroinocu-lation of cotton seeds and application for elucidating functions of GhBI-1 in salt-stress response. Plant Cell Rep. 2018, 37, 1091–1100. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, Y.-F.; Randlett, M.D.; Zhao, X.-C.; Findell, J.L.; Kieber, J.J.; Schaller, G.E. Localization of the Raf-like Kinase CTR1 to the Endoplasmic Reticulum of Arabidopsis through Participation in Ethylene Receptor Signaling Complexes. J. Biol. Chem. 2003, 278, 34725–34732. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, X.; Xu, B.; Li, Y.; Jia, L.; Wang, R.; Ren, X.; Wang, G.; Xia, Q. Identification and expression analysis of EDR1-like genes in tobacco (Nicotiana tabacum) in response to Golovinomyces orontii. PeerJ 2018, 6, e5244. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.C.; Lee, S.-J.; Choi, S.-W.; Na, Y.-J.; Song, M.-R.; Hoang, Q.T.N.; Sim, S.Y.; Kim, M.-S.; Kim, J.-I.; Soh, M.-S.; et al. Arabidopsis Raf-Like Kinase Raf10 Is a Regulatory Component of Core ABA Signaling. Mol Cells 2019, 42, 646–660. [Google Scholar]
- Wang, P.; Song, H.; Li, C.; Li, P.; Li, A.; Guan, H.; Hou, L.; Wang, X. Genome-Wide Dissection of the Heat Shock Transcription Factor Family Genes in Arachis. Front. Plant Sci. 2017, 8, 106. [Google Scholar] [CrossRef]
- Wang, J.; Sun, N.; Deng, T.; Zhang, L.; Zuo, K. Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genom. 2014, 15, 961. [Google Scholar] [CrossRef]
- Cai, R.; Zhang, C.; Zhao, Y.; Zhu, K.; Wang, Y.; Jiang, H.; Xiang, Y.; Cheng, B. Genome-wide analysis of the IQD gene family in maize. Mol. Genet. Genom. 2015, 291, 543–558. [Google Scholar] [CrossRef]
- Liu, Z.; Haider, M.S.; Khan, N.; Fang, J. Comprehensive Sequence Analysis of IQD Gene Family and their Expression Profiling in Grapevine (Vitis vinifera). Genes 2020, 11, 235. [Google Scholar] [CrossRef]
- Feng, L.; Chen, Z.; Ma, H.; Chen, X.; Li, Y.; Wang, Y.; Xiang, Y. The IQD Gene Family in Soybean: Structure, Phylogeny, Evolution and Expression. PLoS ONE 2014, 9, e110896. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Peng, Z.; Li, H.; Qin, G.; Jia, Y.; Pan, Z.; He, S.; Qayyum, A.; Du, X. Genome wide analysis of IQD gene family in diploid and tetraploid species of cotton (Gossypium spp.). Int. J. Biol. Macromol. 2021, 184, 1035–1061. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, K.M.; Gu, Y.; Rodibaugh, N.; Innes, R.W. Negative regulation of defence signalling pathways by the EDR1 protein kinase. Mol. Plant Pathol. 2011, 12, 746–758. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Cai, X.; Zhou, Z.; Wang, X.; Diouf, L.; Xu, Y.; Hou, Y.; Hu, Y.; et al. Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants. Int. J. Mol. Sci. 2018, 19, 2625. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, X.; Sun, C.; Rong, J. Identification and Expression Profiling of the Regulator of Chromosome Condensation 1 (RCC1) Gene Family in Gossypium Hirsutum L. under Abiotic Stress and Hormone Treatments. Int. J. Mol. Sci. 2019, 20, 1727. [Google Scholar] [CrossRef]
- Zhang, H.; Li, G.; Fu, C.; Duan, S.; Hu, D.; Guo, X. Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize. Sci. Rep. 2020, 10, 8073. [Google Scholar] [CrossRef]
- Tang, C.; Qiao, X.; Zhu, X.; Khan, W.; Wu, J.; Zhang, S. Expression and evolutionary analysis of soluble inorganic pyrophosphatase gene family in pear and four other Rosaceae species. Plant Syst. Evol. 2020, 306, 1–15. [Google Scholar] [CrossRef]
- Wu, M.; Li, Y.; Chen, D.; Liu, H.; Zhu, D.; Xiang, Y. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2016, 6, 24520. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, T.; Yu, Z.; Li, Y.; Ren, H.; Hou, X.; Li, Y. Genome-wide analysis of the Chinese cabbage IQD gene family and the response of BrIQD5 in drought resistance. Plant Mol. Biol. 2019, 99, 603–620. [Google Scholar] [CrossRef]
- Lin, Y.; Cheng, Y.; Jin, J.; Jin, X.; Jiang, H.; Yan, H.; Cheng, B. Genome Duplication and Gene Loss Affect the Evolution of Heat Shock Transcription Factor Genes in Legumes. PLoS ONE 2014, 9, e102825. [Google Scholar] [CrossRef]
- Rehman, A.; Wang, N.; Peng, Z.; He, S.; Zhao, Z.; Gao, Q.; Wang, Z.; Li, H.; Du, X. Identification of C2H2 subfamily ZAT genes in Gossypium species reveals GhZAT34 and GhZAT79 enhanced salt tolerance in Arabidopsis and cotton. Int. J. Biol. Macromol. 2021, 184, 967–980. [Google Scholar] [CrossRef]
- He, X.; Luo, X.; Wang, T.; Liu, S.; Zhang, X.; Zhu, L. GhHB12 negatively regulates abiotic stress tolerance in Arabidopsis and cotton. Environ. Exp. Bot. 2020, 176, 104087. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Lin, M.; Lin, Z.; Wang, P.; Yang, Q.; Ye, Z.; Shen, C.; Li, J.; Zhang, L.; et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 2017, 49, 579–587. [Google Scholar] [CrossRef]
- Jiang, W.; Yin, J.; Zhang, H.; He, Y.; Shuai, S.; Chen, S.; Cao, S.; Li, W.; Ma, D.; Chen, H. Genome-wide identification, characterization analysis and expression profiling of auxin-responsive GH3 family genes in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2020, 47, 3885–3907. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim.Biophys. Acta (BBA) Bioenerg. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome du-plications in eukaryotes. Proceedings of the National Academy of Sciences 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Chao, D.-Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids Exhibit Higher Potassium Uptake and Salinity Tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, L.; Zhang, H.; Yang, Z.; Wang, H.; Wen, S.; Zhang, C.; Rustgi, S.; von Wettstein, D.; Liu, B. Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl. Acad. Sci. USA 2014, 111, 11882–11887. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Vaseva, I.; Mishev, K.; Depaepe, T.; Vassileva, V.; Van Der Straeten, D. The Diverse Salt-Stress Response of Arabidopsis ctr1-1 and ein2-1 Ethylene Signaling Mutants Is Linked to Altered Root Auxin Homeostasis. Plants 2021, 10, 452. [Google Scholar] [CrossRef]
- Ibrahim, W.; Qiu, C.-W.; Zhang, C.; Cao, F.; Shuijin, Z.; Wu, F.; Zhu, S. Comparative physiological analysis in the tolerance to salinity and drought individual and combination in two cotton genotypes with contrasting salt tolerance. Physiol. Plant. 2018, 165, 155–168. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Q.-L.; Tian, J.; Dang, Y.; Liu, J.; Li, S.; Shang, J.; Fang, M. Antioxidant and Anticancer Activities of Extracts Derived from Four Kinds of Lichen. Plant Sci. J. 2014, 32, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Sinha, A.K. Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice 2013, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Kato, H.; Imai, R. Biochemical identification of the OsMKK6–OsMPK3 signalling pathway for chilling stress tolerance in rice1. Biochem. J. 2012, 443, 95–102. [Google Scholar] [CrossRef]
- Na, Y.-J.; Choi, H.-K.; Park, M.Y.; Choi, S.-W.; Vo, K.T.X.; Jeon, J.-S.; Kim, S.Y. OsMAPKKK63 is involved in salt stress response and seed dormancy control. Plant Signal. Behav. 2019, 14, e1578633. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14 (Suppl. 1), S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Fu, Y.; Dong, N.; Wang, Q. Effects of Overexpression of Cotton Superoxide Dismutase Genes on Salt Tolerant Capability in Upland Cotton. Acta Agric. Boreali-Sin. 2017, 32, 54–59. [Google Scholar] [CrossRef]
- He, X.; Zhu, L.; Xu, L.; Guo, W.; Zhang, X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 2016, 35, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Xu, F.-C.; Guo, D.-D.; Zhao, J.-R.; Liu, J.; Guo, Y.-W.; Singh, P.K.; Ma, X.-N.; Long, L.; Botella, J.R.; et al. Calcium-dependent protein kinases in cotton: Insights into early plant responses to salt stress. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef]
- Long, L.; Zhao, J.-R.; Guo, D.-D.; Ma, X.-N.; Xu, F.-C.; Yang, W.-W.; Gao, W. Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance. BMC Plant Biol. 2020, 20, 1–13. [Google Scholar] [CrossRef]
- Li, Z.; Li, L.; Zhou, K.; Zhang, Y.; Han, X.; Din, Y.; Ge, X.; Qin, W.; Wang, P.; Li, F.; et al. GhWRKY6 Acts as a Negative Regulator in Both Transgenic Arabidopsis and Cotton During Drought and Salt Stress. Front. Genet. 2019, 10, 392. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Tsugawa, H.; Fukusaki, E. Method for Assessing the Statistical Significance of Mass Spectral Similarities Using Basic Local Alignment Search Tool Statistics. Anal. Chem. 2013, 85, 8291–8297. [Google Scholar] [CrossRef] [PubMed]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular lo-calization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins: Structure, Function, and Bioinformatics 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Bailey, T.L.; Gribskov, M. Concerning the accuracy of MAST E-values. Bioinform. 2000, 16, 488–489. [Google Scholar] [CrossRef][Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An inte-grative toolkit developed for interactive analyses of big biological data. Molecules Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular evolution and phylogenetics; Oxford university press: Oxford, UK, 2000; ISBN 0-19-513584-9. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Weedall, G.D.; Polley, S.D.; Conway, D.J. Gene-Specific Signatures of Elevated Non-Synonymous Substitution Rates Correlate Poorly across the Plasmodium Genus. PLoS ONE 2008, 3, e2281. [Google Scholar] [CrossRef]
- Guéguen, L.; Duret, L. Unbiased Estimate of Synonymous and Nonsynonymous Substitution Rates with Nonstationary Base Composition. Mol. Biol. Evol. 2018, 35, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-culture Method for Growing Plants without Soil. Californian Agricultural Experimental Station. Circular No. 347; University of California: Berkeley, CA, USA, 1950. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bachan, S.; Dinesh-Kumar, S.P. Tobacco Rattle Virus (TRV)-Based Virus-Induced Gene Silencing. In Springer Protocols Handbooks; Springer: Singapore, 2012; Volume 894, pp. 83–92. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Jiang, X.; Wang, Z.; Wang, X.; Li, H.; He, S.; Pan, Z.; Qayyum, A.; Rehman, A.; Du, X. Identification of Raf-Like Kinases B Subfamily Genes in Gossypium Species Revealed GhRAF42 Enhanced Salt Tolerance in Cotton. Int. J. Mol. Sci. 2021, 22, 12649. https://doi.org/10.3390/ijms222312649
Peng Z, Jiang X, Wang Z, Wang X, Li H, He S, Pan Z, Qayyum A, Rehman A, Du X. Identification of Raf-Like Kinases B Subfamily Genes in Gossypium Species Revealed GhRAF42 Enhanced Salt Tolerance in Cotton. International Journal of Molecular Sciences. 2021; 22(23):12649. https://doi.org/10.3390/ijms222312649
Chicago/Turabian StylePeng, Zhen, Xuran Jiang, Zhenzhen Wang, Xiaoyang Wang, Hongge Li, Shoupu He, Zhaoe Pan, Abdul Qayyum, Abdul Rehman, and Xiongming Du. 2021. "Identification of Raf-Like Kinases B Subfamily Genes in Gossypium Species Revealed GhRAF42 Enhanced Salt Tolerance in Cotton" International Journal of Molecular Sciences 22, no. 23: 12649. https://doi.org/10.3390/ijms222312649
APA StylePeng, Z., Jiang, X., Wang, Z., Wang, X., Li, H., He, S., Pan, Z., Qayyum, A., Rehman, A., & Du, X. (2021). Identification of Raf-Like Kinases B Subfamily Genes in Gossypium Species Revealed GhRAF42 Enhanced Salt Tolerance in Cotton. International Journal of Molecular Sciences, 22(23), 12649. https://doi.org/10.3390/ijms222312649





