Transcriptomics Changes in the Peritoneum of Mice with Lipopolysaccharide-Induced Peritonitis
Abstract
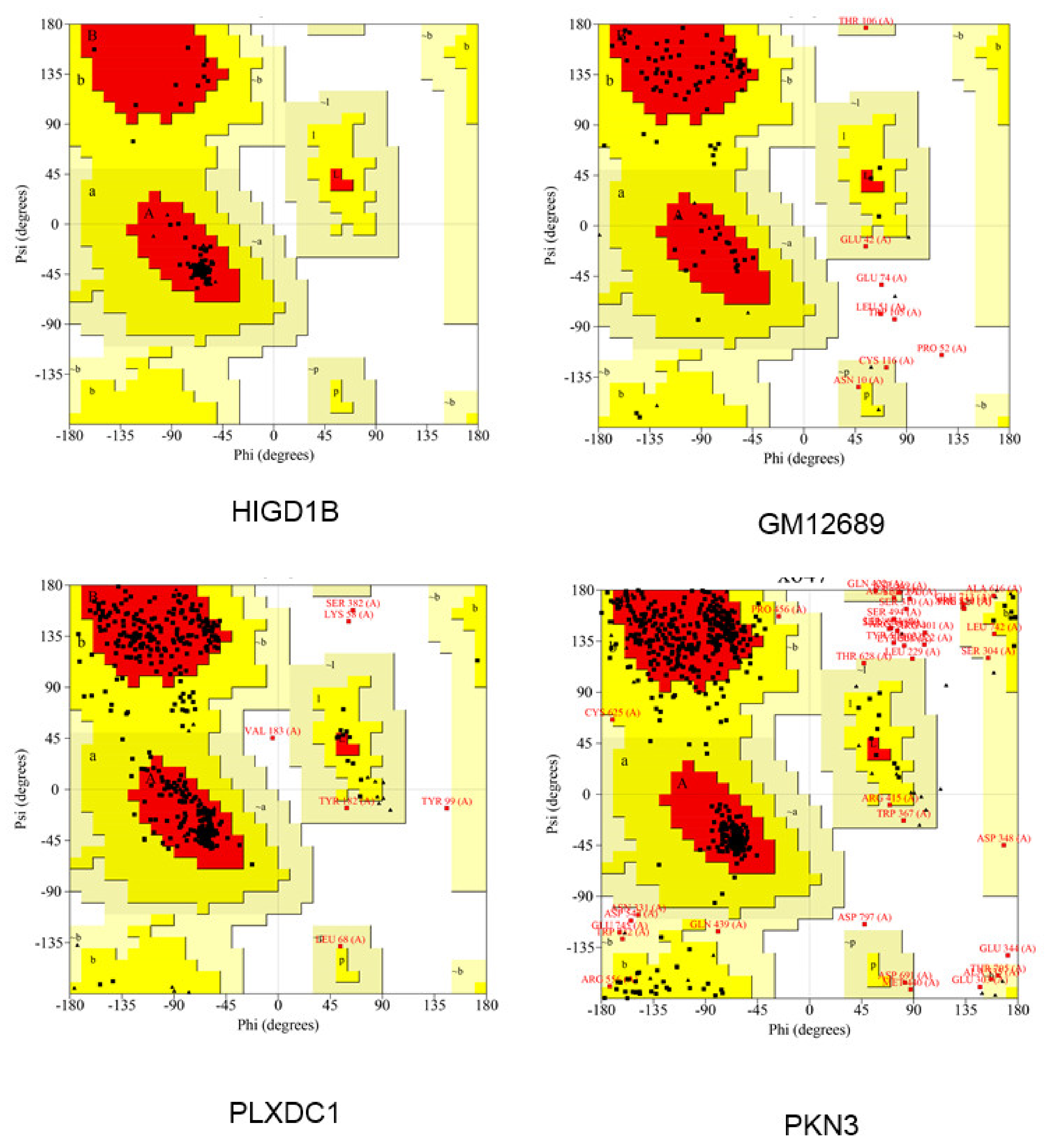
:1. Introduction
2. Results
2.1. LPS-Induced Peritonitis in Mice
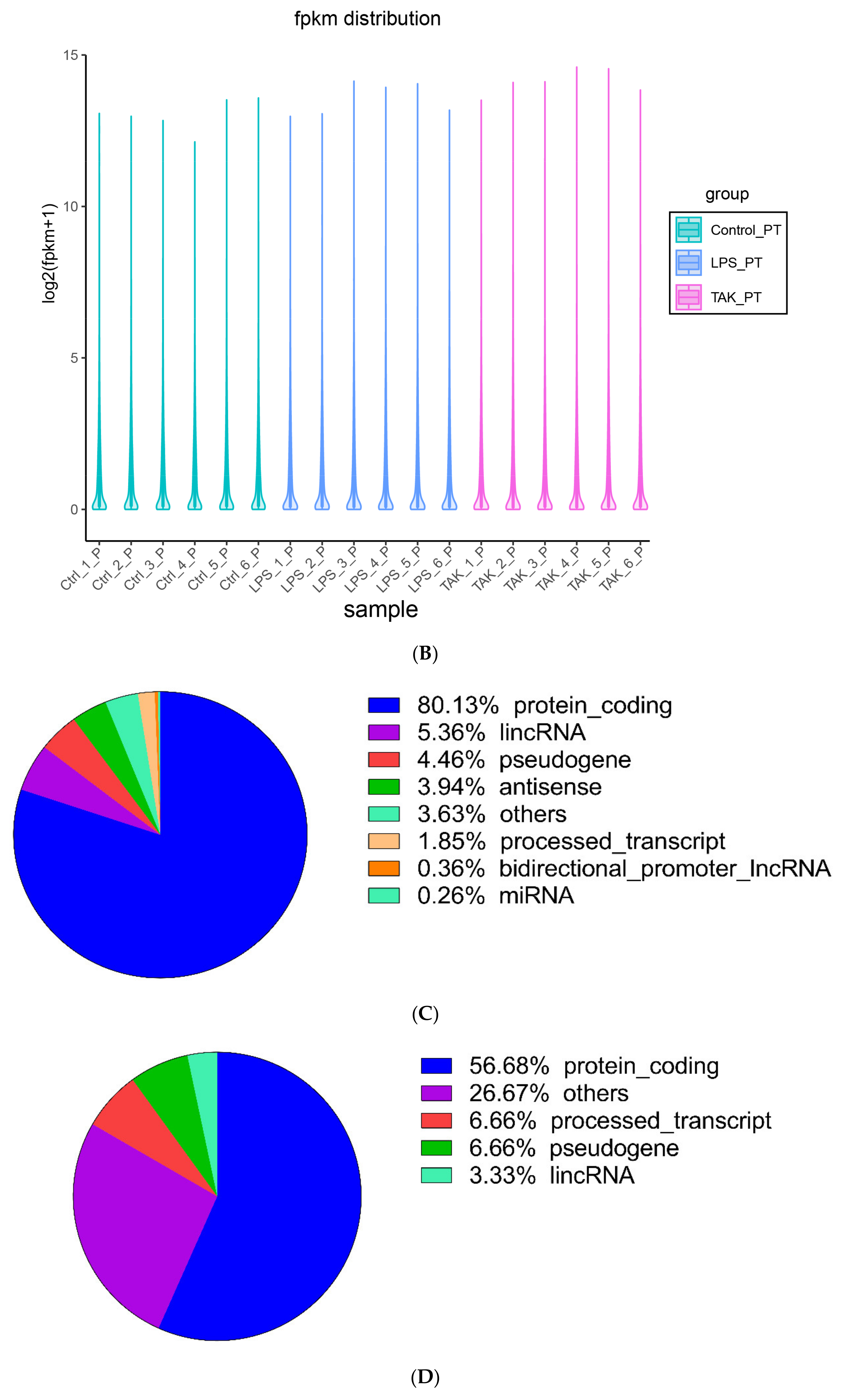
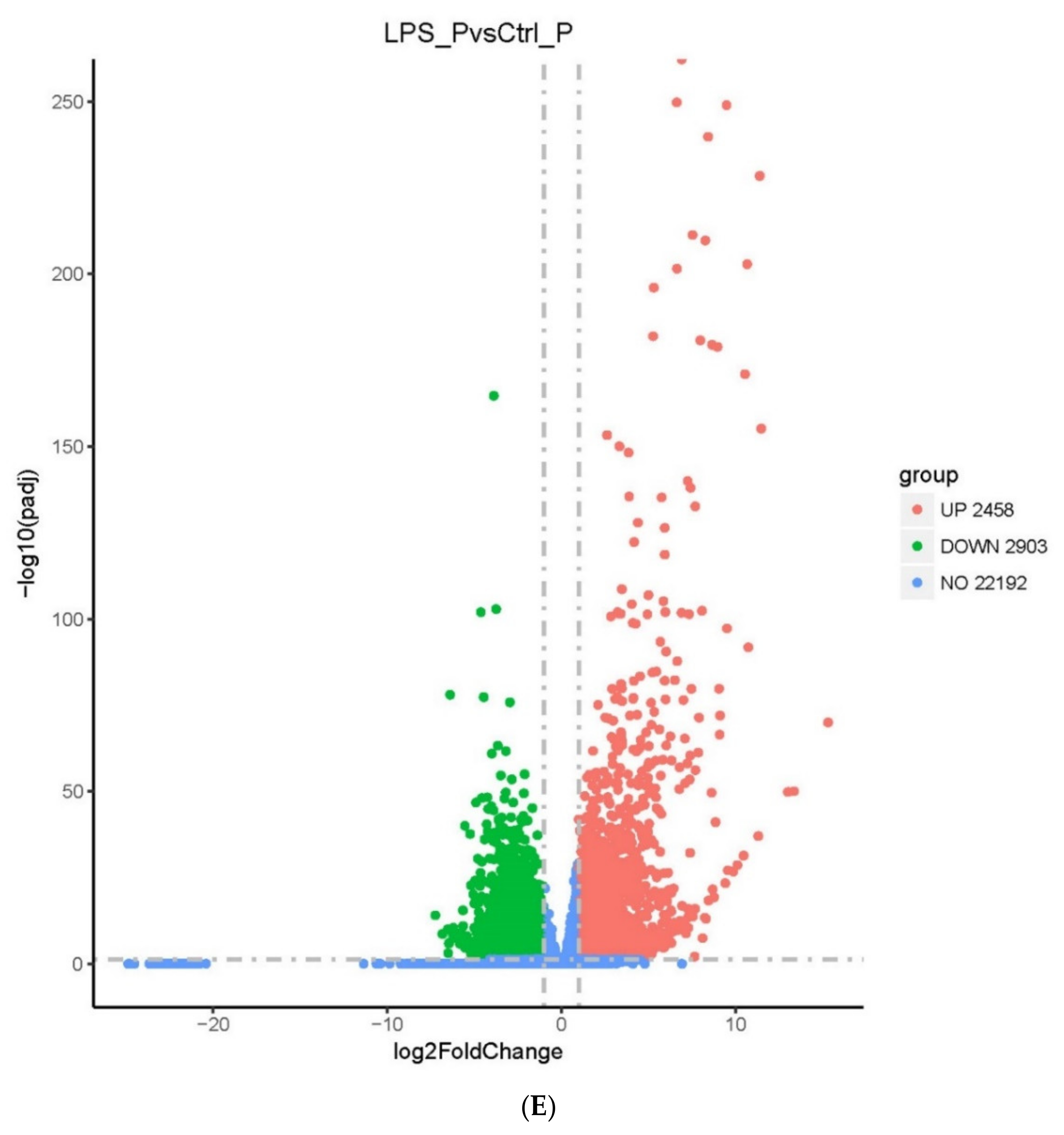
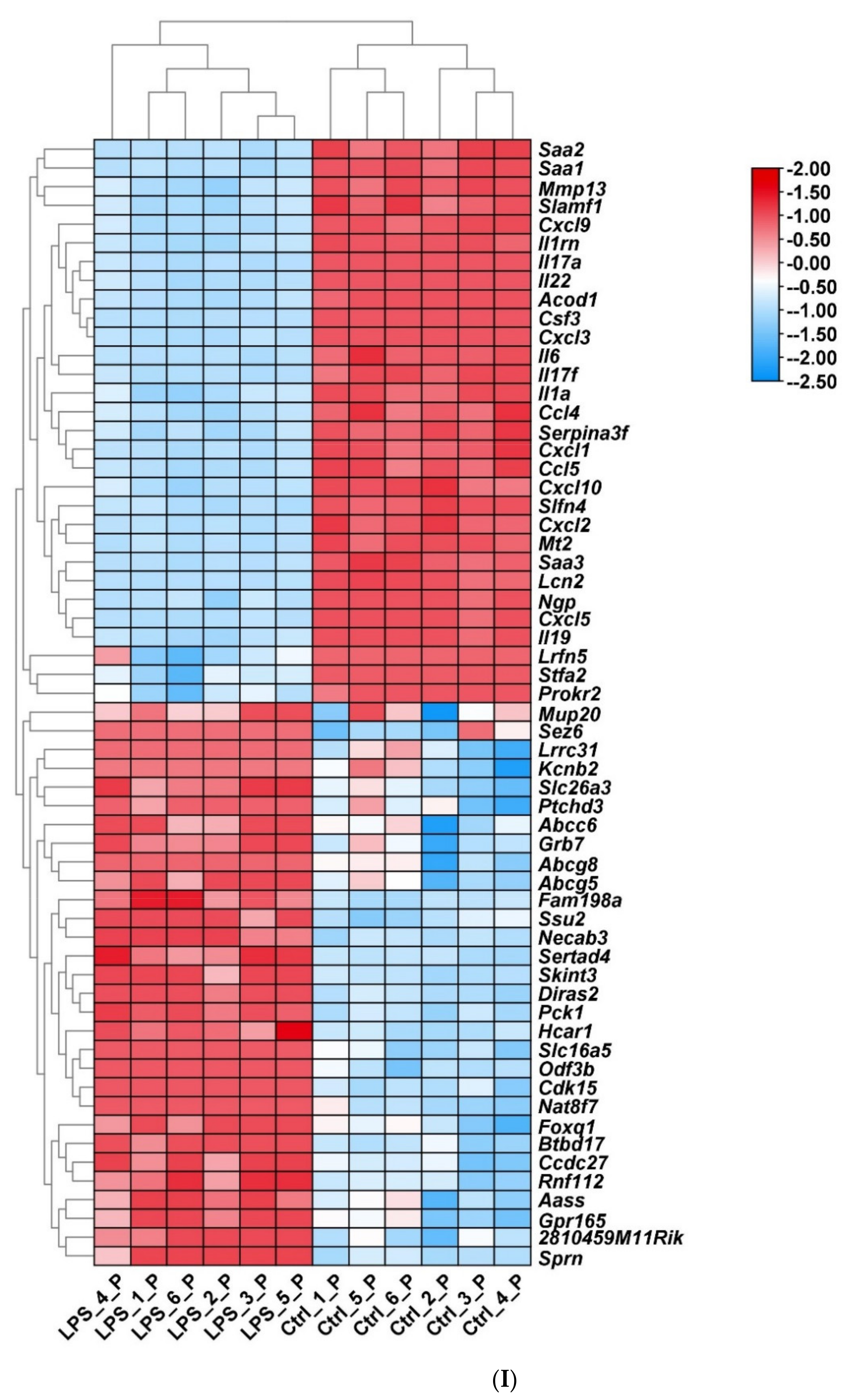
2.2. Screening of DEGs
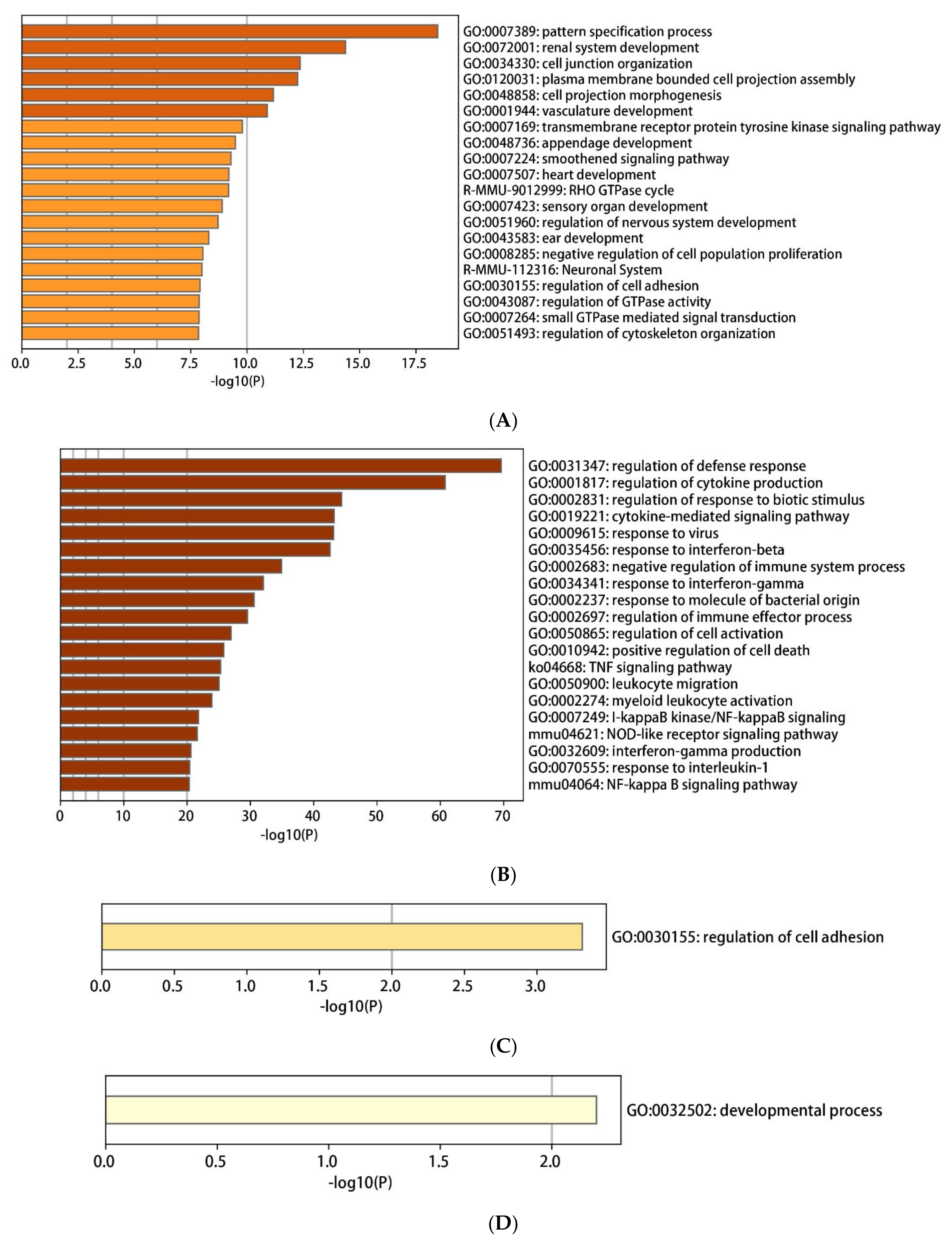
2.3. Enrichment Pathway of DEGs-KEGG
2.4. The Enrichment Pathway of DEGs-Metascape
2.5. Enrichment Pathway of DEGs-PANTHER
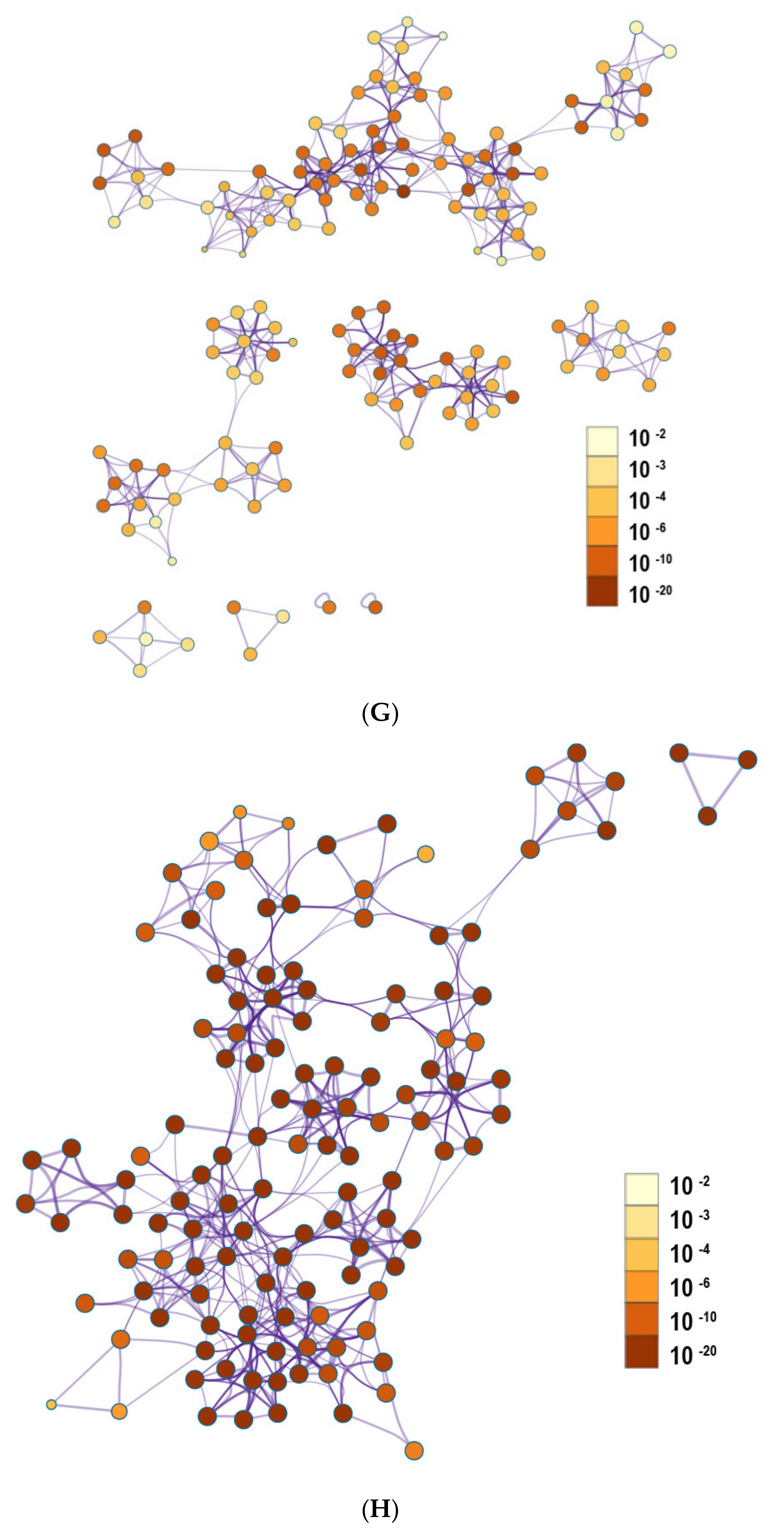
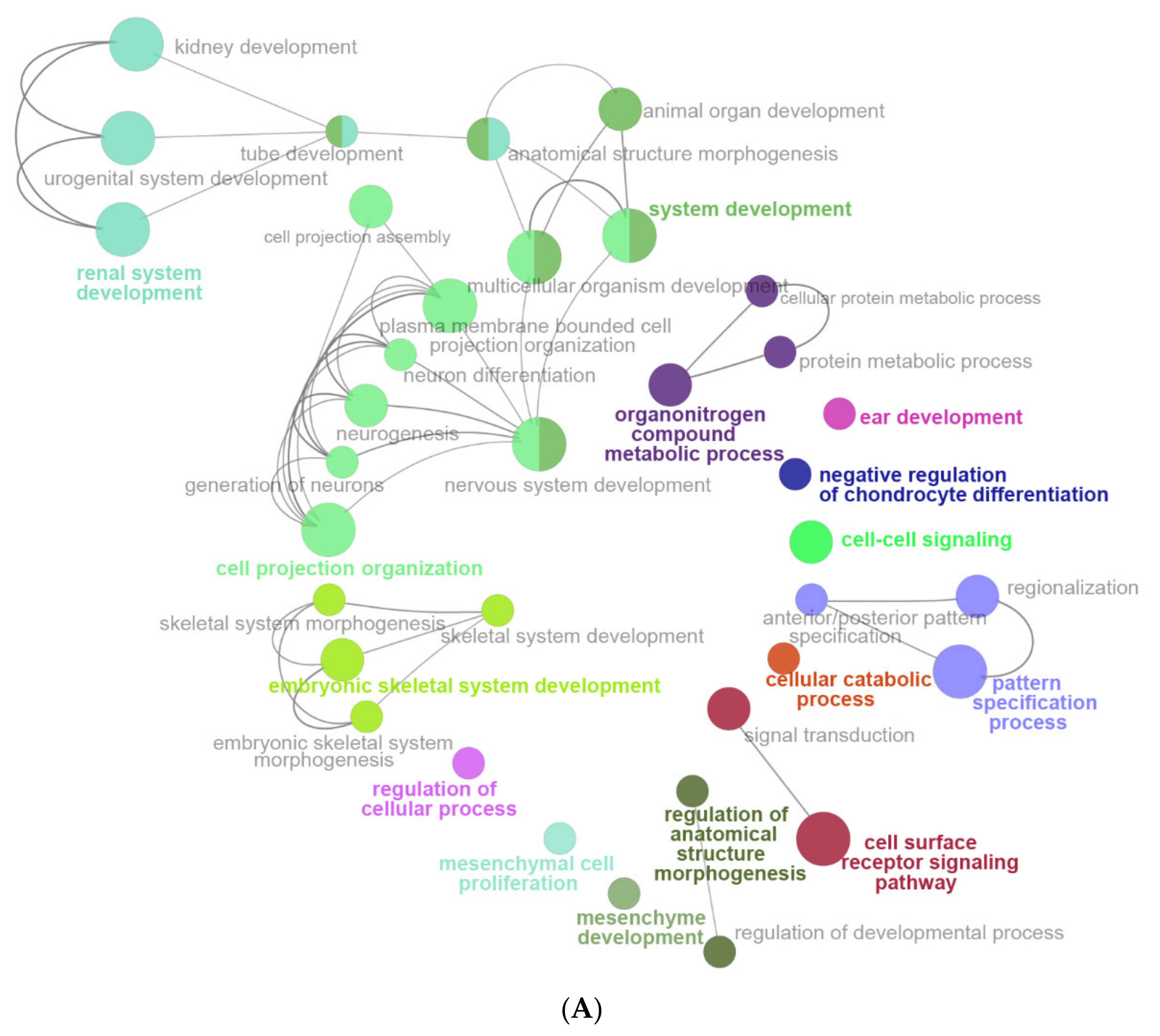
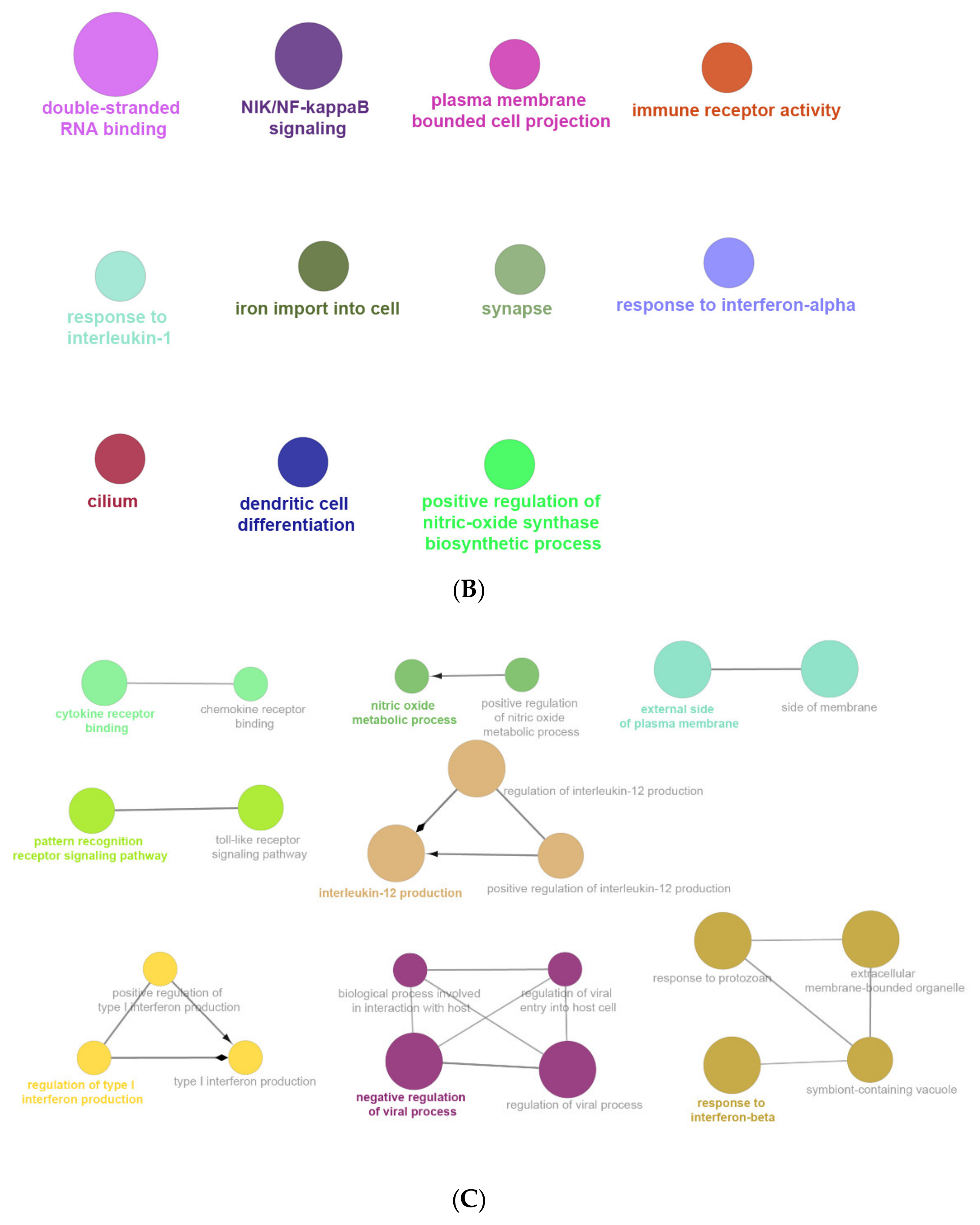
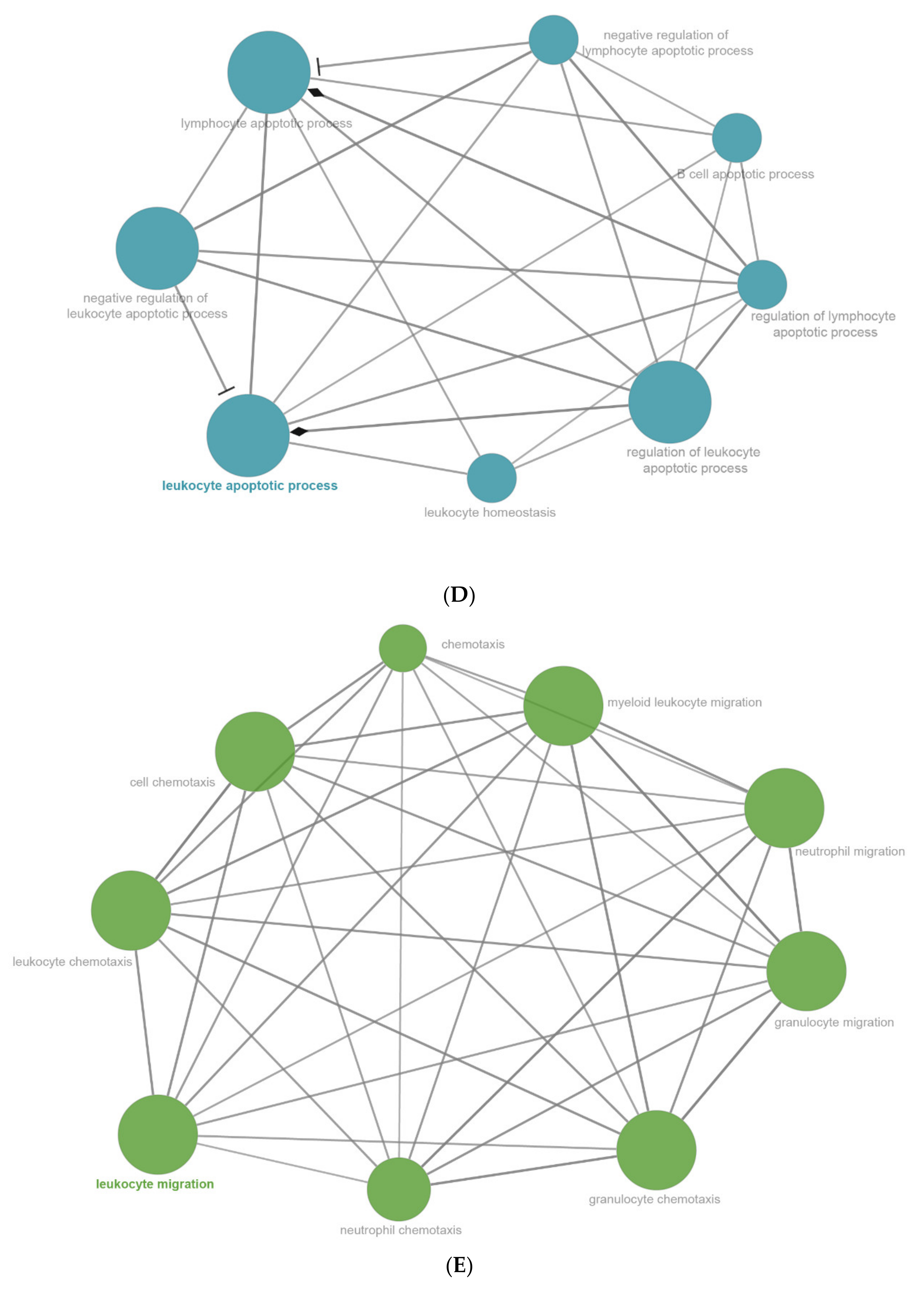
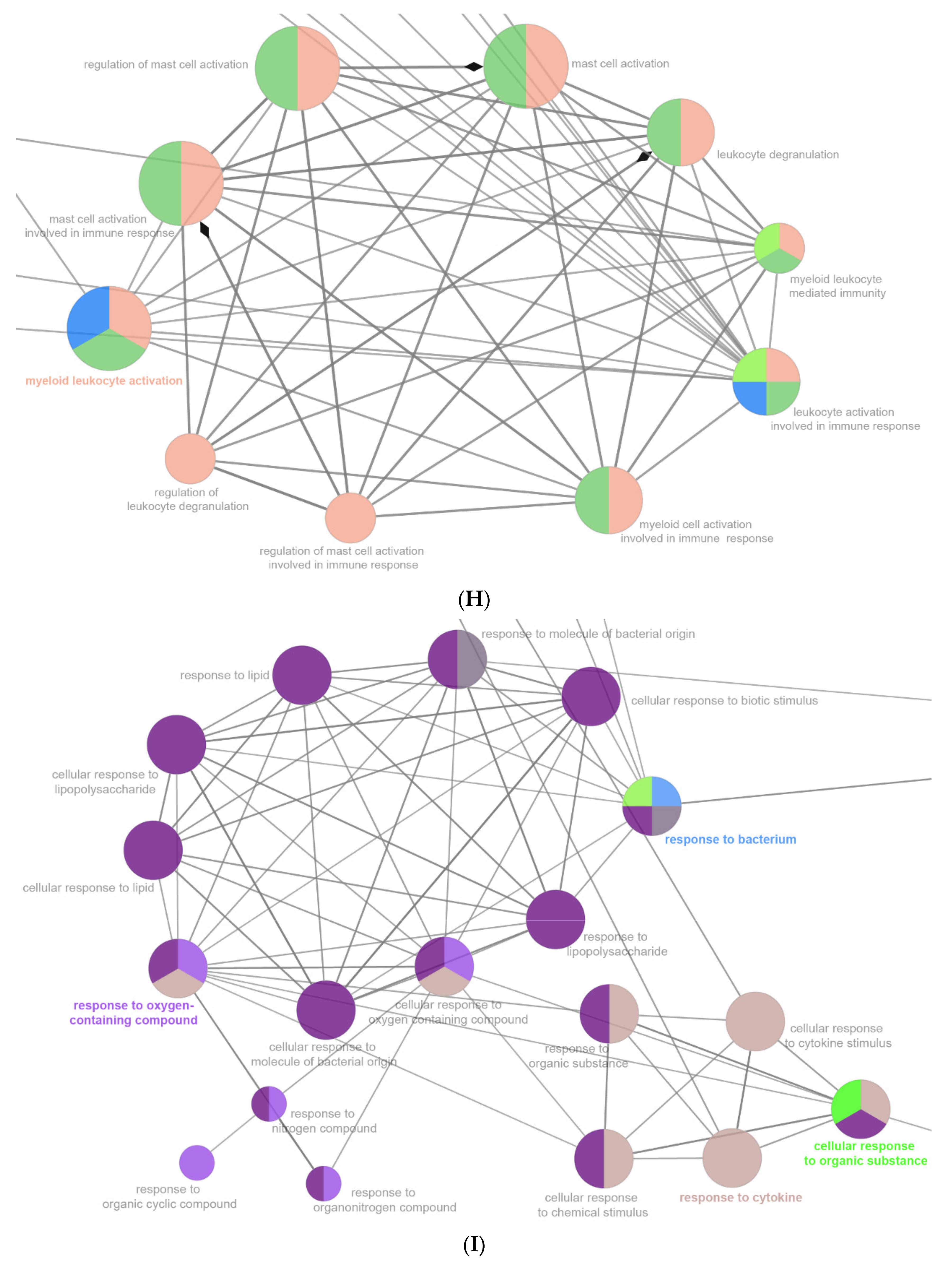
2.6. The Enrichment Pathway of DEGs-Cluego
2.7. Transcriptional Factors Tied to DEGs
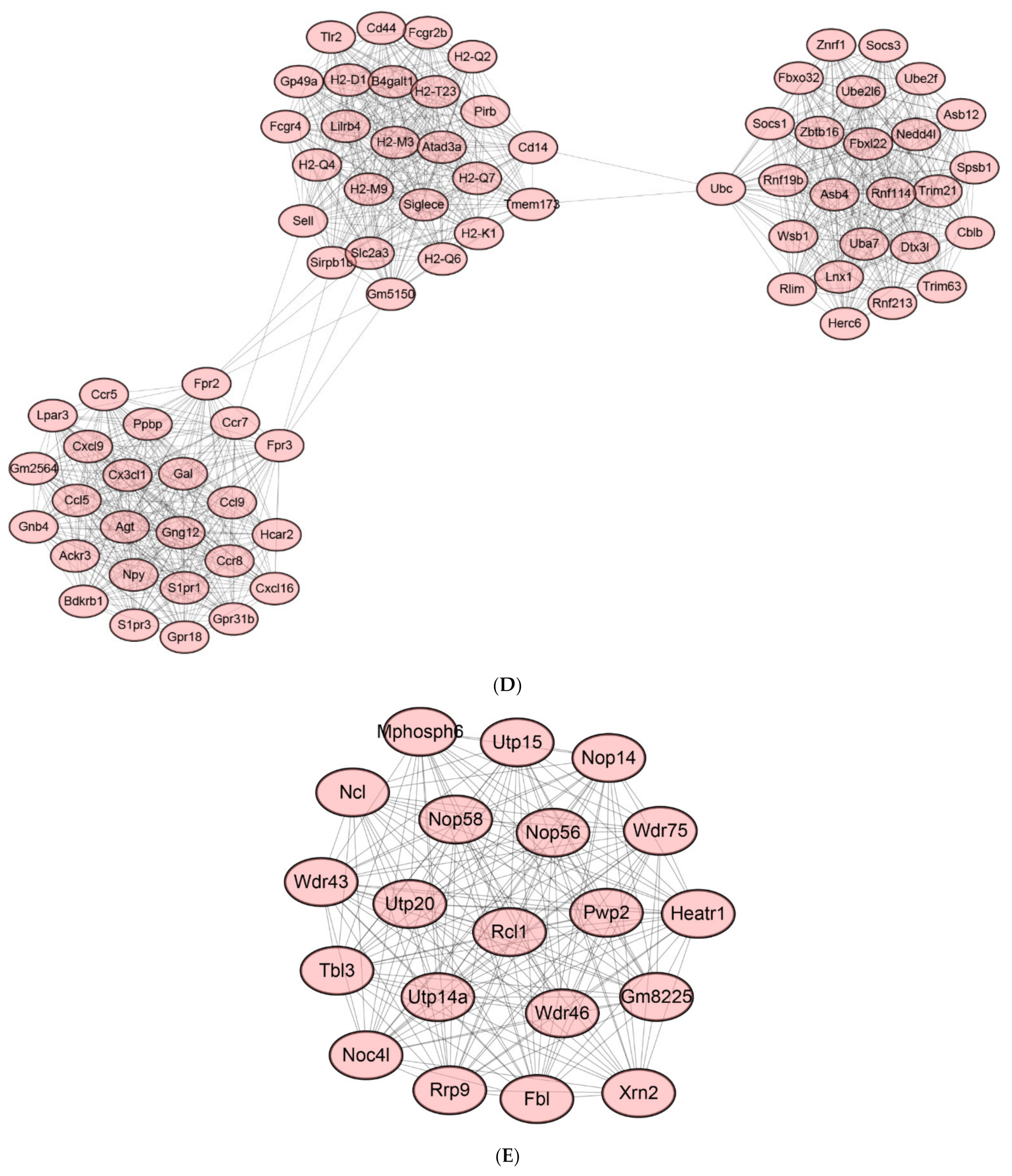
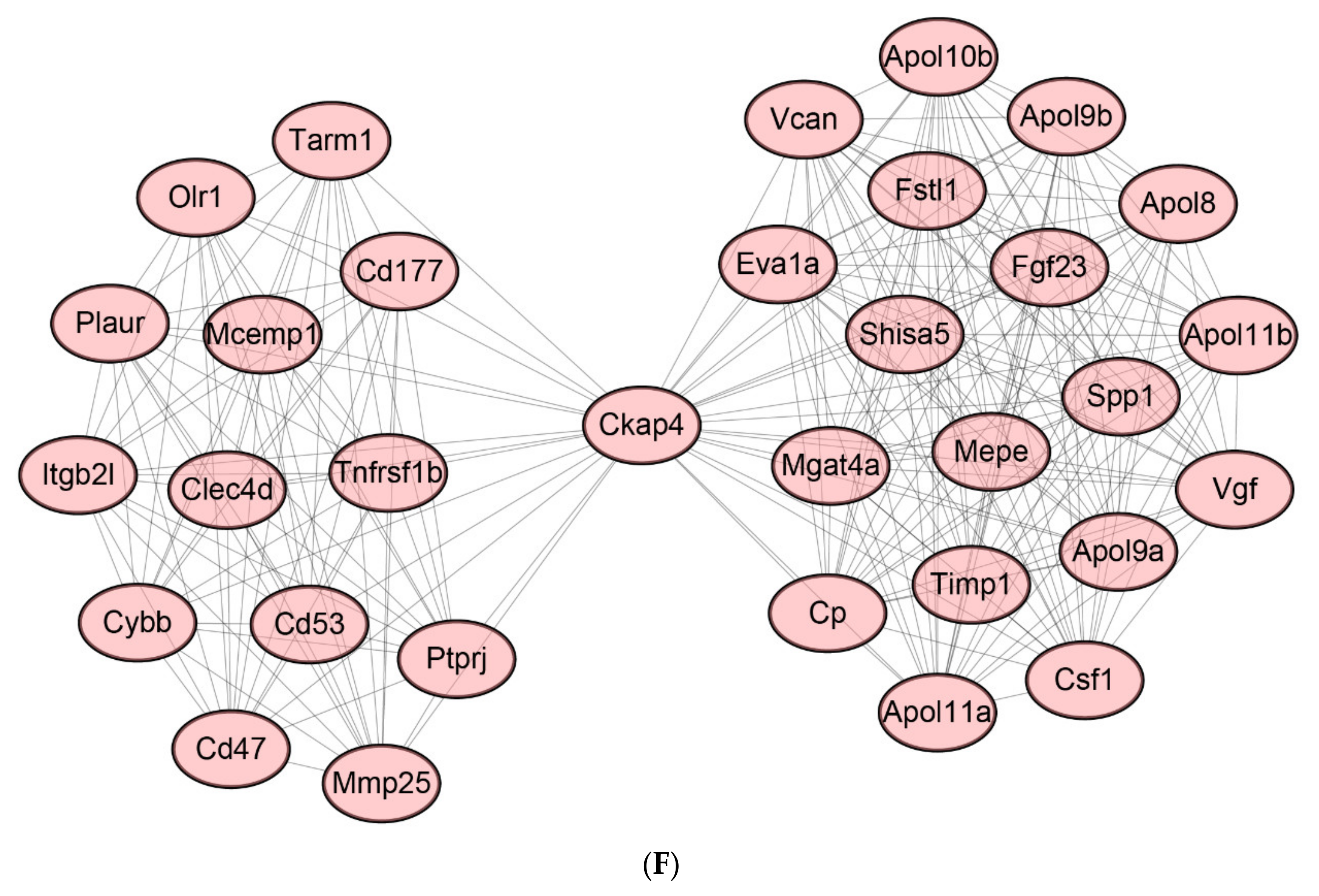
2.8. Protein-Protein Interaction (PPI) Network of DEGs
2.9. Identification of Hub Genes
2.10. qPCR Detection of Hub Genes
2.11. Modeling of Hub Proteins
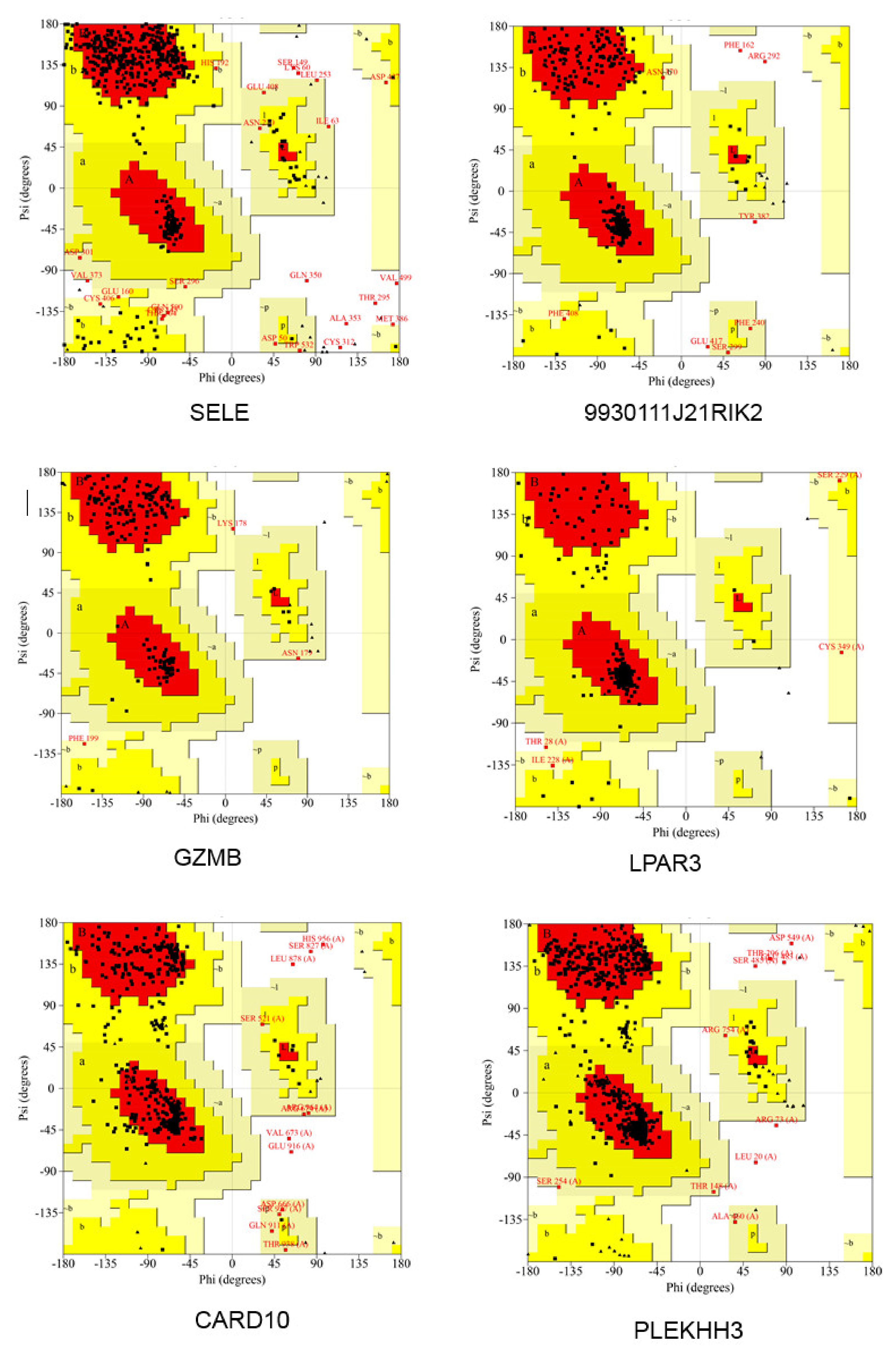
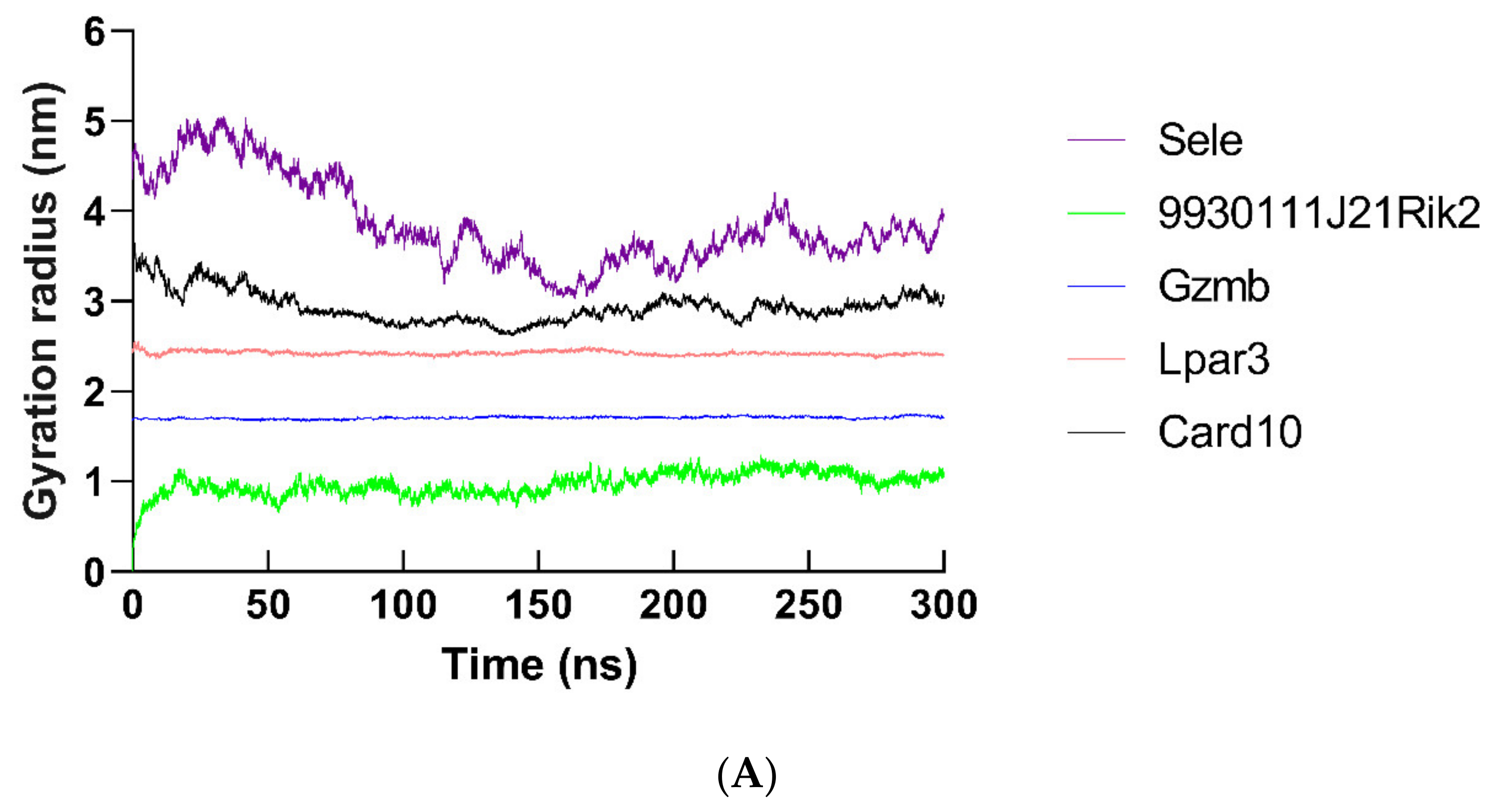
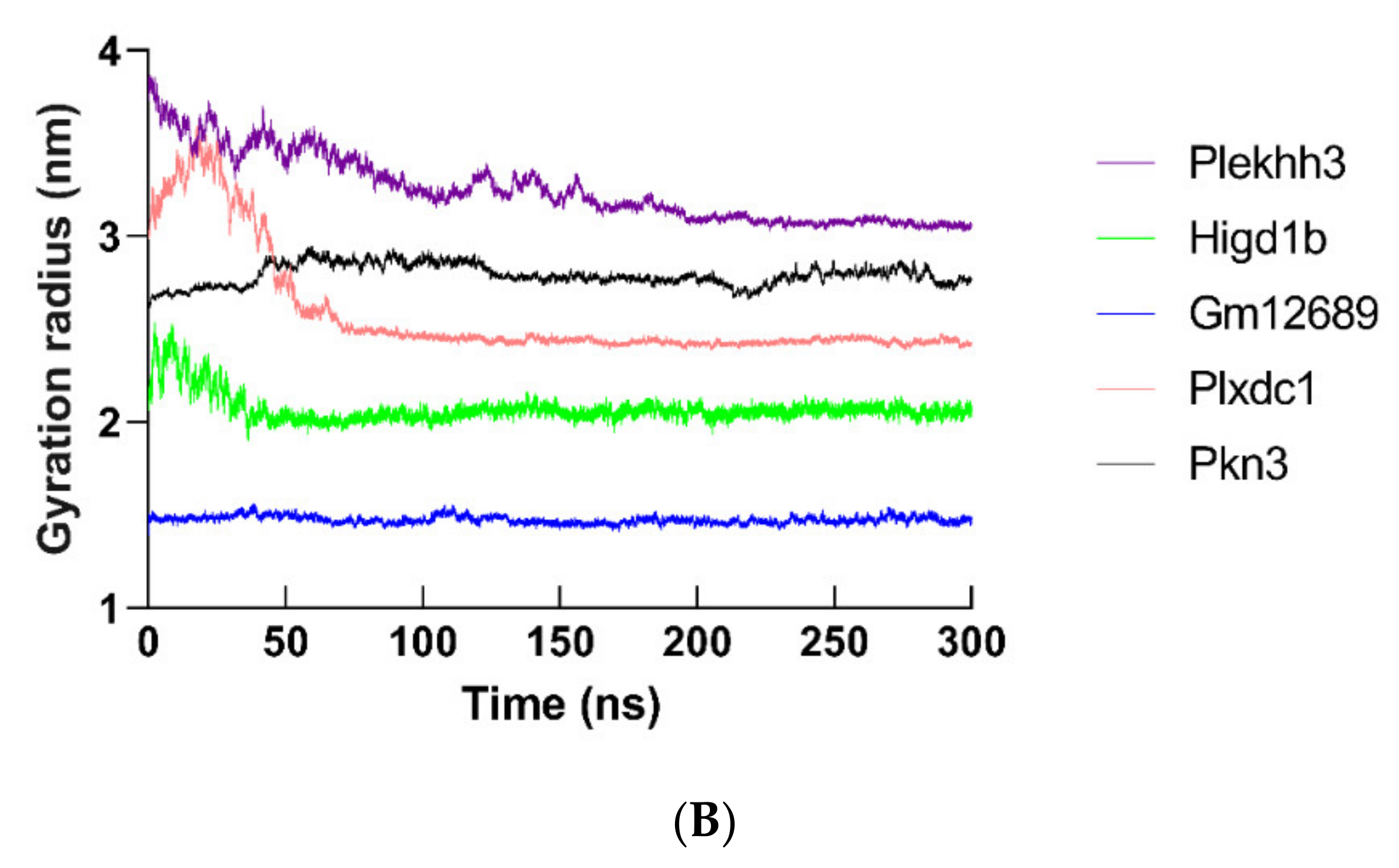
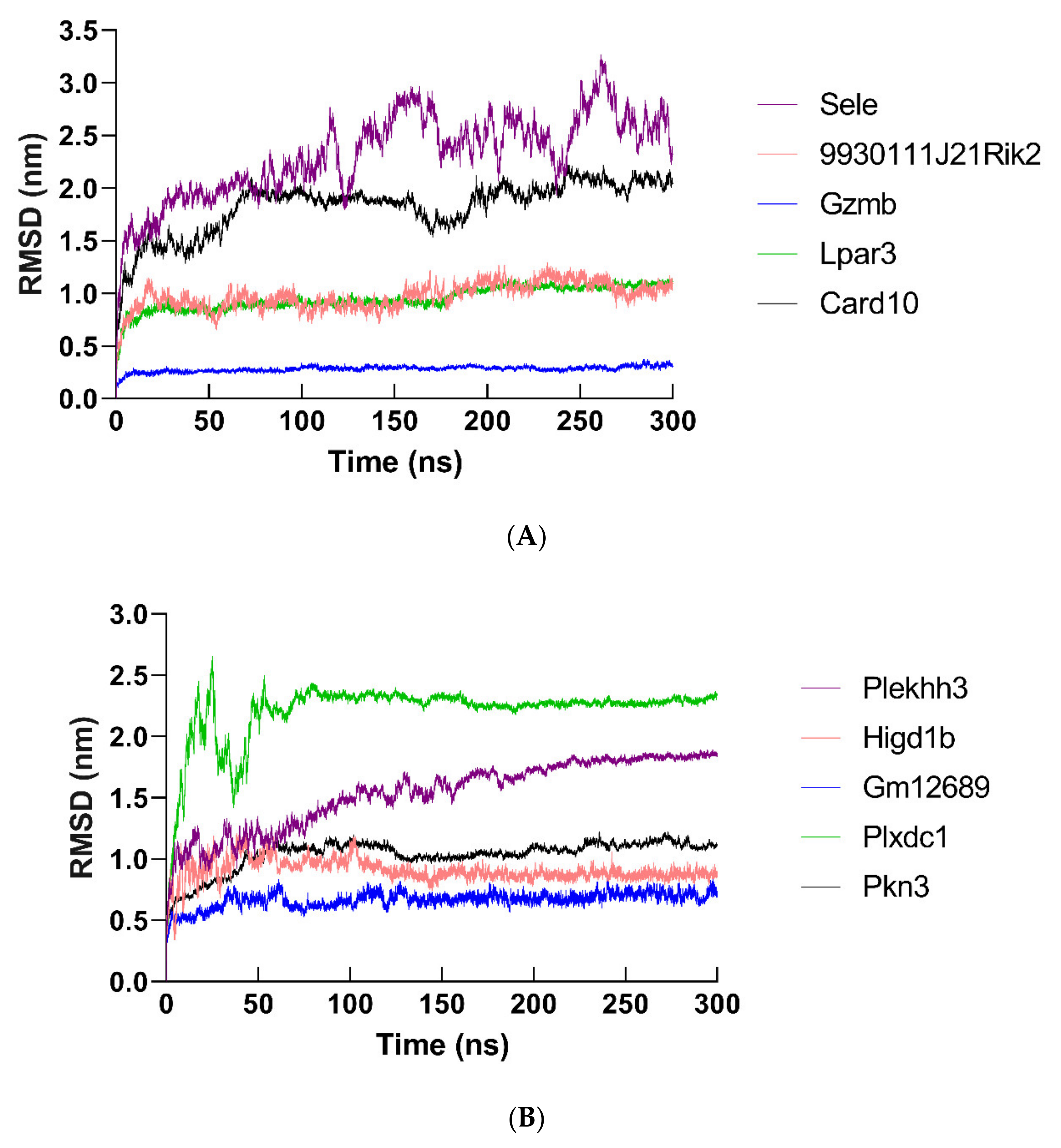
2.12. Molecular Dynamics Simulation of Hub Proteins
2.13. TAK 242-Hub Protein Docking
3. Discussion
3.1. Transcriptomics Study of LPS-Induced Peritonitis
3.2. Transcription Factors for Peritonitis Caused by LPS
3.3. Hub Genes
3.4. Anti-Peritonitis Activity of TAK 242 (TLR4 Inhibitor)
4. Materials and Methods
4.1. Ethical Declaration
4.2. Mice
4.3. Reagents
4.4. LPS-induced Mouse Peritonitis Model
4.5. ELISA Assay
4.6. Mouse Peritoneal Sample Preparation for RNA-Seq
4.7. RNA-Seq
4.8. Bioinformatics Analysis of RNA-Seq Data
4.8.1. Differential Expression Analysis
4.8.2. Enrichment Interpretation
4.8.3. Protein-Protein Interaction (PPI) Analysis
4.9. Prediction of Transcriptional Factors for DEGs
4.10. Real-Time Quantitative PCR (qPCR) Detection
4.11. Modeling of Hub Proteins
4.12. Molecular Dynamics Simulation
4.13. TAK 242-hub Protein Docking
4.14. Statistical Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Montravers, P.; Assadi, M.; Gouel-Cheron, A. Priorities in peritonitis. Curr. Opin. Crit. Care 2021, 27, 201–207. [Google Scholar] [CrossRef]
- Pörner, D.; Von Vietinghoff, S.; Nattermann, J.; Strassburg, C.P.; Lutz, P. Advances in the pharmacological management of bacterial peritonitis. Expert Opin. Pharmacother. 2021, 22, 1567–1578. [Google Scholar] [CrossRef]
- Gao, W.; Xiong, Y.; Li, Q.; Yang, H. Inhibition of Toll-Like Receptor Signaling as a Promising Therapy for Inflammatory Diseases: A Journey from Molecular to Nano Therapeutics. Front. Physiol. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.W.; Sud, K.; Van, C.; Zaidi, S.T.R.; Patel, R.P.; Peterson, G.M.; Castelino, R.L. Pharmacokinetics of culture-directed antibiotics for the treatment of peritonitis in automated peritoneal dialysis: A systematic narrative review. Perit. Dial. Int. 2021, 41, 261–272. [Google Scholar] [CrossRef]
- Sun, W.; Liu, C.; Zhang, Y.; Qiu, X.; Zhang, L.; Zhao, H.; Rong, Y.; Sun, Y. Ilexgenin A, a novel pentacyclic triterpenoid extracted from Aquifoliaceae shows reduction of LPS-induced peritonitis in mice. Eur. J. Pharmacol. 2017, 797, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Chuang, H.C.; Tsai, C.Y.; Xiao, Y.Z.; Yang, J.Y.; Huang, R.H.; Shih, Y.C.; Tan, T.H. DUSP11 Attenuates Lipopolysaccharide-Induced Macrophage Activation by Targeting TAK1. J. Immunol. 2020, 205, 1644–1652. [Google Scholar] [CrossRef]
- Orimo, T.; Sasaki, I.; Hemmi, H.; Ozasa, T.; Fukuda-Ohta, Y.; Ohta, T.; Morinaka, M.; Kitauchi, M.; Yamaguchi, T.; Sato, Y.; et al. Cholera toxin B induces interleukin-1β production from resident peritoneal macrophages through the pyrin inflammasome as well as the NLRP3 inflammasome. Int. Immunol. 2019, 31, 657–668. [Google Scholar] [CrossRef] [Green Version]
- Ajayi, A.M.; Martins, D.T.O.; Balogun, S.O.; Oliveira, R.G.; Ascêncio, S.D.; Soares, I.M.; Barbosa, R.D.S.; Ademowo, O.G. Ocimum gratissimum L. leaf flavonoid-rich fraction suppress LPS-induced inflammatory response in RAW 264.7 macrophages and peritonitis in mice. J. Ethnopharmacol. 2017, 204, 169–178. [Google Scholar] [CrossRef]
- He, L.; Yan, B.; Yao, C.; Chen, X.; Li, L.; Wu, Y.; Song, Z.; Song, S.; Zhang, Z.; Luo, P. Oligosaccharides from Polygonatum Cyrtonema Hua: Structural characterization and treatment of LPS-induced peritonitis in mice. Carbohydr. Polym. 2021, 255, 117392. [Google Scholar] [CrossRef]
- Lamping, N.; Dettmer, R.; Schröder, N.W.; Pfeil, D.; Hallatschek, W.; Burger, R.; Schumann, R.R. LPS-binding protein protects mice from septic shock caused by LPS or gram-negative bacteria. J. Clin. Investig. 1998, 101, 2065–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, H.; Zhang, Q.; Lin, Y.; Gao, Y.; Zhang, P. The Ovotransferrin-Derived Peptide IRW Attenuates Lipopolysaccharide-Induced Inflammatory Responses. BioMed Res. Int. 2019, 2019, 8676410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.X.; Tang, X.; Zhu, J.Y.; Zhang, W.; Tang, W.Q.; Yan, J.; Xu, X.; Liang, H.P. Cytochrome P450 1A1 enhances Arginase-1 expression, which reduces LPS-induced mouse peritonitis by targeting JAK1/STAT6. Cell. Immunol. 2020, 349, 104047. [Google Scholar] [CrossRef]
- Ha, A.T.; Kim, M.Y.; Cho, J.Y. TAK1/AP-1-Targeted Anti-Inflammatory Effects of Barringtonia augusta Methanol Extract. Molecules 2021, 26, 3053. [Google Scholar] [CrossRef]
- Borghi, S.M.; Mizokami, S.S.; Carvalho, T.T.; Rasquel-Oliveira, F.S.; Ferraz, C.R.; Fattori, V.; Hayashida, T.H.; Peron, J.P.S.; Camilios-Neto, D.; Ambrosio, S.R.; et al. The diterpene from Sphagneticola trilobata (L.) Pruski, kaurenoic acid, reduces lipopolysaccharide-induced peritonitis and pain in mice. J. Ethnopharmacol. 2021, 273, 113980. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Meng, N.; Chang, B.; Quan, X.; Yuan, R.; Li, B. Anti-Inflammatory Activity of Epimedium brevicornu Maxim Ethanol Extract. J. Med. Food 2018, 21, 726–733. [Google Scholar] [CrossRef]
- Hara, T.; Hiratsuka, T.; Etoh, T.; Itai, Y.; Kono, Y.; Shiroshita, H.; Shiraishi, N.; Inomata, M. Intraperitoneal Phototherapy Suppresses Inflammatory Reactions in a Surgical Model of Peritonitis. J. Surg. Res. 2020, 252, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Correa, F.; Lopes Martins, R.A.; Correa, J.C.; Iversen, V.V.; Joenson, J.; Bjordal, J.M. Low-level laser therapy (GaAs lambda = 904 nm) reduces inflammatory cell migration in mice with lipopolysaccharide-induced peritonitis. Photomed. Laser Surg. 2007, 25, 245–249. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Vaure, C.; Liu, Y. A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 2014, 5, 316. [Google Scholar] [CrossRef] [Green Version]
- Ii, M.; Matsunaga, N.; Hazeki, K.; Nakamura, K.; Takashima, K.; Seya, T.; Hazeki, O.; Kitazaki, T.; Iizawa, Y. A novel cyclohexene derivative, ethyl (6R)-6-[N-(2-Chloro-4-fluorophenyl)sulfamoyl]cyclohex-1-ene-1-carboxylate (TAK-242), selectively inhibits toll-like receptor 4-mediated cytokine production through suppression of intracellular signaling. Mol. Pharmacol. 2006, 69, 1288–1295. [Google Scholar] [CrossRef] [Green Version]
- Yamada, M.; Ichikawa, T.; Yamano, T.; Kikumoto, F.; Nishikimi, Y.; Tamura, N.; Kitazaki, T. Optically active cyclohexene derivative as a new antisepsis agent: An efficient synthesis of ethyl (6R)-6-[N-(2-chloro-4-fluorophenyl)sulfamoyl]cyclohex-1-ene-1-carboxylate (TAK-242). Chem. Pharm. Bull. 2006, 54, 58–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sha, T.; Sunamoto, M.; Kitazaki, T.; Sato, J.; Ii, M.; Iizawa, Y. Therapeutic effects of TAK-242, a novel selective Toll-like receptor 4 signal transduction inhibitor, in mouse endotoxin shock model. Eur. J. Pharmacol. 2007, 571, 231–239. [Google Scholar] [CrossRef]
- Kawamoto, T.; Ii, M.; Kitazaki, T.; Iizawa, Y.; Kimura, H. TAK-242 selectively suppresses Toll-like receptor 4-signaling mediated by the intracellular domain. Eur. J. Pharmacol. 2008, 584, 40–48. [Google Scholar] [CrossRef]
- Rice, T.W.; Wheeler, A.P.; Bernard, G.R.; Vincent, J.L.; Angus, D.C.; Aikawa, N.; Demeyer, I.; Sainati, S.; Amlot, N.; Cao, C.; et al. A randomized, double-blind, placebo-controlled trial of TAK-242 for the treatment of severe sepsis. Crit. Care Med. 2010, 38, 1685–1694. [Google Scholar] [CrossRef]
- Matsunaga, N.; Tsuchimori, N.; Matsumoto, T.; Ii, M. TAK-242 (resatorvid), a small-molecule inhibitor of Toll-like receptor (TLR) 4 signaling, binds selectively to TLR4 and interferes with interactions between TLR4 and its adaptor molecules. Mol. Pharmacol. 2011, 79, 34–41. [Google Scholar] [CrossRef] [Green Version]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Chalikonda, G.; Lee, H.; Sheik, A.; Huh, Y.S. Targeting key transcriptional factor STAT3 in colorectal cancer. Mol. Cell. Biochem. 2021, 476, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.C.; Yu, W.W.; Zhou, H.C.; Lan, Z.C.; Wu, T.; Xiong, S.M.; Yan, L.; Liu, H.B. Lycium barbarum polysaccharides ameliorate LPS-induced inflammation of RAW264.7 cells and modify the behavioral score of peritonitis mice. J. Food Biochem. 2021, 45, e13889. [Google Scholar] [CrossRef]
- Pastwińska, J.; Żelechowska, P.; Walczak-Drzewiecka, A.; Brzezińska-Błaszczyk, E.; Dastych, J. The Art of Mast Cell Adhesion. Cells 2020, 9, 2664. [Google Scholar] [CrossRef] [PubMed]
- Zeglinski, M.R.; Granville, D.J. Granzymes in cardiovascular injury and disease. Cell. Signal. 2020, 76, 109804. [Google Scholar] [CrossRef]
- Lin, Y.N.; Audira, G.; Malhotra, N.; Ngoc Anh, N.T.; Siregar, P.; Lu, J.H.; Lee, H.; Hsiao, C.D. A Novel Function of the Lysophosphatidic Acid Receptor 3 (LPAR3) Gene in Zebrafish on Modulating Anxiety, Circadian Rhythm Locomotor Activity, and Short-Term Memory. Int. J. Mol. Sci. 2020, 21, 2837. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Lin, X. CARMA3: Scaffold Protein Involved in NF-κB Signaling. Front. Immunol 2019, 10, 176. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, K.D.; Su, M.C.; Chin, C.H.; Chen, C.J.; Liou, C.W.; Chen, T.W.; Chang, Y.C.; Huang, K.T.; Wang, C.C.; et al. Genome-wide gene expression array identifies novel genes related to disease severity and excessive daytime sleepiness in patients with obstructive sleep apnea. PLoS ONE 2017, 12, e0176575. [Google Scholar]
- Zhou, Y.; Xu, B.; Zhou, Y.; Liu, J.; Zheng, X.; Liu, Y.; Deng, H.; Liu, M.; Ren, X.; Xia, J.; et al. Identification of Key Genes With Differential Correlations in Lung Adenocarcinoma. Front. Cell Dev. Biol. 2021, 9, 675438. [Google Scholar] [CrossRef] [PubMed]
- Van Beijnum, J.R.; Petersen, K.; Griffioen, A.W. Tumor endothelium is characterized by a matrix remodeling signature. Front. Biosci. 2009, 1, 216–225. [Google Scholar] [CrossRef]
- Uehara, S.; Udagawa, N.; Kobayashi, Y. Regulation of osteoclast function via Rho-Pkn3-c-Src pathways. J. Oral Biosci. 2019, 61, 135–140. [Google Scholar] [CrossRef]
- Ono, Y.; Maejima, Y.; Saito, M.; Sakamoto, K.; Horita, S.; Shimomura, K.; Inoue, S.; Kotani, J. TAK-242, a specific inhibitor of Toll-like receptor 4 signalling, prevents endotoxemia-induced skeletal muscle wasting in mice. Sci. Rep. 2020, 10, 694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takashima, K.; Matsunaga, N.; Yoshimatsu, M.; Hazeki, K.; Kaisho, T.; Uekata, M.; Hazeki, O.; Akira, S.; Iizawa, Y.; Ii, M. Analysis of binding site for the novel small-molecule TLR4 signal transduction inhibitor TAK-242 and its therapeutic effect on mouse sepsis model. Br. J. Pharmacol. 2009, 157, 1250–1262. [Google Scholar] [CrossRef] [Green Version]
- Fenhammar, J.; Rundgren, M.; Forestier, J.; Kalman, S.; Eriksson, S.; Frithiof, R. Toll-like receptor 4 inhibitor TAK-242 attenuates acute kidney injury in endotoxemic sheep. Anesthesiology 2011, 114, 1130–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gárate, I.; García-Bueno, B.; Madrigal, J.L.; Caso, J.R.; Alou, L.; Gómez-Lus, M.L.; Leza, J.C. Toll-like 4 receptor inhibitor TAK-242 decreases neuroinflammation in rat brain frontal cortex after stress. J. Neuroinflamm. 2014, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Hua, F.; Tang, H.; Wang, J.; Prunty, M.C.; Hua, X.; Sayeed, I.; Stein, D.G. TAK-242, an antagonist for Toll-like receptor 4, protects against acute cerebral ischemia/reperfusion injury in mice. J. Cereb. Blood Flow Metab. 2015, 35, 536–542. [Google Scholar] [CrossRef]
- Kashani, B.; Zandi, Z.; Karimzadeh, M.R.; Bashash, D.; Nasrollahzadeh, A.; Ghaffari, S.H. Blockade of TLR4 using TAK-242 (resatorvid) enhances anti-cancer effects of chemotherapeutic agents: A novel synergistic approach for breast and ovarian cancers. Immunol. Res. 2019, 67, 505–516. [Google Scholar] [CrossRef]
- Samarpita, S.; Kim, J.Y.; Rasool, M.K.; Kim, K.S. Investigation of toll-like receptor (TLR) 4 inhibitor TAK-242 as a new potential anti-rheumatoid arthritis drug. Arthritis Res. Ther. 2020, 22, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Chen, X.; Chen, Z.; Ma, X.; Li, D.; Shang, P.; Qian, A. Neuropeptide FF attenuates RANKL-induced differentiation of macrophage-like cells into osteoclast-like cells. Arch. Oral Biol. 2015, 60, 282–292. [Google Scholar] [CrossRef]
- Sun, Y.; Zuo, Z.; Kuang, Y. Prolactin-Releasing Peptide Differentially Regulates Gene Transcriptomic Profiles in Mouse Bone Marrow-Derived Macrophages. Int. J. Mol. Sci. 2021, 22, 4456. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kuang, Y.; Zuo, Z.; Zhang, J.; Ma, X.; Xing, X.; Liu, L.; Miao, Y.; Ren, T.; Li, H.; et al. Cellular processes involved in RAW 264.7 macrophages exposed to NPFF: A transcriptional study. Peptides 2021, 136, 170469. [Google Scholar] [CrossRef]
- Sun, Y.; Kuang, Y.; Zuo, Z. Transcriptomic Changes in Mouse Bone Marrow-Derived Macrophages Exposed to Neuropeptide FF. Genes 2021, 12, 705. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Ahn, J.H.; Hwang, S.H.; Cho, H.S.; Lee, M. Differential Gene Expression Common to Acquired and Intrinsic Resistance to BRAF Inhibitor Revealed by RNA-Seq Analysis. Biomol. Ther. 2019, 27, 302–310. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Sun, Y.L.; Chen, Z.H.; Chen, X.H.; Yin, C.; Li, D.J.; Ma, X.L.; Zhao, F.; Zhang, G.; Shang, P.; Qian, A.R. Diamagnetic Levitation Promotes Osteoclast Differentiation from RAW264.7 Cells. IEEE Trans. Biomed. Eng. 2015, 62, 900–908. [Google Scholar] [CrossRef]
- Li, H.; Liu, S.; Du, Y.; Tan, J.; Luo, J.; Sun, Y. Hub Proteins Involved in RAW 264.7 Macrophages Exposed to Direct Current Electric Field. Int. J. Mol. Sci. 2020, 21, 4505. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E.J.S. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Suppl. 2), W270–W277. [Google Scholar] [CrossRef] [PubMed] [Green Version]



























Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zhang, S.; Sun, Y.; Zhou, W. Transcriptomics Changes in the Peritoneum of Mice with Lipopolysaccharide-Induced Peritonitis. Int. J. Mol. Sci. 2021, 22, 13008. https://doi.org/10.3390/ijms222313008
Liu S, Zhang S, Sun Y, Zhou W. Transcriptomics Changes in the Peritoneum of Mice with Lipopolysaccharide-Induced Peritonitis. International Journal of Molecular Sciences. 2021; 22(23):13008. https://doi.org/10.3390/ijms222313008
Chicago/Turabian StyleLiu, Shaoguang, Shaotong Zhang, Yulong Sun, and Wence Zhou. 2021. "Transcriptomics Changes in the Peritoneum of Mice with Lipopolysaccharide-Induced Peritonitis" International Journal of Molecular Sciences 22, no. 23: 13008. https://doi.org/10.3390/ijms222313008





