Pathogenesis and Treatment of Cytokine Storm Induced by Infectious Diseases
Abstract
1. Introduction
1.1. The Definition of Cytokine Storm
1.2. Cytokines Associated with Cytokine Storm
1.3. The Clinical Effects, Therapeutic Approaches, and Prognosis of Cytokine Storms
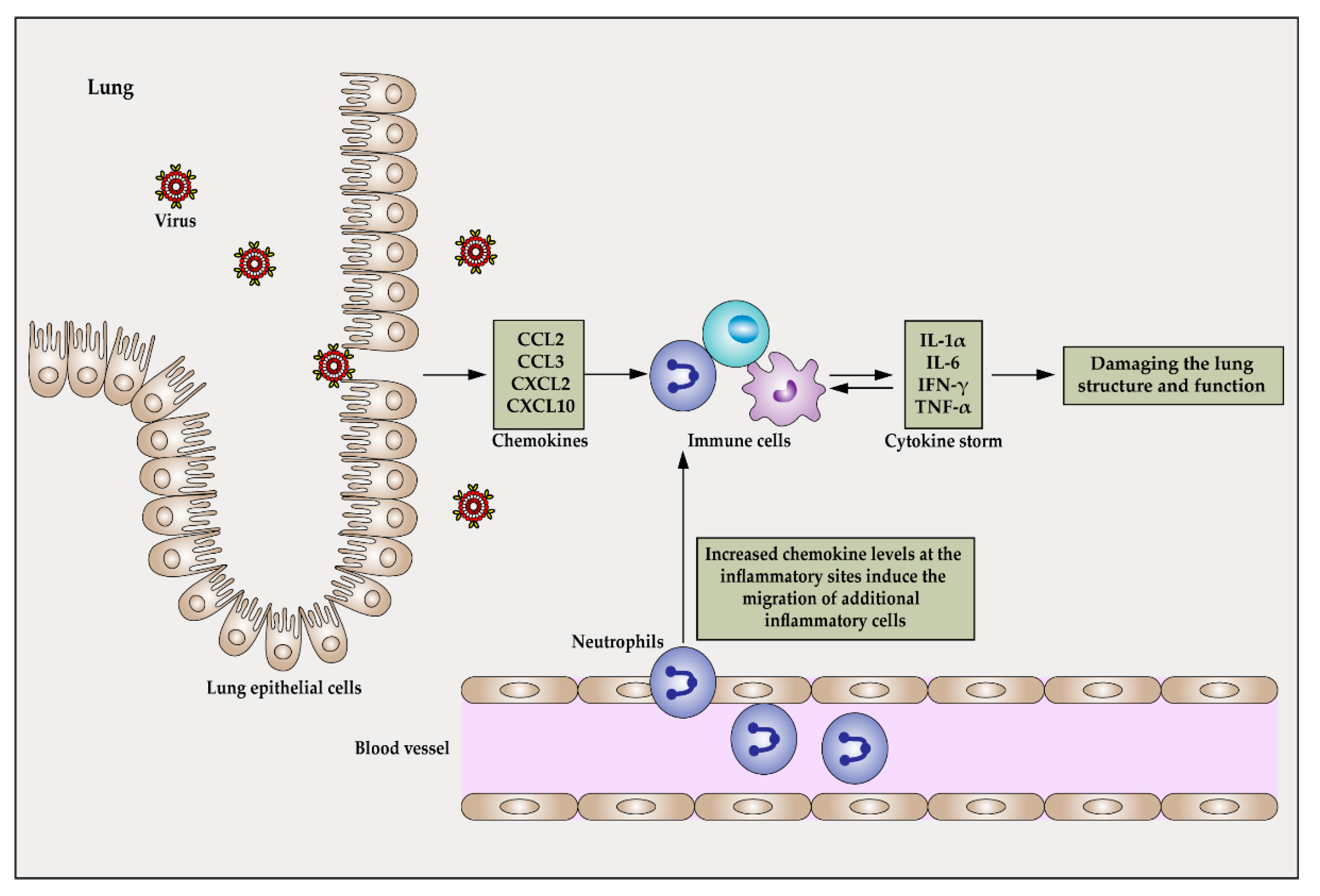
2. Pathophysiological Mechanisms of Cytokine Storms
2.1. The Type of Pathogen
2.1.1. The Pathophysiological Mechanism of CRS Caused by Influenza Virus Infection
2.1.2. The Pathophysiological Mechanism of Bacterial Sepsis-Induced CRS
2.1.3. The Pathophysiological Mechanism of CRS Caused by SARS-CoV-2 Infection
2.2. The Amount of Pathogen Infection and the Level of Pathogen Growth
3. Treatment Strategies for CRS Induced by Infectious Diseases
3.1. Targeting the Pathogen
3.2. Targeting the Host
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef]
- Mangalmurti, N.; Hunter, C.A. Cytokine Storms: Understanding COVID-19. Immunity 2020, 53, 19–25. [Google Scholar] [CrossRef]
- Moore, J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef]
- Ferrara, J.L.; Abhyankar, S.; Gilliland, D.G. Cytokine storm of graft-versus-host disease: A critical effector role for interleukin-1. Transplant. Proc. 1993, 25, 1216–1217. [Google Scholar]
- Koyama, M.; Hill, G.R. The primacy of gastrointestinal tract antigen-presenting cells in lethal graft-versus-host disease. Blood 2019, 134, 2139–2148. [Google Scholar] [CrossRef]
- Yuen, K.Y.; Wong, S.S. Human infection by avian influenza A H5N1. Hong Kong Med. J. = Xianggang Yi Xue Za Zhi 2005, 11, 189–199. [Google Scholar]
- A Morgan, R.; Yang, J.C.; Kitano, M.; E Dudley, M.; Laurencot, C.M.; A Rosenberg, S. Case Report of a Serious Adverse Event Following the Administration of T Cells Transduced with a Chimeric Antigen Receptor Recognizing ERBB2. Mol. Ther. 2010, 18, 843–851. [Google Scholar] [CrossRef]
- Hay, K.A. Cytokine release syndrome and neurotoxicity after CD19 chimeric antigen receptor-modified (CAR-) T cell therapy. Br. J. Haematol. 2018, 183, 364–374. [Google Scholar] [CrossRef]
- Coley, W.B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases, 1893. Clin. Orthop. Relat. Res. 1991, 262, 3–11. [Google Scholar] [CrossRef]
- Pechous, R.D.; Sivaraman, V.; Price, P.A.; Stasulli, N.M.; Goldman, W.E. Early Host Cell Targets of Yersinia pestis during Primary Pneumonic Plague. PLoS Pathog. 2013, 9, e1003679. [Google Scholar] [CrossRef]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Y.-H.; Yang, Z.-Q. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2015, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R. Cytokine storms in infectious diseases. Semin. Immunopathol. 2017, 39, 501–503. [Google Scholar] [CrossRef]
- Shimabukuro-Vornhagen, A.; Gödel, P.; Subklewe, M.; Stemmler, H.J.; Schlößer, H.A.; Schlaak, M.; Kochanek, M.; Böll, B.; Von Bergwelt-Baildon, M.S. Cytokine release syndrome. J. Immunother. Cancer 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Vogel, W.H. Infusion Reactions. Clin. J. Oncol. Nurs. 2010, 14, E10–E21. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 2016, 8, 959–970. [Google Scholar] [CrossRef]
- Said, E.A.; Al-Reesi, M.I.; Al-Shizawi, M.N.; Jaju, S.; Al-Balushi, M.S.; Koh, C.Y.; Al-Jabri, P.A.A.; Jeyaseelan, P.L. Defining IL-6 levels in healthy individuals: A meta-analysis. J. Med. Virol. 2020, 93, 3915–3924. [Google Scholar] [CrossRef]
- Ulhaq, Z.S.; Soraya, G.V. Anti-IL-6 receptor antibody treatment for severe COVID-19 and the potential implication of IL-6 gene polymorphisms in novel coronavirus pneumonia. Med. Clin. 2020, 155, 548–556. [Google Scholar] [CrossRef]
- Lee, D.W.; A Gardner, R.; Porter, D.L.; Louis, C.U.; Ahmed, N.; Jensen, M.C.; Grupp, S.A.; Mackall, C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood 2014, 124, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Galván-Román, J.M.; Rodríguez-García, S.C.; Roy-Vallejo, E.; Marcos-Jiménez, A.; Sánchez-Alonso, S.; Fernández-Díaz, C.; Alcaraz-Serna, A.; Mateu-Albero, T.; Rodríguez-Cortes, P.; Sánchez-Cerrillo, I.; et al. IL-6 serum levels predict severity and response to tocilizumab in COVID-19: An observational study. J. Allergy Clin. Immunol. 2020, 147, 72–80.e8. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhao, X.; Qi, Y.; Li, H.; Ye, G.; Liu, Y.; Zhang, Y.; Gou, J. Sepsis-associated severe interleukin-6 storm in critical coronavirus disease 2019. Cell. Mol. Immunol. 2020, 17, 1092–1094. [Google Scholar] [CrossRef] [PubMed]
- Hay, K.A.; Hanafi, L.-A.; Li, D.; Gust, J.; Liles, W.C.; Wurfel, M.M.; López, J.A.; Chen, J.; Chung, D.; Harju-Baker, S.; et al. Kinetics and biomarkers of severe cytokine release syndrome after CD19 chimeric antigen receptor–modified T-cell therapy. Blood 2017, 130, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Norelli, M.; Camisa, B.; Barbiera, G.; Falcone, L.; Purevdorj, A.; Genua, M.; Sanvito, F.; Ponzoni, M.; Doglioni, C.; Cristofori, P.; et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 2018, 24, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fehr, A.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef]
- Kindler, E.; Thiel, V.; Weber, F. Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response. Adv. Virus Res. 2016, 96, 219–243. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Lee, J.Y.; Yang, J.W.; Lee, K.H.; Effenberger, M.; Szpirt, W.; Kronbichler, A.; Shin, J.I. Immunopathogenesis and treatment of cytokine storm in COVID-19. Theranostics 2021, 11, 316–329. [Google Scholar] [CrossRef]
- Grupp, S.A.; Kalos, M.; Barrett, D.; Aplenc, R.; Porter, D.L.; Rheingold, S.R.; Teachey, D.T.; Chew, A.; Hauck, B.; Wright, J.F.; et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. N. Engl. J. Med. 2013, 18, 1509–1518. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- England, J.T.; Abdulla, A.; Biggs, C.; Lee, A.Y.; Hay, K.; Hoiland, R.L.; Wellington, C.L.; Sekhon, M.; Jamal, S.; Shojania, K.; et al. Weathering the COVID-19 storm: Lessons from hematologic cytokine syndromes. Blood Rev. 2020, 45, 100707. [Google Scholar] [CrossRef] [PubMed]
- The Writing Committee for the REMAP-CAP Investigators; Angus, D.C.; Derde, L.; Al-Beidh, F.; Annane, D.; Arabi, Y.; Beane, A.; Van Bentum-Puijk, W.; Berry, L.; Bhimani, Z.; et al. Effect of Hydrocortisone on Mortality and Organ Support in Patients With Severe COVID-19: The REMAP-CAP COVID-19 Corticosteroid Domain Randomized Clinical Trial. JAMA 2020, 324, 1317–1329. [Google Scholar] [CrossRef] [PubMed]
- Inchingolo, A.; Inchingolo, A.; Bordea, I.; Malcangi, G.; Xhajanka, E.; Scarano, A.; Lorusso, F.; Farronato, M.; Tartaglia, G.; Isacco, C.; et al. SARS-CoV-2 Disease Adjuvant Therapies and Supplements Breakthrough for the Infection Prevention. Microorganisms 2021, 9, 525. [Google Scholar] [CrossRef]
- de Ligt, M.; Hesselink, M.K.C.; Jorgensen, J.; Hoebers, N.; Blaak, E.E.; Goossens, G.H. Resveratrol supplementation reduces ACE2 expression in human adipose tissue. Adipocyte 2021, 10, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Rathi, H.; Haq, A.; Wimalawansa, S.J.; Sharma, A. Putative roles of vitamin D in modulating immune response and immunopathology associated with COVID-19. Virus Res. 2020, 292, 198235. [Google Scholar] [CrossRef]
- Shah, M.A.; Rasul, A.; Yousaf, R.; Haris, M.; Faheem, H.I.; Hamid, A.; Khan, H.; Khan, A.H.; Aschner, M.; Batiha, G.E. Combination of natural antivirals and potent immune invigorators: A natural remedy to combat COVID-19. Phytotherapy Res. 2021. [Google Scholar] [CrossRef]
- Liu, F.; Zhu, Y.; Zhang, J.; Li, Y.; Peng, Z. Intravenous high-dose vitamin C for the treatment of severe COVID-19: Study protocol for a multicentre randomised controlled trial. BMJ Open 2020, 10, e039519. [Google Scholar] [CrossRef]
- Dravid, A.; Kashiva, R.; Khan, Z.; Memon, D.; Kodre, A.; Potdar, P.; Mane, M.; Borse, R.; Pawar, V.; Patil, D.; et al. Combination therapy of Tocilizumab and steroid for management of COVID-19 associated cytokine release syndrome. Medicine 2021, 100, e26705. [Google Scholar] [CrossRef]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; Van De Veen, W.; Brüggen, M.-C.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Elalamy, I.; Kastritis, E.; Sergentanis, T.N.; Politou, M.; Psaltopoulou, T.; Gerotziafas, G.; Dimopoulos, M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020, 95, 834–847. [Google Scholar] [CrossRef]
- Kempkes, R.; Joosten, I.; Koenen, H.J.P.M.; He, X. Metabolic Pathways Involved in Regulatory T Cell Functionality. Front. Immunol. 2019, 10, 2839. [Google Scholar] [CrossRef] [PubMed]
- Giganti, G.; Atif, M.; Mohseni, Y.; Mastronicola, D.; Grageda, N.; Povoleri, G.A.; Miyara, M.; Scottà, C. Treg cell therapy: How cell heterogeneity can make the difference. Eur. J. Immunol. 2020, 51, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-K.; Zhang, W.-H.; Chen, W.-X.; Wang, X.; Mu, X.-W. Effects of early enteral nutrition on Th17/Treg cells and IL-23/IL-17 in septic patients. World J. Gastroenterol. 2019, 25, 2799–2808. [Google Scholar] [CrossRef]
- Dasgupta, S.; Dasgupta, S.; Bandyopadhyay, M. Regulatory B cells in infection, inflammation, and autoimmunity. Cell. Immunol. 2020, 352, 104076. [Google Scholar] [CrossRef]
- Gong, Y.; Zhao, C.; Zhao, P.; Wang, M.; Zhou, G.; Han, F.; Cui, Y.; Qian, J.; Zhang, H.; Xiong, H.; et al. Role of IL-10-Producing Regulatory B Cells in Chronic Hepatitis B Virus Infection. Dig. Dis. Sci. 2014, 60, 1308–1314. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Ellis, G.; Pallant, C.; Lopes, A.R.; Khanna, P.; Peppa, D.; Chen, A.; A Blair, P.; Dusheiko, G.M.; Gill, U.; et al. IL-10–Producing Regulatory B Cells in the Pathogenesis of Chronic Hepatitis B Virus Infection. J. Immunol. 2012, 189, 3925–3935. [Google Scholar] [CrossRef]
- Duruel, O.; Berker, E.; Özşin-Özler, C.; Gharibzadeh-Hızal, M.; Gürpınar, O.; Eryılmaz-Polat, S.; Ataman-Duruel, E.T.; Tan, C.; Karabulut, E.; Tekçiçek, M.; et al. Levels of pro- and anti-inflammatory cytokines in cystic fibrosis patients with or without gingivitis. Cytokine 2020, 127, 154987. [Google Scholar] [CrossRef]
- Hofmann, S.R.; Kapplusch, F.; Girschick, H.J.; Morbach, H.; Pablik, J.; Ferguson, P.J.; Hedrich, C.M. Chronic Recurrent Multifocal Osteomyelitis (CRMO): Presentation, Pathogenesis, and Treatment. Curr. Osteoporos. Rep. 2017, 15, 542–554. [Google Scholar] [CrossRef]
- Gorji, Z.; Modaresi, M.; Yekanni-Nejad, S.; Rezaei, N.; Mahmoudi, M. Comparing effects of low glycemic index/high-fat, high-calorie diet and high-fat, high-calorie diet on cytokine levels of patients with cystic fibrosis: A randomized controlled clinical trial. Eur. Cytokine Netw. 2020, 31, 32–38. [Google Scholar] [PubMed]
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Langer, V.; Vivi, E.; Regensburger, D.; Winkler, T.H.; Waldner, M.J.; Rath, T.; Schmid, B.; Skottke, L.; Lee, S.; Jeon, N.L.; et al. IFN-γ drives inflammatory bowel disease pathogenesis through VE-cadherin–directed vascular barrier disruption. J. Clin. Investig. 2019, 129, 4691–4707. [Google Scholar] [CrossRef]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Tartaro, D.L.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef]
- Wu, D.; Yang, X.O. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020, 53, 368–370. [Google Scholar] [CrossRef]
- Behrens, E.M.; Koretzky, G.A. Review: Cytokine Storm Syndrome: Looking Toward the Precision Medicine Era. Arthritis Rheumatol. 2017, 69, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Hsu, A.C.-Y.; Pang, Z.; Pan, H.; Zuo, X.; Wang, G.; Zheng, J.; Wang, F. Role of the Innate Cytokine Storm Induced by the Influenza A Virus. Viral Immunol. 2019, 32, 244–251. [Google Scholar] [CrossRef]
- Gu, Y.; Zuo, X.; Zhang, S.; Ouyang, Z.; Jiang, S.; Wang, F.; Wang, G. The Mechanism behind Influenza Virus Cytokine Storm. Viruses 2021, 13, 1362. [Google Scholar] [CrossRef]
- Gao, H.; Lu, H.-Z.; Cao, B.; Du, B.; Shang, H.; Gan, J.-H.; Lu, S.-H.; Yang, Y.-D.; Fang, Q.; Shen, Y.-Z.; et al. Clinical Findings in 111 Cases of Influenza A (H7N9) Virus Infection. N. Engl. J. Med. 2013, 368, 2277–2285. [Google Scholar] [CrossRef]
- Maignan, M.; Viglino, D.; Hablot, M.; Masson, N.T.; Lebeugle, A.; Muret, R.C.; Makele, P.M.; Guglielmetti, V.; Morand, P.; Lupo, J.; et al. Diagnostic accuracy of a rapid RT-PCR assay for point-of-care detection of influenza A/B virus at emergency department admission: A prospective evaluation during the 2017/2018 influenza season. PLoS ONE 2019, 14, e0216308. [Google Scholar] [CrossRef]
- Oldstone, M.B.A.; Rosen, H. Cytokine Storm Plays a Direct Role in the Morbidity and Mortality from Influenza Virus Infection and is Chemically Treatable with a Single Sphingosine-1-Phosphate Agonist Molecule. Curr. Top. Microbiol. Immunol. 2014, 378, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Haran, J.P.; Buglione-Corbett, R.; Lu, S. Cytokine markers as predictors of type of respiratory infection in patients during the influenza season. Am. J. Emerg. Med. 2013, 31, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Jochems, S.P.; Marcon, F.; Carniel, B.F.; Holloway, M.; Mitsi, E.; Smith, E.; Gritzfeld, J.F.; Solórzano, C.; Reiné, J.; Pojar, S.; et al. Inflammation induced by influenza virus impairs human innate immune control of pneumococcus. Nat. Immunol. 2018, 19, 1299–1308. [Google Scholar] [CrossRef]
- Teijaro, J.R.; Walsh, K.B.; Cahalan, S.; Fremgen, D.M.; Roberts, E.; Scott, F.; Martinborough, E.; Peach, R.; Oldstone, M.B.; Rosen, H. Endothelial Cells Are Central Orchestrators of Cytokine Amplification during Influenza Virus Infection. Cell 2011, 146, 980–991. [Google Scholar] [CrossRef]
- Ryabkova, V.A.; Churilov, L.P.; Shoenfeld, Y. Influenza infection, SARS, MERS and COVID-19: Cytokine storm—The common denominator and the lessons to be learned. Clin. Immunol. 2020, 223, 108652. [Google Scholar] [CrossRef] [PubMed]
- La Gruta, N.L.; Kedzierska, K.; Stambas, J.; Doherty, P.C. A question of self-preservation: Immunopathology in influenza virus infection. Immunol. Cell Biol. 2007, 85, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Tanaka, T.; Inoue, H.; Ono, C.; Hashimoto, S.; Kioi, Y.; Matsumoto, H.; Matsuura, H.; Matsubara, T.; Shimizu, K.; et al. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc. Natl. Acad. Sci. USA 2020, 117, 22351–22356. [Google Scholar] [CrossRef]
- Michaelis, M.; Sithisarn, P.J.C., Jr. Effects of flavonoid-induced oxidative stress on anti-H5N1 influenza a virus activity exerted by baicalein and biochanin A. BMC Res. Notes 2014, 7, 384. [Google Scholar] [CrossRef]
- Gopal, R.; Mendy, A.; Marinelli, M.A.; Richwalls, L.J.; Seger, P.J.; Patel, S.; McHugh, K.J.; Rich, H.E.; Grousd, J.A.; Forno, E.; et al. Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) Suppresses Inflammation and Bacterial Clearance during Influenza-Bacterial Super-Infection. Viruses 2019, 11, 505. [Google Scholar] [CrossRef]
- Cao, W.; Liu, X.; Bai, T.; Fan, H.; Hong, K.; Song, H.; Han, Y.; Lin, L.; Ruan, L.; Li, T. High-Dose Intravenous Immunoglobulin as a Therapeutic Option for Deteriorating Patients with Coronavirus Disease 2019. Open Forum Infect. Dis. 2020, 7, ofaa102. [Google Scholar] [CrossRef]
- Clark, I.A. The advent of the cytokine storm. Immunol. Cell Biol. 2007, 85, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Feuerecker, M.; Sudhoff, L.; Crucian, B.; Pagel, J.-I.; Sams, C.; Strewe, C.; Guo, A.; Schelling, G.; Briegel, J.; Kaufmann, I.; et al. Early immune anergy towards recall antigens and mitogens in patients at onset of septic shock. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.; Filbin, M.R.; Bhattacharyya, R.P.; Billman, K.; Eisenhaure, T.; Hung, D.T.; Levy, B.D.; Baron, R.M.; Blainey, P.C.; Goldberg, M.B.; et al. An immune-cell signature of bacterial sepsis. Nat. Med. 2020, 26, 333–340. [Google Scholar] [CrossRef]
- Chousterman, B.G.; Swirski, F.; Weber, G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017, 39, 517–528. [Google Scholar] [CrossRef]
- Weber, G.F.; Chousterman, B.G.; He, S.; Fenn, A.M.; Nairz, M.; Anzai, A.; Brenner, T.; Uhle, F.; Iwamoto, Y.; Robbins, C.S.; et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science 2015, 347, 1260–1265. [Google Scholar] [CrossRef]
- Cavaillon, J.-M. Exotoxins and endotoxins: Inducers of inflammatory cytokines. Toxicon Off. J. Int. Soc. Toxinol. 2018, 149, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.-H.; Chao, C.-H.; Yeh, T.-M. Inhibition of autophagy protects against sepsis by concurrently attenuating the cytokine storm and vascular leakage. J. Infect. 2019, 78, 178–186. [Google Scholar] [CrossRef]
- van der Poll, T.; Opal, S.M. Host–pathogen interactions in sepsis. Lancet Infect. Dis. 2008, 8, 32–43. [Google Scholar] [CrossRef]
- Karbian, N.; Abutbul, A.; El-Amore, R.; Eliaz, R.; Beeri, R.; Reicher, B.; Mevorach, D. Apoptotic cell therapy for cytokine storm associated with acute severe sepsis. Cell Death Dis. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Kumar, V. Toll-like receptors in sepsis-associated cytokine storm and their endogenous negative regulators as future immunomodulatory targets. Int. Immunopharmacol. 2020, 89, 107087. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Schmitz, S.; De Santis, R.; Doemming, S.; Haase, H.; Hoeger, J.; Heinbockel, L.; Brandenburg, K.; Marx, G.; Schuerholz, T. Peptide 19-2.5 Inhibits Heparan Sulfate-Triggered Inflammation in Murine Cardiomyocytes Stimulated with Human Sepsis Serum. PLoS ONE 2015, 10, e0127584. [Google Scholar] [CrossRef]
- AbdAllah, N.B.; Toraih, E.A.; Al Ageeli, E.; Elhagrasy, H.; Gouda, N.S.; Fawzy, M.S.; Helal, G.M. MYD88, NFKB1, and IL6 transcripts overexpression are associated with poor outcomes and short survival in neonatal sepsis. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Pan, B.; Gao, J.; Chen, W.; Liu, C.; Shang, L.; Xu, M.; Fu, C.; Zhu, S.; Niu, M.; Xu, K. Selective inhibition of interleukin-1 receptor-associated kinase 1 ameliorates lipopolysaccharide-induced sepsis in mice. Int. Immunopharmacol. 2020, 85, 106597. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, Y.-T.; Zhang, S.-X.; Fu, S.-Z.; Ye, X.-Z. MiR-590-3p Attenuates Acute Kidney Injury by Inhibiting Tumor Necrosis Factor Receptor-Associated Factor 6 in Septic Mice. Inflammation 2018, 42, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Ogura, H.; Shimizu, K.; Ikeda, M.; Hirose, T.; Matsuura, H.; Kang, S.; Takahashi, K.; Tanaka, T.; Shimazu, T. The clinical importance of a cytokine network in the acute phase of sepsis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mera, S.; Tatulescu, D.; Cismaru, C.; Bondor, C.I.; Slavcovici, A.; Zanc, V.; Carstina, D.; Oltean, M. Multiplex cytokine profiling in patients with sepsis. APMIS 2010, 119, 155–163. [Google Scholar] [CrossRef] [PubMed]
- A Bozza, F.; I Salluh, J.; Japiassu, A.M.; Soares, M.; Assis, E.F.; Gomes, R.N.; Bozza, M.T.; Castro-Faria-Neto, H.C.; Bozza, P.T. Cytokine profiles as markers of disease severity in sepsis: A multiplex analysis. Crit. Care 2007, 11, R49. [Google Scholar] [CrossRef]
- Meyer, N.J.; Reilly, J.P.; Anderson, B.; Palakshappa, J.; Jones, T.K.; Dunn, T.G.; Shashaty, M.G.S.; Feng, R.; Christie, J.; Opal, S.M. Mortality Benefit of Recombinant Human Interleukin-1 Receptor Antagonist for Sepsis Varies by Initial Interleukin-1 Receptor Antagonist Plasma Concentration. Crit. Care Med. 2018, 46, 21–28. [Google Scholar] [CrossRef]
- Shakoory, B.; Carcillo, J.A.; Chatham, W.W.; Amdur, R.L.; Zhao, H.; Dinarello, C.A.; Cron, R.Q.; Opal, S.M. Interleukin-1 Receptor Blockade Is Associated with Reduced Mortality in Sepsis Patients with Features of Macrophage Activation Syndrome. Reanalysis of a Prior Phase III Trial. Crit. Care Med. 2016, 44, 275–281. [Google Scholar] [CrossRef]
- Qiu, D.; Chu, X.; Hua, L.; Yang, Y.; Li, K.; Han, Y.; Yin, J.; Zhu, M.; Mu, S.; Sun, Z.; et al. Gpr174-deficient regulatory T cells decrease cytokine storm in septic mice. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Chen, X.; Feng, Y.; Shen, X.; Pan, G.; Fan, G.; Gao, X.; Han, J.; Zhu, Y. Anti-sepsis protection of Xuebijing injection is mediated by differential regulation of pro- and anti-inflammatory Th17 and T regulatory cells in a murine model of polymicrobial sepsis. J. Ethnopharmacol. 2018, 211, 358–365. [Google Scholar] [CrossRef]
- Hotchkiss, R.S.; Swanson, P.E.; Cobb, J.P.; Jacobson, A.; Buchman, T.G.; Karl, I.E. Apoptosis in lymphoid and parenchymal cells during sepsis: Findings in normal and T- and B-cell-deficient mice. Crit. Care Med. 1997, 25, 1298–1307. [Google Scholar] [CrossRef]
- Hirano, T.; Murakami, M. COVID-19: A New Virus, but a Familiar Receptor and Cytokine Release Syndrome. Immunity 2020, 52, 731–733. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.-H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X.; et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wei, H. Tocilizumab is recommended for the treatment of severe COVID-19. EBioMedicine 2020, 61, 103045. [Google Scholar] [CrossRef] [PubMed]
- Investigators, R.-C.; Gordon, A.C.; Mouncey, P.R.; Al-Beidh, F.; Rowan, K.M.; Nichol, A.D.; Arabi, Y.M.; Annane, D.; Beane, A.; van Bentum-Puijk, W.; et al. The REMAP-CAP Investigators. Interleukin-6 Receptor Antagonists in Critically Ill Patients with COVID-19. N. Engl. J. Med. 2021, 384, 1491–1502. [Google Scholar] [CrossRef]
- Franzetti, M.; Forastieri, A.; Borsa, N.; Pandolfo, A.; Molteni, C.; Borghesi, L.; Pontiggia, S.; Evasi, G.; Guiotto, L.; Erba, M.; et al. IL-1 Receptor Antagonist Anakinra in the Treatment of COVID-19 Acute Respiratory Distress Syndrome: A Retrospective, Observational Study. J. Immunol. 2021, 206, 1569–1575. [Google Scholar] [CrossRef]
- Cavalli, G.; De Luca, G.; Campochiaro, C.; Della-Torre, E.; Ripa, M.; Canetti, D.; Oltolini, C.; Castiglioni, B.; Din, C.T.; Boffini, N.; et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e325–e331. [Google Scholar] [CrossRef]
- Cauchois, R.; Koubi, M.; Delarbre, D.; Manet, C.; Carvelli, J.; Blasco, V.B.; Jean, R.; Fouche, L.; Bornet, C.; Pauly, V.; et al. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 18951–18953. [Google Scholar] [CrossRef]
- Luo, W.; Li, Y.-X.; Jiang, L.-J.; Chen, Q.; Wang, T.; Ye, D.-W. Targeting JAK-STAT Signaling to Control Cytokine Release Syndrome in COVID-19. Trends Pharmacol. Sci. 2020, 41, 531–543. [Google Scholar] [CrossRef]
- Stebbing, J.; Nievas, G.S.; Falcone, M.; Youhanna, S.; Richardson, P.; Ottaviani, S.; Shen, J.X.; Sommerauer, C.; Tiseo, G.; Ghiadoni, L.; et al. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci. Adv. 2021, 7, eabe4724. [Google Scholar] [CrossRef] [PubMed]
- Gucyetmez, B.; Atalan, H.K.; Sertdemir, I.; Cakir, U.; Telci, L.; Ogan, A.; Ayyildiz, A.C.; Yalcin, B.; Oren, B.; Havas, F.; et al. Therapeutic plasma exchange in patients with COVID-19 pneumonia in intensive care unit: A retrospective study. Crit. Care 2020, 24, 492. [Google Scholar] [CrossRef] [PubMed]
- Gluck, W.L.; Callahan, S.P.; Brevetta, R.A.; Stenbit, A.E.; Smith, W.M.; Martin, J.C.; Blenda, A.V.; Arce, S.; Edenfield, W.J. Efficacy of therapeutic plasma exchange in the treatment of penn class 3 and 4 cytokine release syndrome complicating COVID-19. Respir. Med. 2020, 175, 106188. [Google Scholar] [CrossRef]
- Kamran, S.M.; Mirza, Z.-E.; Naseem, A.; Liaqat, J.; Fazal, I.; Alamgir, W.; Saeed, F.; Saleem, S.; Nisar, S.; Yousaf, M.A.; et al. Therapeutic plasma exchange for coronavirus disease-2019 triggered cytokine release syndrome; a retrospective propensity matched control study. PLoS ONE 2021, 16, e0244853. [Google Scholar] [CrossRef]
- Umar, S.; Guerin, J.L.; Ducatez, M.F. Low Pathogenic Avian Influenza and Coinfecting Pathogens: A Review of Experimental Infections in Avian Models. Avian Dis. 2017, 61, 3–15. [Google Scholar] [CrossRef]
- Asehnoune, K.; Villadangos, J.; Hotchkiss, R.S. Understanding host–pathogen interaction. Intensiv. Care Med. 2016, 42, 2084–2086. [Google Scholar] [CrossRef]
- Bezemer, G.F.G.; Garssen, J. TLR9 and COVID-19: A Multidisciplinary Theory of a Multifaceted Therapeutic Target. Front. Pharmacol. 2021, 11, 601685. [Google Scholar] [CrossRef]
- Lee, I.-H.; Lee, J.-W.; Kong, S.W. A survey of genetic variants in SARS-CoV-2 interacting domains of ACE2, TMPRSS2 and TLR3/7/8 across populations. Infect. Genet. Evol. 2020, 85, 104507. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.Y.; Lin, M.Y.; Coffman, R.L.; Campbell, J.D.; Traquina, P.; Lin, Y.J.; Liu, L.T.C.; Cheng, J.Y.; Wu, Y.C.; Wu, C.C.; et al. Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci. Rep. 2020, 10, 20085. [Google Scholar]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-19. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, A.; Braga, T.; Correa-Costa, M.; De Aguiar, C.F.; Bassi, J.; Correa-Silva, R.; Elias, R.M.; Salvador, F.; Vieira, P.; Cenedeze, M.A.; et al. TLR2, TLR4 and the MYD88 Signaling Pathway Are Crucial for Neutrophil Migration in Acute Kidney Injury Induced by Sepsis. PLoS ONE 2012, 7, e37584. [Google Scholar] [CrossRef] [PubMed]
- Redondo, A.C.; Ceccon, M.E.J.R.; Silveira-Lessa, A.L.; Quinello, C.; Palmeira, P.; Carvalho, W.B.; Carneiro-Sampaio, M. TLR-2 and TLR-4 expression in monocytes of newborns with late-onset sepsis. J. Pediatr. 2014, 90, 472–478. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Chen, J.-J.; Zhang, L.-N.; Hou, H.; Xu, L.; Ji, K. Interleukin-6 signaling blockade treatment for cytokine release syndrome in COVID-19 (Review). Exp. Ther. Med. 2020, 21, 1. [Google Scholar] [CrossRef]
- Conti, P.; Caraffa, A.; Gallenga, C.E.; Ross, R.; Kritas, S.K.; Frydas, I.; Younes, A.; Di Emidio, P.; Ronconi, G.; Toniato, E. Biolife IL-1 induces throboxane-A2 (TxA2) in COVID-19 causing inflammation and micro-thrombi: Inhibitory effect of the IL-1 receptor antagonist (IL-1Ra). J. Biol. Regul. Homeost. Agents 2020, 34, 1623–1627. [Google Scholar] [CrossRef]
- Ramiro, S.; Mostard, R.L.M.; Magro-Checa, C.; Van Dongen, C.M.P.; Dormans, T.; Buijs, J.; Gronenschild, M.; De Kruif, M.D.; Van Haren, E.H.J.; Van Kraaij, T.; et al. Historically controlled comparison of glucocorticoids with or without tocilizumab versus supportive care only in patients with COVID-19-associated cytokine storm syndrome: Results of the CHIC study. Ann. Rheum. Dis. 2020, 79, 1143–1151. [Google Scholar] [CrossRef]
- Group, A.-T.L.-C.S.; Lundgren, J.D.; Grund, B.; Barkauskas, C.E.; Holland, T.L.; Gottlieb, R.L.; Sandkovsky, U.; Brown, S.M.; Knowlton, K.U.; Self, W.H.; et al. ACTIV-3/TICO LY-CoV555 Study Group. A Neutralizing Monoclonal Antibody for Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 905–914. [Google Scholar] [CrossRef]
- Fabijan, A.P.; Lin, R.C.; Ho, J.; Maddocks, S.; Zakour, N.L.b.; Iredell, J.R. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 2020, 5, 465–472. [Google Scholar] [CrossRef]
- D’Elia, R.V.; Harrison, K.; Oyston, P.C.; Lukaszewski, R.A.; Clark, G.C. Targeting the “Cytokine Storm” for Therapeutic Benefit. Clin. Vaccine Immunol. 2013, 20, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Erbil, B.; Ersoy, G.; Ozkutuk, A.; Akarca, F.K.; Korkmaz, T.; Demir, O.F.; Kiyan, S. The Effects of Oral Antibiotics on Infection Prophylaxis in Traumatic Wounds. Turk. J. Trauma Emerg. Surg. 2014, 20, 231–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dudareva, M.; Kümin, M.; Vach, W.; Kaier, K.; Ferguson, J.; McNally, M.; Scarborough, M. Short or Long Antibiotic Regimes in Orthopaedics (SOLARIO): A randomised controlled open-label non-inferiority trial of duration of systemic antibiotics in adults with orthopaedic infection treated operatively with local antibiotic therapy. Trials 2019, 20, 1–9. [Google Scholar] [CrossRef]
- Coates, A.R.M.; Hu, Y. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 2007, 152, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Kutateladze, M.; Adamia, R. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 2010, 28, 591–595. [Google Scholar] [CrossRef]
- Hansel, T.T.; Kropshofer, H.; Singer, T.; Mitchell, J.A.; George, A. The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Discov. 2010, 9, 325–338. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, S.; Dong, X.; Li, Z.; Xu, Q.; Feng, H.; Cai, J.; Huang, S.; Guo, J.; Zhang, L.; et al. Corticosteroid treatment in severe COVID-19 patients with acute respiratory distress syndrome. J. Clin. Investig. 2020, 130, 6417–6428. [Google Scholar] [CrossRef]
- Pierson, S.; Stonestrom, A.J.; Shilling, D.; Ruth, J.; Nabel, C.S.; Singh, A.; Ren, Y.; Stonestrom, A.; Li, H.; Van Rhee, F.; et al. Plasma proteomics identifies a ‘chemokine storm’ in idiopathic multicentric Castleman disease. Am. J. Hematol. 2018, 93, 902–912. [Google Scholar] [CrossRef]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Lim, K.; Hyun, Y.-M.; Lambert-Emo, K.; Capece, T.; Bae, S.; Miller, R.; Topham, D.J.; Kim, M. Neutrophil trails guide influenza-specific CD8 + T cells in the airways. Science 2015, 349, aaa4352. [Google Scholar] [CrossRef]
- Tan, Q.; Zhu, Y.; Li, J.; Chen, Z.; Han, G.W.; Kufareva, I.; Li, T.; Ma, L.; Fenalti, G.; Li, J.; et al. Structure of the CCR5 Chemokine Receptor–HIV Entry Inhibitor Maraviroc Complex. Science 2013, 341, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.U.; Saadabadi, A.; Vanmeert, M.; Salo-Ahen, O.M.; Abdullah, I.; Claes, S.; De Jonghe, S.; Schols, D.; Ahmad, S.; Froeyen, M. Discovery of HIV entry inhibitors via a hybrid CXCR4 and CCR5 receptor pharmacophore-based virtual screening approach. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2020, 155, 105537. [Google Scholar] [CrossRef] [PubMed]
- Charo, I.F.; Ransohoff, R.M. The Many Roles of Chemokines and Chemokine Receptors in Inflammation. N. Engl. J. Med. 2006, 354, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wan, M.; Lyon, C.J.; Hu, T.Y. Nanomedicine therapies modulating Macrophage Dysfunction: A potential strategy to attenuate Cytokine Storms in severe infections. Theranostics 2020, 10, 9591–9600. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Isolauri, E.; Kirjavainen, P.V.; Salminen, S. Probiotics: A role in the treatment of intestinal infection and inflammation? Gut 2002, 50, iii54–iii59. [Google Scholar] [CrossRef] [PubMed]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron 2019, 101, 246–259.e6. [Google Scholar] [CrossRef] [PubMed]
- Leite, M.D.M.; Gonzalez-Galarza, F.F.; da Silva, B.C.C.; Middleton, D.; dos Santos, E.J.M. Predictive immunogenetic markers in COVID-19. Hum. Immunol. 2021, 82, 247–254. [Google Scholar] [CrossRef]
- Xu, P.-B.; Lou, J.-S.; Ren, Y.; Miao, C.-H.; Deng, X.-M. Gene expression profiling reveals the defining features of monocytes from septic patients with compensatory anti-inflammatory response syndrome. J. Infect. 2012, 65, 380–391. [Google Scholar] [CrossRef]


| Incentive | Cytokines |
|---|---|
| CAR-T | IFN-γ, IL-2, IL-2Ra, IL-6, sIL-6R, GM-CSF, IL-1α, IL-1β, IL-10, IL-12, TNF-α, IFN-α, MCP-1, MIP-1α |
| H5N1 | MCP-1, CXCL10, CXCL9, IL-8 |
| H1N1 | IL-8, IL-9, IL-17, IL-6, TNF-α, IL-15, IL-12p70 |
| SARS | IL-1β, IL-6, IL-12, IFN-γ, IP10, MCP-1 |
| MERS | IFN-γ, TNF-α, IL-15, IL-17 |
| COVID-19 | IL-2, IL-7, IL-10, G-SCF, IP10, MCP-1, MIP-1α, TNF-α |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.-D.; Ji, T.-T.; Dong, J.-R.; Feng, H.; Chen, F.-Q.; Chen, X.; Zhao, H.-Y.; Chen, D.-K.; Ma, W.-T. Pathogenesis and Treatment of Cytokine Storm Induced by Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 13009. https://doi.org/10.3390/ijms222313009
Tang X-D, Ji T-T, Dong J-R, Feng H, Chen F-Q, Chen X, Zhao H-Y, Chen D-K, Ma W-T. Pathogenesis and Treatment of Cytokine Storm Induced by Infectious Diseases. International Journal of Molecular Sciences. 2021; 22(23):13009. https://doi.org/10.3390/ijms222313009
Chicago/Turabian StyleTang, Xi-Dian, Tian-Tian Ji, Jia-Rui Dong, Hao Feng, Feng-Qiang Chen, Xi Chen, Hui-Ying Zhao, De-Kun Chen, and Wen-Tao Ma. 2021. "Pathogenesis and Treatment of Cytokine Storm Induced by Infectious Diseases" International Journal of Molecular Sciences 22, no. 23: 13009. https://doi.org/10.3390/ijms222313009
APA StyleTang, X.-D., Ji, T.-T., Dong, J.-R., Feng, H., Chen, F.-Q., Chen, X., Zhao, H.-Y., Chen, D.-K., & Ma, W.-T. (2021). Pathogenesis and Treatment of Cytokine Storm Induced by Infectious Diseases. International Journal of Molecular Sciences, 22(23), 13009. https://doi.org/10.3390/ijms222313009





