KLK4T2 Is a Hormonally Regulated Transcript from the KLK4 Locus
Abstract
:1. Introduction
2. Results
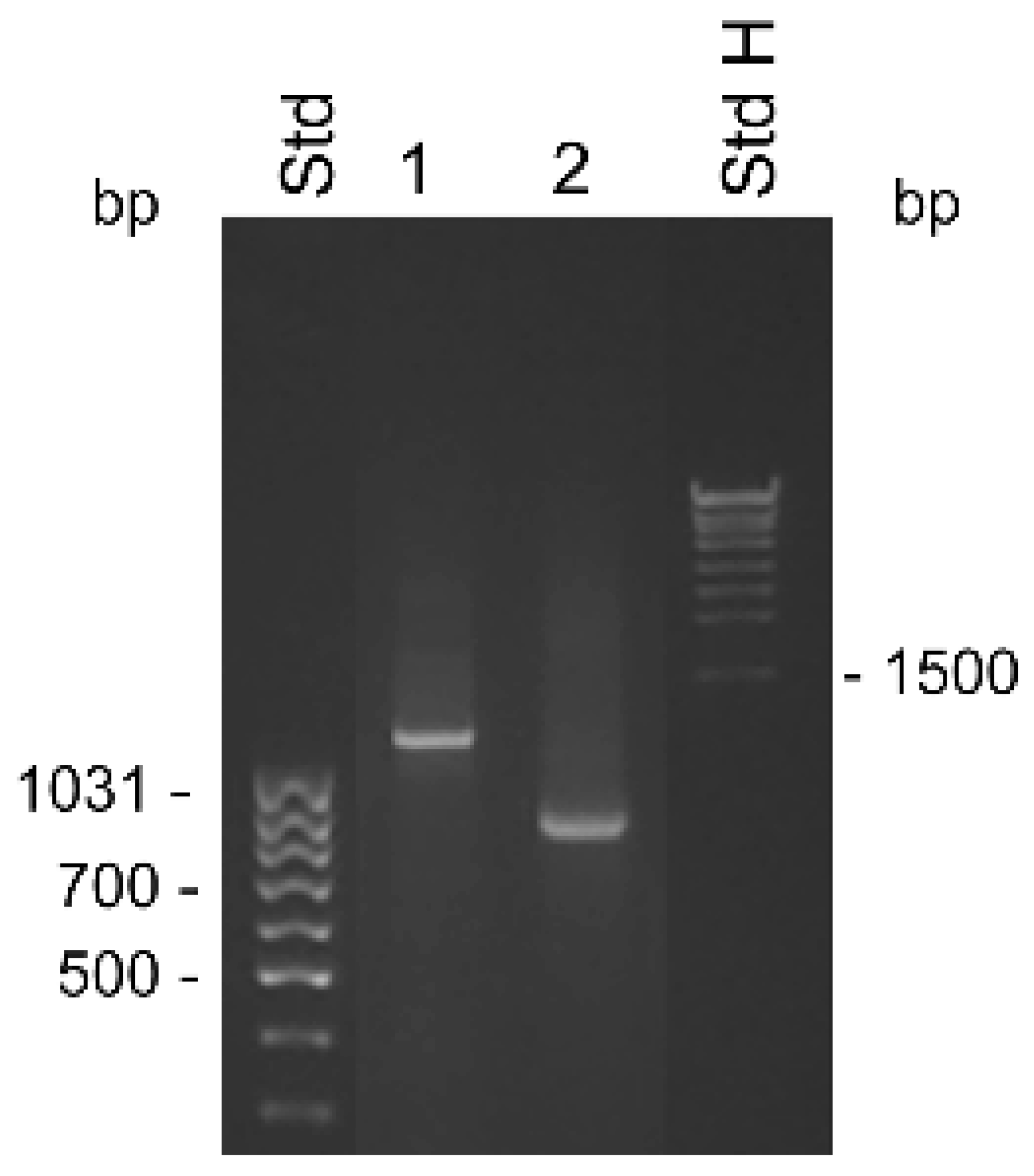
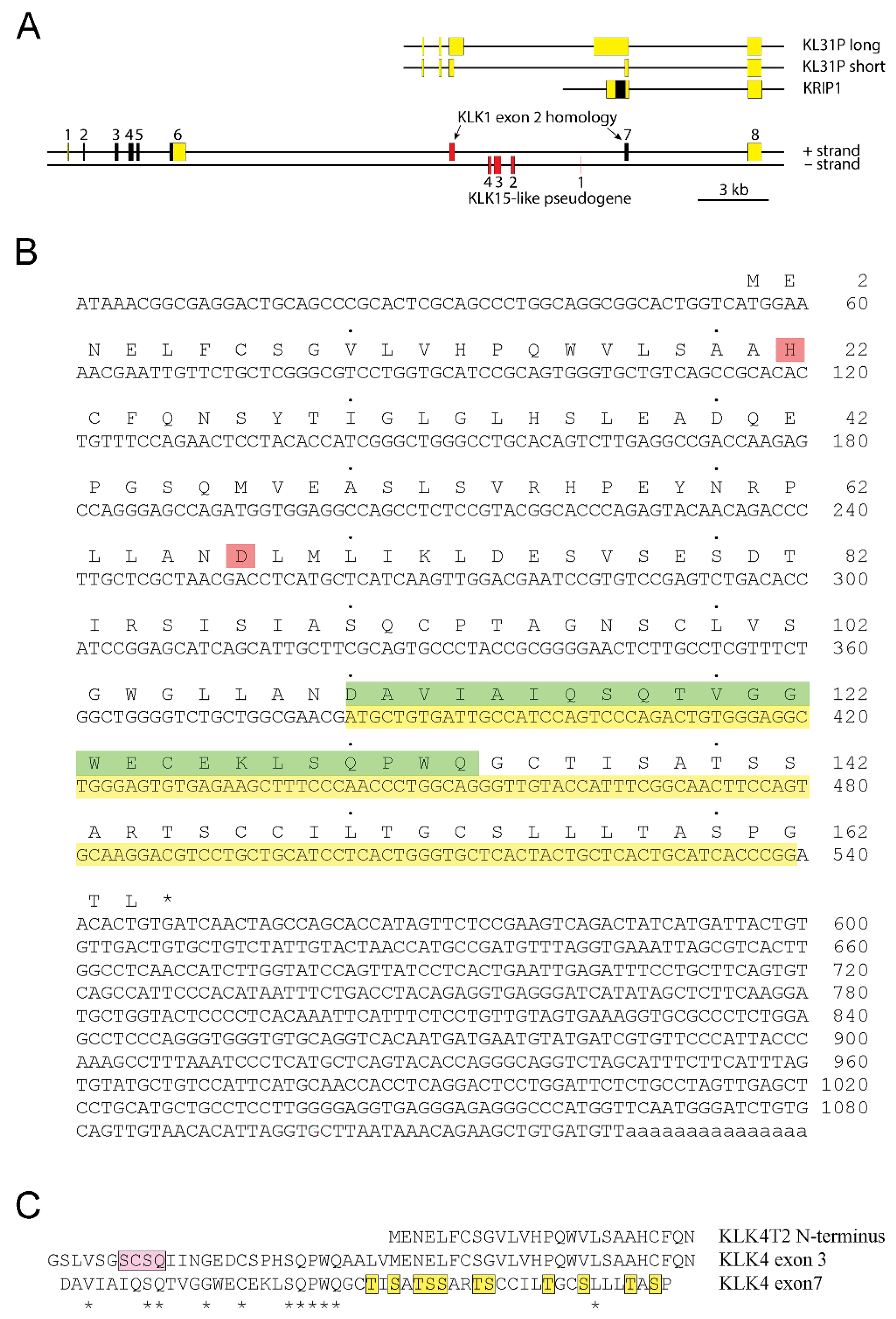
2.1. Identification of a Transcript from the KLKP1 Locus as a Splice Variant of KLK4
2.2. Analysis of Transcripts from the KLK4/KLKP1 Locus by RACE
2.3. Analysis of Transcripts from the KLK4/KLKP1 Locus by RT-qPCR
2.4. Expression Pattern of KLK4-Related Transcripts in the Prostate and during Prostate Cancer Disease Progression
2.5. Tissue Expression
3. Discussion
4. Materials and Methods
4.1. Nomenclature
4.2. Cell Culture
4.3. RACE PCR
4.4. RNA Preparation, RT-PCR, and cDNA Cloning
4.5. Patient Samples and Gene Expression Analysis
4.6. Statistical Analysis
4.7. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gan, L.; Lee, I.; Smith, R.; Argonza-Barrett, R.; Lei, H.; McCuaig, J.; Moss, P.; Paeper, B.; Wang, K. Sequencing and expression analysis of the serine protease gene cluster located in chromosome 19q13 region. Gene 2000, 257, 119–130. [Google Scholar] [CrossRef]
- Yousef, G.M.; Chang, A.; Scorilas, A.; Diamandis, E.P. Genomic Organization of the Human Kallikrein Gene Family on Chromosome 19q13.3–q13.4. Biochem. Biophys. Res. Commun. 2000, 276, 125–133. [Google Scholar] [CrossRef]
- Clements, J.; Hooper, J.; Dong, Y.; Harvey, T. The Expanded Human Kallikrein (KLK) Gene Family: Genomic Organisation, Tissue-Specific Expression and Potential Functions. Biol. Chem. 2001, 382, 5–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kryza, T.; Silva, M.L.; Loessner, D.; Heuzé-Vourc’h, N.; Clements, J.A. The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie 2016, 122, 283–299. [Google Scholar] [CrossRef]
- Fuhrman-Luck, R.A.; Loessner, D.; Clements, J.A. Kallikrein-Related Peptidases in Prostate Cancer: From Molecular Function to Clinical Application. EJIFCC 2014, 25, 269–281. [Google Scholar] [PubMed]
- Hollenberg, M.D. KLKs and their hormone-like signaling actions: A new life for the PSA-KLK family. Biol. Chem. 2014, 395, 915–929. [Google Scholar] [CrossRef]
- Avgeris, M.; Mavridis, K.; Scorilas, A. Kallikrein-related peptidases in prostate, breast, and ovarian cancers: From pathobiology to clinical relevance. Biol. Chem. 2012, 393, 301–317. [Google Scholar] [CrossRef]
- Borgono, C.A.; Michael, P.I.; Diamandis, E.P. Human tissue kallikreins: Physiologic roles and applications in cancer. Mol. Cancer Res. 2004, 2, 257–280. [Google Scholar]
- Schmitt, M.; Magdolen, V.; Yang, F.; Kiechle, M.; Bayani, J.; Yousef, G.M.; Scorilas, A.; Diamandis, E.P.; Dorn, J. Emerging clinical importance of the cancer biomarkers kallikrein-related peptidases (KLK) in female and male reproductive organ malignancies. Radiol. Oncol. 2013, 47, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Diamandis, E.P.; Yousef, G.M. Human tissue kallikreins: A family of new cancer biomarkers. Clin. Chem. 2002, 48, 1198–1205. [Google Scholar] [CrossRef] [Green Version]
- Clements, J.A.; Willemsen, N.M.; Myers, S.A.; Dong, Y. The Tissue Kallikrein Family of Serine Proteases: Functional Roles in Human Disease and Potential as Clinical Biomarkers. Crit. Rev. Clin. Lab. Sci. 2004, 41, 265–312. [Google Scholar] [CrossRef]
- Lundwall, A.; Lilja, H. Molecular cloning of human prostate specific antigen cDNA. FEBS Lett. 1987, 214, 317–322. [Google Scholar] [CrossRef] [Green Version]
- Steuber, T.; Vickers, A.J.; Serio, A.M.; Vaisanen, V.; Haese, A.; Pettersson, K.; Eastham, J.A.; Scardino, P.T.; Huland, H.; Lilja, H. Comparison of Free and Total Forms of Serum Human Kallikrein 2 and Prostate-Specific Antigen for Prediction of Locally Advanced and Recurrent Prostate Cancer. Clin. Chem. 2007, 53, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, D.; Vijai, J.; Cronin, A.M.; Bhatia, J.; Vickers, A.; Gaudet, M.M.; Fine, S.; Reuter, V.; Scher, H.I.; Halldén, C.; et al. Susceptibility Loci Associated with Prostate Cancer Progression and Mortality. Clin. Cancer Res. 2010, 16, 2819–2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, R.J.; Halldén, C.; Cronin, A.M.; Ploner, A.; Wiklund, F.; Bjartell, A.S.; Stattin, P.; Xu, J.; Scardino, P.T.; Offit, K.; et al. Blood Biomarker Levels to Aid Discovery of Cancer-Related Single-Nucleotide Polymorphisms: Kallikreins and Prostate Cancer. Cancer Prev. Res. 2010, 3, 611–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmer, J.; Fukae, M.; Tanabe, T.; Yamakoshi, Y.; Uchida, T.; Xue, J.; Margolis, H.; Shimizu, M.; Dehart, B.; Hu, C.-C.; et al. Purification, Characterization, and Cloning of Enamel Matrix Serine Proteinase 1. J. Dent. Res. 1998, 77, 377–386. [Google Scholar] [CrossRef]
- Bartlett, J.D.; Ryu, O.H.; Xue, J.; Simmer, J.P.; Margolis, H.C. Enamelysin mRNA Displays a Developmental Defined Pattern of Expression and Encodes a Protein which Degrades Amelogenin. Connect. Tissue Res. 1998, 39, 101–109. [Google Scholar] [CrossRef]
- Yousef, G.M.; Obiezu, C.V.; Luo, L.Y.; Black, M.H.; Diamandis, E.P. Prostase/KLK-L1 is a new member of the human kallikrein gene family, is expressed in prostate and breast tissues, and is hormonally regulated. Cancer Res. 1999, 59, 4252–4256. [Google Scholar]
- Hu, J.C.-C.; Zhang, C.; Sun, X.; Yang, Y.; Cao, X.; Ryu, O.; Simmer, J.P. Characterization of the mouse and human PRSS17 genes, their relationship to other serine proteases, and the expression of PRSS17 in developing mouse incisors. Gene 2000, 251, 1–8. [Google Scholar] [CrossRef]
- Nelson, P.S.; Gan, L.; Ferguson, C.; Moss, P.; Gelinas, R.; Hood, L.; Wang, K. Molecular cloning and characterization of prostase, an androgen-regulated serine protease with prostate-restricted expression. Proc. Natl. Acad. Sci. USA 1999, 96, 3114–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korkmaz, K.S.; Korkmaz, C.G.; Pretlow, T.G.; Saatcioglu, F. Distinctly different gene structure of KLK4/KLK-L1/prostase/ARM1 compared with other members of the kallikrein family: Intracellular localization, alternative cDNA forms, and Regulation by multiple hormones. DNA Cell Biol. 2001, 20, 435–445. [Google Scholar] [CrossRef]
- Yousef, G.M.; Borgono, C.A.; Michael, I.P.; Diamandis, E.P. Cloning of a kallikrein pseudogene. Clin. Biochem. 2004, 37, 961–967. [Google Scholar] [CrossRef]
- Kaushal, A.; Myers, S.; Dong, Y.; Lai, J.; Tan, O.L.; Bui, L.T.; Hunt, M.L.; Digby, M.R.; Samaratunga, H.; Gardiner, R.A.; et al. A novel transcript from theKLKP1 gene is androgen regulated, down-regulated during prostate cancer progression and encodes the first non-serine protease identified from the human kallikrein gene locus. Prostate 2008, 68, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zhou, D.; Glusman, G.; Utleg, A.G.; White, J.T.; Nelson, P.S.; Vasicek, T.J.; Hood, L.; Lin, B. KLK31P is a novel androgen regulated and transcribed pseudogene of kallikreins that is expressed at lower levels in prostate cancer cells than in normal prostate cells. Prostate 2006, 66, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Lehman, M.L.; Dinger, M.E.; Hendy, S.C.; Mercer, T.R.; Seim, I.; Lawrence, M.G.; Mattick, J.S.; Clements, J.A.; Nelson, C.C. A variant of the KLK4 gene is expressed as a cis sense–antisense chimeric transcript in prostate cancer cells. RNA 2010, 16, 1156–1166. [Google Scholar] [CrossRef] [Green Version]
- Chakravarthi, B.V.; Dedigama-Arachchige, P.; Carskadon, S.; Sundaram, S.K.; Li, J.; Wu, K.-H.H.; Chandrashekar, D.S.; Peabody, J.O.; Stricker, H.; Hwang, C.; et al. Pseudogene Associated Recurrent Gene Fusion in Prostate Cancer. Neoplasia 2019, 21, 989–1002. [Google Scholar] [CrossRef]
- Kryza, T.; Silva, L.M.; Bock, N.; Fuhrman-Luck, R.A.; Stephens, C.R.; Gao, J.; Clements, J.A. Kallikrein-related peptidase 4 induces cancer-associated fibroblast features in prostate-derived stromal cells. Mol. Oncol. 2017, 11, 1307–1329. [Google Scholar] [CrossRef] [Green Version]
- Avgeris, M.; Stravodimos, K.; Scorilas, A. Kallikrein-related peptidase 4 gene (KLK4) in prostate tumors: Quantitative expression analysis and evaluation of its clinical significance. Prostate 2011, 71, 1780–1789. [Google Scholar] [CrossRef]
- Klokk, T.I.; Kilander, A.; Xi, Z.; Waehre, H.; Risberg, B.; Danielsen, H.E.; Saatcioglu, F. Kallikrein 4 Is a Proliferative Factor that Is Overexpressed in Prostate Cancer. Cancer Res. 2007, 67, 5221–5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, Z.; Klokk, T.I.; Korkmaz, K.; Kurys, P.; Elbi, C.; Risberg, B.; Danielsen, H.; Loda, M.; Saatcioglu, F. Kallikrein 4 is a Predominantly Nuclear Protein and Is Overexpressed in Prostate Cancer. Cancer Res. 2004, 64, 2365–2370. [Google Scholar] [CrossRef] [Green Version]
- Abrahamsson, P.-A.; Lilja, H.; Falkmer, S.; Wadströ, L.B. Immunohistochemical distribution of the three predominant secretory proteins in the parenchyma of hyperplastic and neoplastic prostate glands. Prostate 1988, 12, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Bui, L.T.; Odorico, D.M.; Tan, O.L.; Myers, S.A.; Samaratunga, H.; Clements, J.A. Compartmentalized expression of kallikrein 4 (KLK4/hK4) isoforms in prostate cancer: Nuclear, Cytoplasmic and secreted forms. Endocr. Relat. Cancer 2005, 12, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Hörnberg, E.; Ylitalo, E.B.; Crnalic, S.; Antti, H.; Stattin, P.; Widmark, A.; Bergh, A.; Wikström, P. Expression of Androgen Receptor Splice Variants in Prostate Cancer Bone Metastases is Associated with Castration-Resistance and Short Survival. PLoS ONE 2011, 6, e19059. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Primer | Sequence 5′–3′ | Purpose |
|---|---|---|
| KLKP9R | tggcagcggccgcAACATCACAGCTTCTGTTTATTAAGCA | 5′ RACE, cloning |
| KLKP8F | tcgcagaattcATGCTGTGATTGCCATCCAGTCCCAGA | 3′ RACE, cloning |
| KLK4r18 | gatcggtcgactgACTGGCCTGGACGGTTTTCTCTAT | 5′ RACE, cloning |
| KLKPf10 | agctgaattcTAAACGGCGAGGACTGCAGCCCGCACT | cloning |
| KLK4Ne2f | GGCGGCACTGGTCATGGAAAACGAA | 3′ RACE |
| KLK4Ne3f | CGGAGCATCAGCATTGCTTCGCAGT | 3′ RACE |
| K4T2e23f | CTGCTGGCGAACGATGCTGT | qPCR |
| K4T2e3r | GACGTCCTTGCACTGGAAGT | qPCR |
| KL4e34f | CTGCTGGCGAACGGCAGAAT | qPCR |
| KL4e4r | GTGGTACAGCGGGTCATAGA | qPCR |
| KPT2e1f | ACTCCCTCTGCAGATGCTGT | qPCR |
| KLKPF1 | CCCAACCCTGGCAGGGTTGTACCATT | DNA sequencing, cloning |
| KLKPR4 | ACTTCGGAGAACTATGGTGCTGGCTA | DNA sequencing |
| KLK4Ne4r | AGCTTACTGCAGACCTCCTCAGACA | qPCR |
| HPRTf1 | AGGGGACATAAAAGTAATTGGTGGAGAT | qPCR |
| HPRTr2 | TGACCAAGGAAAGCAAAGTCTGCATTGT | qPCR |
| rpl13a-f | GTACGCTGTGAAGGCATCAA | qPCR |
| rpl13a-r | GTTGGTGTTCATCCGCTTG | qPCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lundwall, Å.; Ylitalo, E.B.; Wikström, P.; Brattsand, M. KLK4T2 Is a Hormonally Regulated Transcript from the KLK4 Locus. Int. J. Mol. Sci. 2021, 22, 13023. https://doi.org/10.3390/ijms222313023
Lundwall Å, Ylitalo EB, Wikström P, Brattsand M. KLK4T2 Is a Hormonally Regulated Transcript from the KLK4 Locus. International Journal of Molecular Sciences. 2021; 22(23):13023. https://doi.org/10.3390/ijms222313023
Chicago/Turabian StyleLundwall, Åke, Erik Bovinder Ylitalo, Pernilla Wikström, and Maria Brattsand. 2021. "KLK4T2 Is a Hormonally Regulated Transcript from the KLK4 Locus" International Journal of Molecular Sciences 22, no. 23: 13023. https://doi.org/10.3390/ijms222313023






