The Respiratory Burst Oxidase Homolog Protein D (GhRbohD) Positively Regulates the Cotton Resistance to Verticillium dahliae
Abstract
:1. Introduction
2. Results
2.1. Gene Clone and Bioinformatic Analysis of GhRbohD
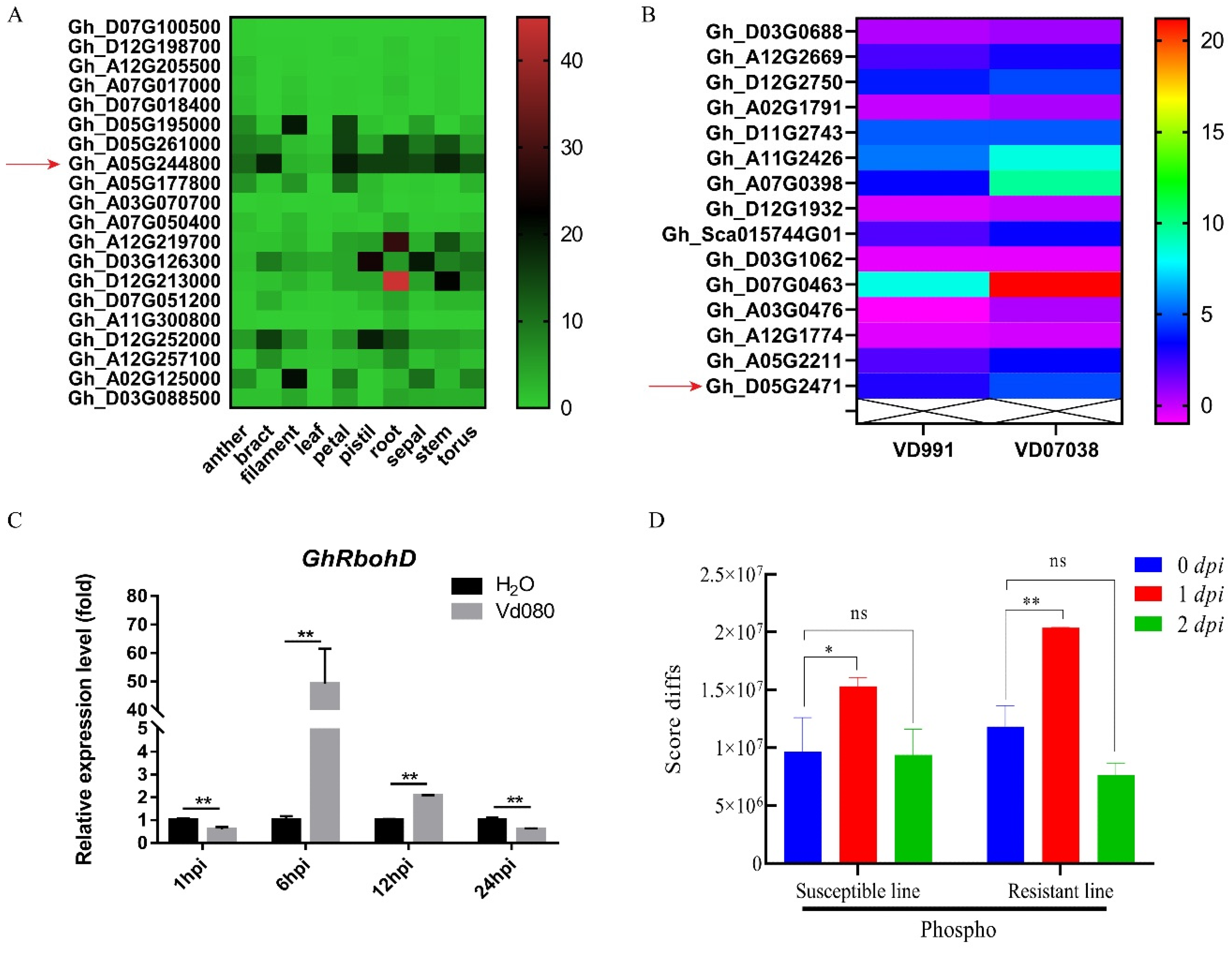
2.2. Expression Pattern of GhRbohD Gene
2.3. Silencing of GhRbohD Reduced the Basal Resistance of Cotton against V. dahliae
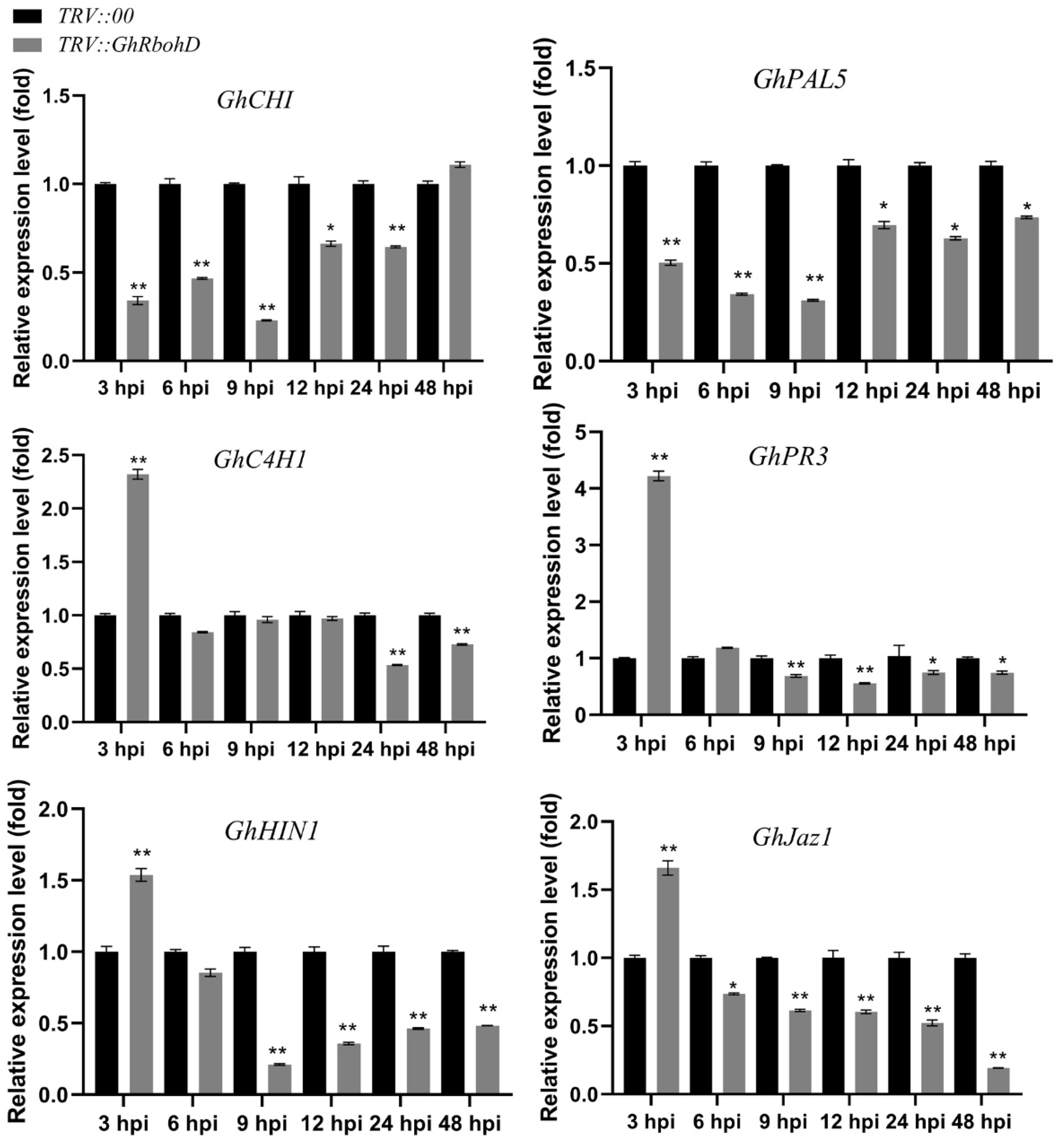
2.4. Silencing of GhRbohD Attenuated the Expression of Resistance-Related Genes
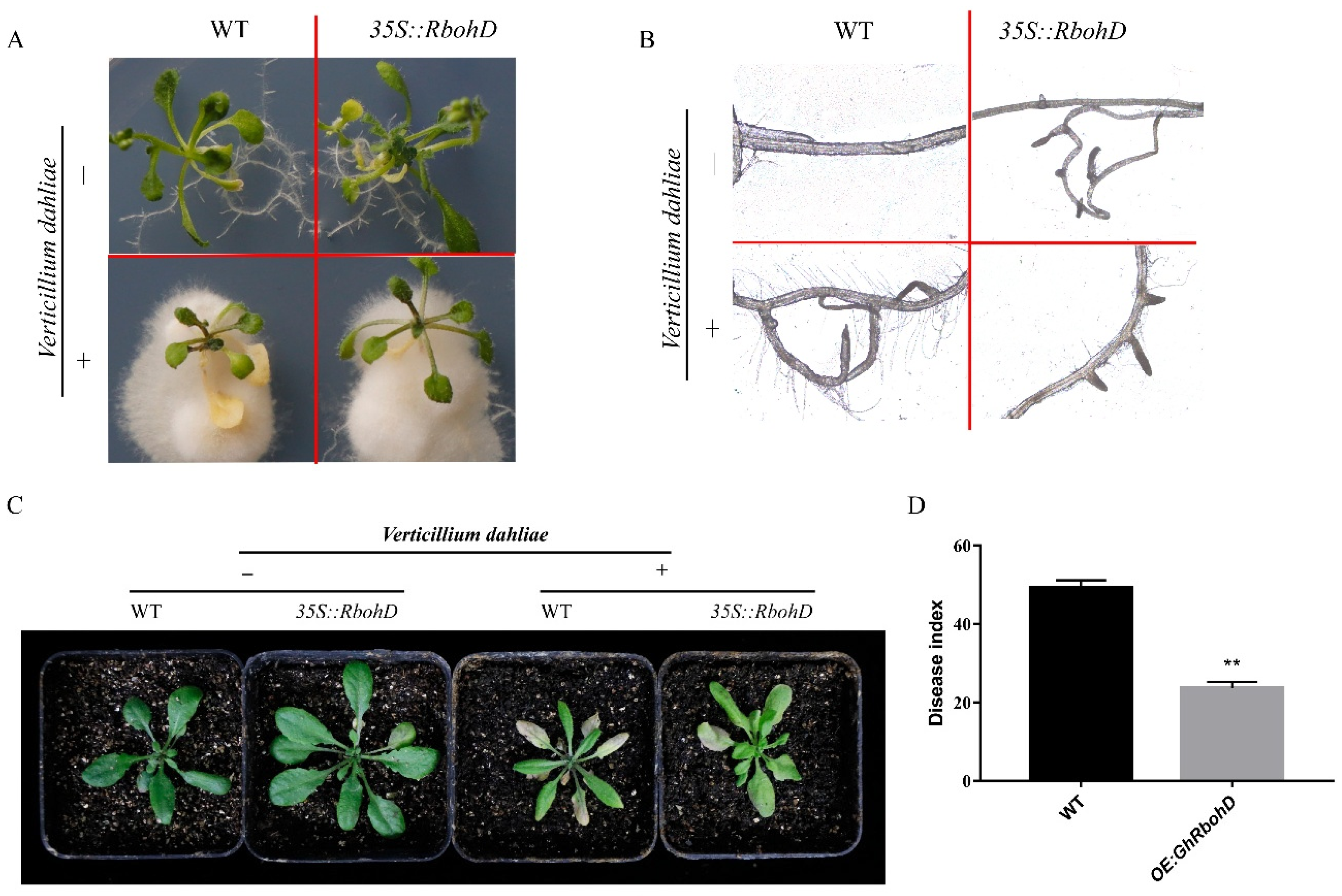
2.5. Overexpression of GhRbohD Enhanced Resistance to V. dahliae in Transgenic Arabidopsis
2.6. Subcellular Localization of GhRbohD
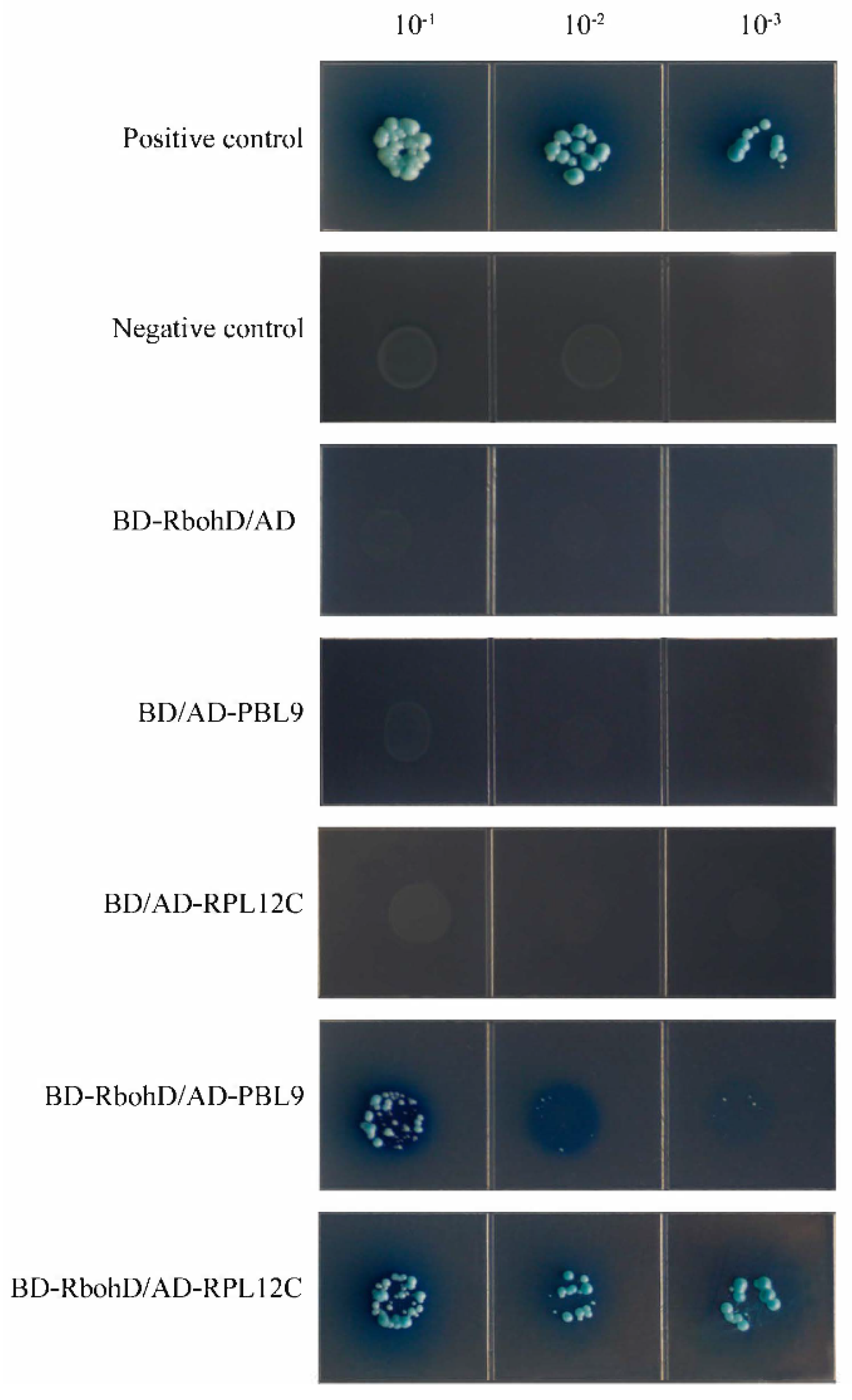
2.7. GhPBL9 and GhRPL12C May Interact with GhRbohD in Cotton
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Fungal Strain and Growth Condition
4.3. Gene Cloning and Bioinformatic Analyses
4.4. Virus-Induced Gene Silencing (VIGS)
4.5. qRT-PCR Analysis
4.6. Plant Disease Resistance Assess
4.7. Observation of ROS, Lignin Synthesis and Callose Accumulation
4.8. Visualization of H2O2, NO and Ca2+
4.9. Subcellular Localization
4.10. Arabidopsis thaliana Transformation
4.11. Yeast Two-Hybrid Assays (Y2H)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Z.; Qanmber, G.; Wang, Z.; Yang, Z.; Li, F. Gossypium Genomics: Trends, Scope, and Utilization for Cotton Improvement. Trends Plant Sci. 2020, 25, 488–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Sanogo, S.; Flynn, R.; Baral, J.B.; Bajaj, S.; Hughs, S.E.; Percy, R.G. Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 2011, 187, 147–160. [Google Scholar] [CrossRef]
- Ayele, A.; Wheeler, T.; Dever, J. Gossypium hirsutum impacts of Verticillium wilt on photosynthesis rate, lint production, and fiber quality of greenhouse-grown Cotton. Plants 2020, 9, 857. [Google Scholar] [CrossRef]
- Fradin, E.; Thomma, B. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Feng, Z.; Liu, S.; Wei, F.; Shi, Y.; Zhao, L.; Huang, W.; Zhou, Y.; Feng, H.; Zhu, H. CGTase, a novel antimicrobial protein from Bacillus cereus YUPP-10, suppresses Verticillium dahliae and mediates plant defence responses. Mol. Plant Pathol. 2021, 22, 130–144. [Google Scholar] [CrossRef]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.Q.; Menghwar, H.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018, 125, 193–204. [Google Scholar] [CrossRef]
- Song, R.; Li, J.; Xie, C.; Jian, W.; Yang, X. An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae. Int. J. Mol. Sci. 2020, 21, 1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Feng, Z.; Zhao, L.; Liu, S.; Wei, F.; Shi, Y.; Feng, H.; Zhu, H. Succinate dehydrogenase SDH1–1 positively regulates cotton resistance to Verticillium dahliae through a salicylic acid pathway. J. Cotton Res. 2020, 3, 1–12. [Google Scholar] [CrossRef]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Katano, K. Coordination Between ROS Regulatory Systems and Other Pathways Under Heat Stress and Pathogen Attack. Front. Plant Sci. 2018, 9, 490. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Kehrer, J.P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Kaya, H.; Takeda, S.; Kobayashi, M.; Kimura, S.; Iizuka, A.; Imai, A.; Hishinuma, H.; Kawarazaki, T.; Mori, K.; Yamamoto, Y.; et al. Comparative analysis of the reactive oxygen species-producing enzymatic activity of Arabidopsis NADPH oxidases. Plant J. 2018, 98, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008, 275, 3249–3277. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct Regulation of the NADPH Oxidase RBOHD by the PRR-Associated Kinase BIK1 during Plant Immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef] [Green Version]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-Associated Kinase BIK1 Directly Phosphorylates the NADPH Oxidase RbohD to Control Plant Immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Wang, Y. Post-translational Modifications of Serine/Threonine and Histidine Kinases and Their Roles in Signal Transductions in Synechocystis Sp. PCC 6803. Appl. Biochem. Biotechnol. 2021, 193, 687–716. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lal, N.; Lin, Z.-J.D.; Ma, S.; Liu, J.; Castro, B.; Toruño, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T. En Garde: CRK2 Preassociates with RBOHD and Regulates ROS Production. Plant Cell 2020, 32, 801–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Shi, Y.; Zhao, L.; Wei, F.; Feng, Z.; Feng, H. Phosphoproteomics Profiling of Cotton (Gossypium hirsutum L.) Roots in Response to Verticillium dahliae Inoculation. ACS Omega 2019, 4, 18434–18443. [Google Scholar] [CrossRef] [Green Version]
- Beliën, T.; Van Campenhout, S.; Robben, J.; Volckaert, G. Microbial Endoxylanases: Effective Weapons to Breach the Plant Cell-Wall Barrier or, Rather, Triggers of Plant Defense Systems? Mol. Plant-Microbe Interact. 2006, 19, 1072–1081. [Google Scholar] [CrossRef]
- Norman, C.; Howell, K.A.; Millar, A.H.; Whelan, J.M.; Day, D.A. Salicylic Acid Is an Uncoupler and Inhibitor of Mitochondrial Electron Transport. Plant Physiol. 2004, 134, 492–501. [Google Scholar] [CrossRef] [Green Version]
- Moreau, M.; Tian, M.; Klessig, D.F. Salicylic acid binds NPR3 and NPR4 to regulate NPR1-dependent defense responses. Cell Res. 2012, 22, 1631–1633. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Doke, N. Generation of superoxide anion by potato tuber protoplasts during the hypersensitive response to hyphal wall components of Phytophthora infestans and specific inhibition of the reaction by suppressors of hypersensitivity. Physiol. Plant Pathol. 1983, 23, 359–367. [Google Scholar] [CrossRef]
- Kadota, Y.; Goh, T.; Tomatsu, H.; Tamauchi, R.; Higashi, K.; Muto, S.; Kuchitsu, K. Cryptogein-induced initial events in tobacco BY-2 cells: Pharmacological characterization of molecular relationship among cytosolic Ca(2+) transients, anion efflux and production of reactive oxygen species. Plant Cell Physiol. 2004, 45, 160–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecourieux, D.; Ranjeva, R.; Pugin, A. Calcium in plant defence-signalling pathways. New Phytol. 2006, 171, 249–269. [Google Scholar] [CrossRef]
- Segonzac, C.; Zipfel, C. Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 2011, 14, 54–61. [Google Scholar] [CrossRef]
- Pogany, M.; von Rad, U.; Grun, S.; Dongó, A.; Pintye, A.; Simoneau, P.; Bahnweg, G.; Kiss, L.; Barna, B.; Durner, J. Dual Roles of Reactive Oxygen Species and NADPH Oxidase RBOHD in an Arabidopsis-Alternaria Pathosystem. Plant Physiol. 2009, 151, 1459–1475. [Google Scholar] [CrossRef] [Green Version]
- Chaouch, S.; Queval, G.; Noctor, G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012, 69, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, H.; Zhu, X.; Wang, W.; He, X.; Shi, Y.; Cai, Y. Analysis of sea-island cotton and upland cotton in response to Verticillium dahliae infection by RNA sequencing. BMC Genom. 2013, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kwak, J.M.; Mori, I.C.; Pei, Z.-M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.; Schroeder, J. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Fichman, Y.; Mittler, R. Vascular Bundles Mediate Systemic Reactive Oxygen Signaling during Light Stress. Plant Cell 2020, 32, 3425–3435. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, Q.; Zhou, S.; Sun, Y.; Li, X.; Li, Y. H2Bub1 regulates RbohD-dependent hydrogen peroxide signal pathway in the defense responses to Verticillium dahliae toxins. Plant Physiol. 2020, 182, 640–657. [Google Scholar] [CrossRef]
- Liu, S.; Sun, R.; Zhang, X.; Feng, Z.; Wei, F.; Zhao, L.; Zhang, Y.; Zhu, L.; Feng, H.; Zhu, H. Genome-Wide Analysis of OPR Family Genes in Cotton Identified a Role for GhOPR9 in Verticillium dahliae Resistance. Genes 2020, 11, 1134. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Li, B.; Shi, Q.; Geng, R.; Geng, S.; Liu, J.; Zhang, Y.; Cai, Y. Comprehensive analysis of respiratory burst oxidase homologs (Rboh) gene family and function of GbRboh5/18 on Verticillium wilt resistance in Gossypium barbadense. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Zhang, Z.; Lei, Y.; Hu, G.; Liu, J.; Hao, M.; Chen, A.; Peng, Q.; Wu, J. Cotton WATs Modulate SA Biosynthesis and Local Lignin Deposition Participating in Plant Resistance Against Verticillium dahliae. Front. Plant Sci. 2019, 10, 526. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.-X.; Zhao, L.-H.; Klosterman, S.J.; Feng, H.-J.; Feng, Z.-L.; Wei, F.; Shi, Y.-Q.; Li, Z.-F.; Zhu, H.-Q. The endochitinase VDECH from Verticillium dahliae inhibits spore germination and activates plant defense responses. Plant Sci. 2017, 259, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Min, L.; Yang, X.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H.; et al. Laccase GhLac1 Modulates Broad-Spectrum Biotic Stress Tolerance via Manipulating Phenylpropanoid Pathway and Jasmonic Acid Synthesis. Plant Physiol. 2018, 176, 1808–1823. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Feng, H.; Wang, L.; Li, Z.; Shi, Y.; Zhao, L.; Feng, Z.; Zhu, H. Potential of Endophytic Fungi Isolated from Cotton Roots for Biological Control against Verticillium Wilt Disease. PLoS ONE 2017, 12, e0170557. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Zhu, L.; Zhang, X.; Guan, Q.; Xiao, S.; Min, L.; Zhang, X. GhCPK33 Negatively Regulates Defense against Verticillium dahliae by Phosphorylating GhOPR3. Plant Physiol. 2018, 178, 876–889. [Google Scholar] [CrossRef] [Green Version]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Gong, Q.; Yang, Z.; Wang, X.; Butt, H.I.; Chen, E.; He, S.; Zhang, C.; Zhang, X.; Li, F. Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 2017, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Zhang, Y.; Zhou, J.; Wei, F.; Feng, Z.; Zhao, L.; Shi, Y.; Feng, H.; Zhu, H. The Respiratory Burst Oxidase Homolog Protein D (GhRbohD) Positively Regulates the Cotton Resistance to Verticillium dahliae. Int. J. Mol. Sci. 2021, 22, 13041. https://doi.org/10.3390/ijms222313041
Huang W, Zhang Y, Zhou J, Wei F, Feng Z, Zhao L, Shi Y, Feng H, Zhu H. The Respiratory Burst Oxidase Homolog Protein D (GhRbohD) Positively Regulates the Cotton Resistance to Verticillium dahliae. International Journal of Molecular Sciences. 2021; 22(23):13041. https://doi.org/10.3390/ijms222313041
Chicago/Turabian StyleHuang, Wanting, Yalin Zhang, Jinglong Zhou, Feng Wei, Zili Feng, Lihong Zhao, Yongqiang Shi, Hongjie Feng, and Heqin Zhu. 2021. "The Respiratory Burst Oxidase Homolog Protein D (GhRbohD) Positively Regulates the Cotton Resistance to Verticillium dahliae" International Journal of Molecular Sciences 22, no. 23: 13041. https://doi.org/10.3390/ijms222313041
APA StyleHuang, W., Zhang, Y., Zhou, J., Wei, F., Feng, Z., Zhao, L., Shi, Y., Feng, H., & Zhu, H. (2021). The Respiratory Burst Oxidase Homolog Protein D (GhRbohD) Positively Regulates the Cotton Resistance to Verticillium dahliae. International Journal of Molecular Sciences, 22(23), 13041. https://doi.org/10.3390/ijms222313041






