Assessment of Normal Tissue Radiosensitivity by Evaluating DNA Damage and Repair Kinetics in Human Brain Organoids
Abstract
:1. Introduction
2. Results
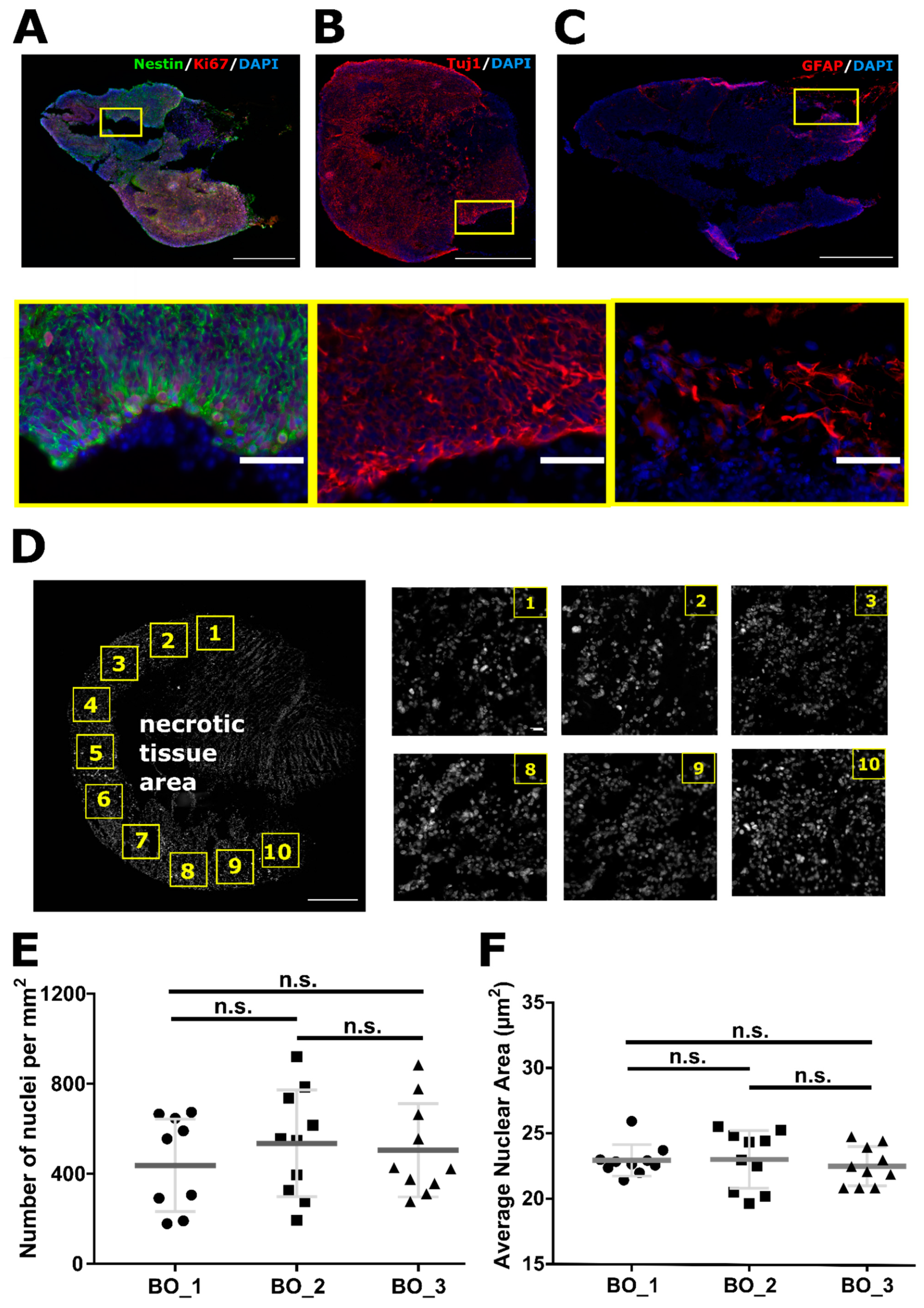
2.1. Organoids Characterization
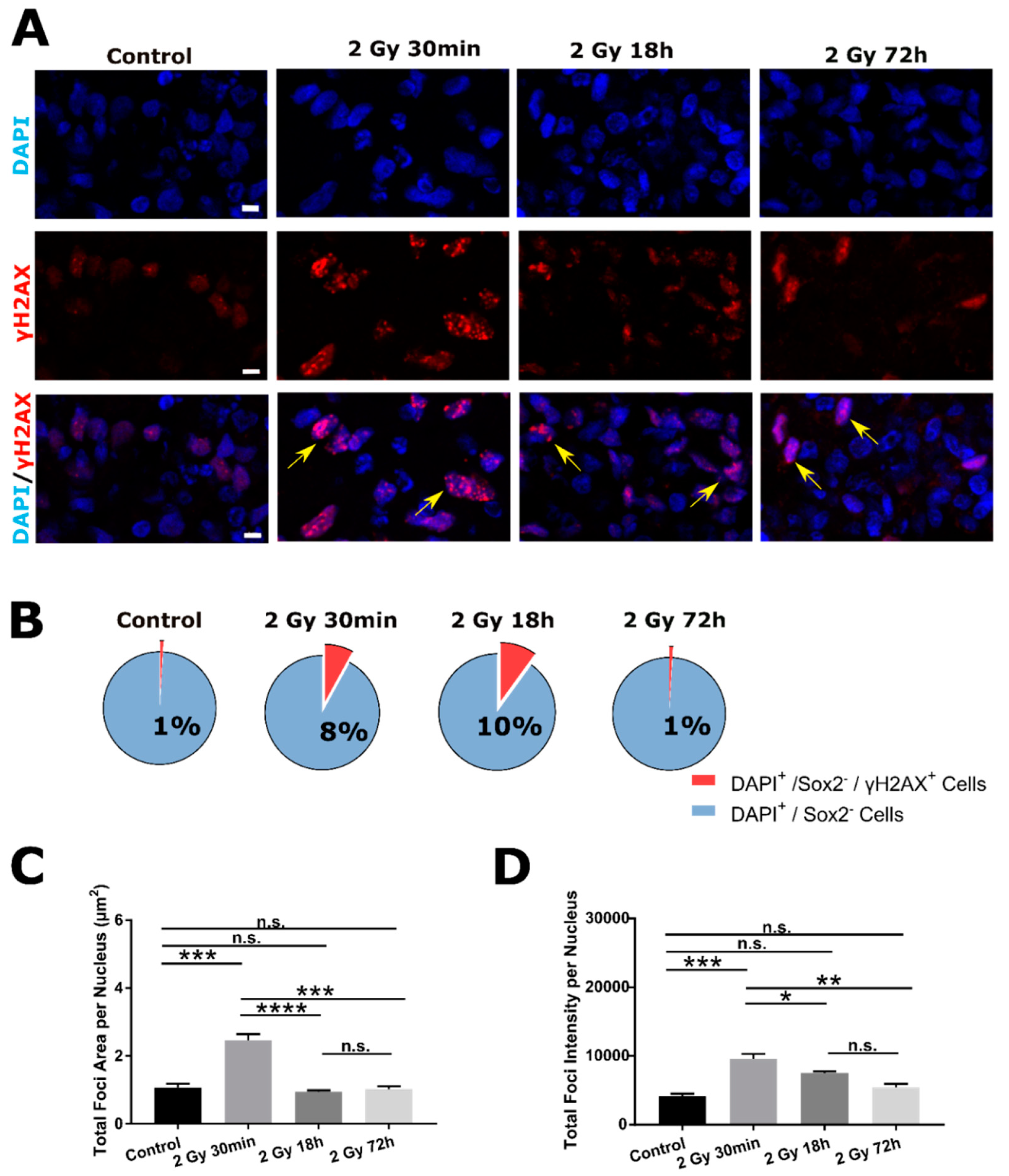
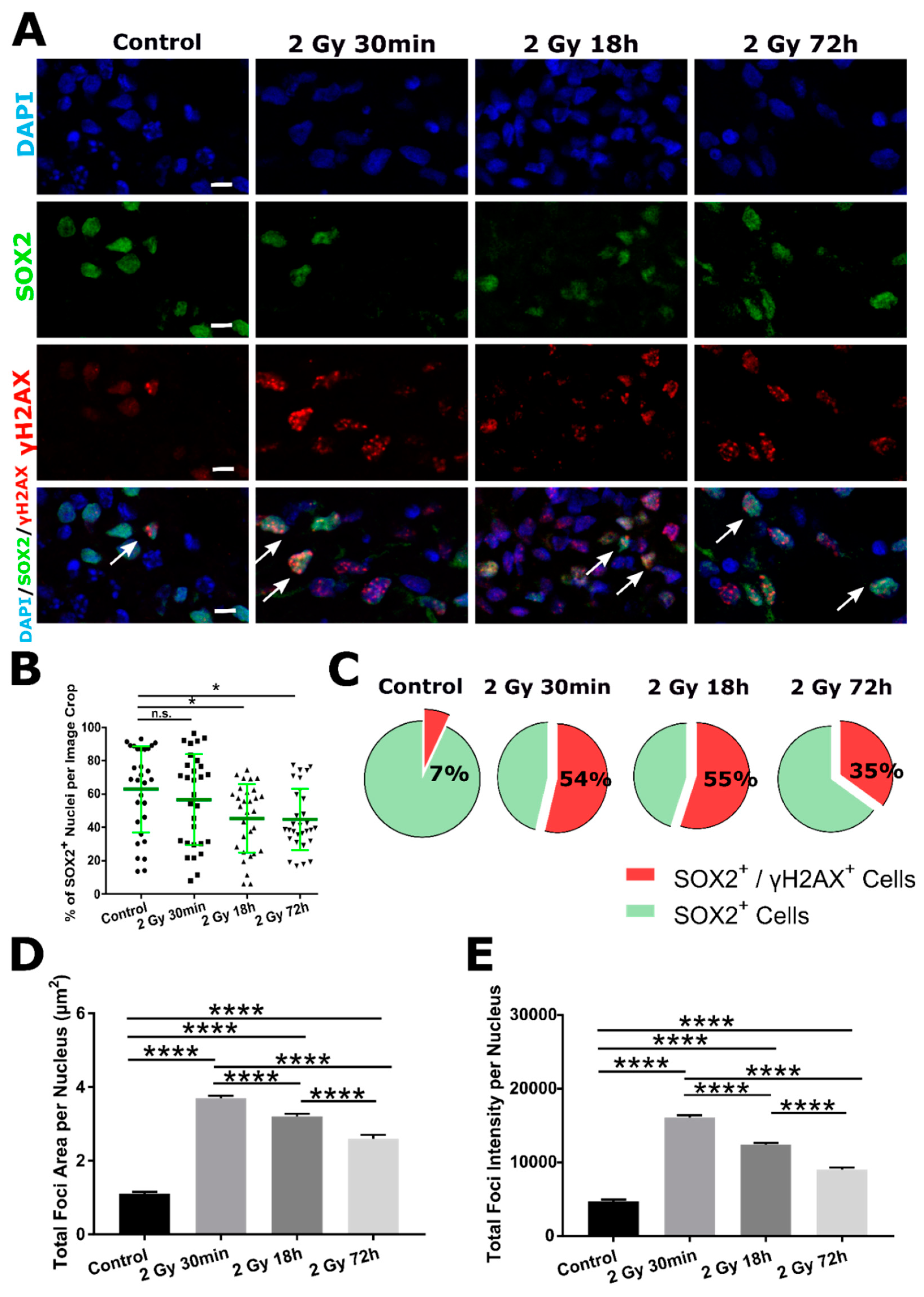
2.2. DSB DNA Damage Response Following 2 Gy Irradiation in SOX2− Cells
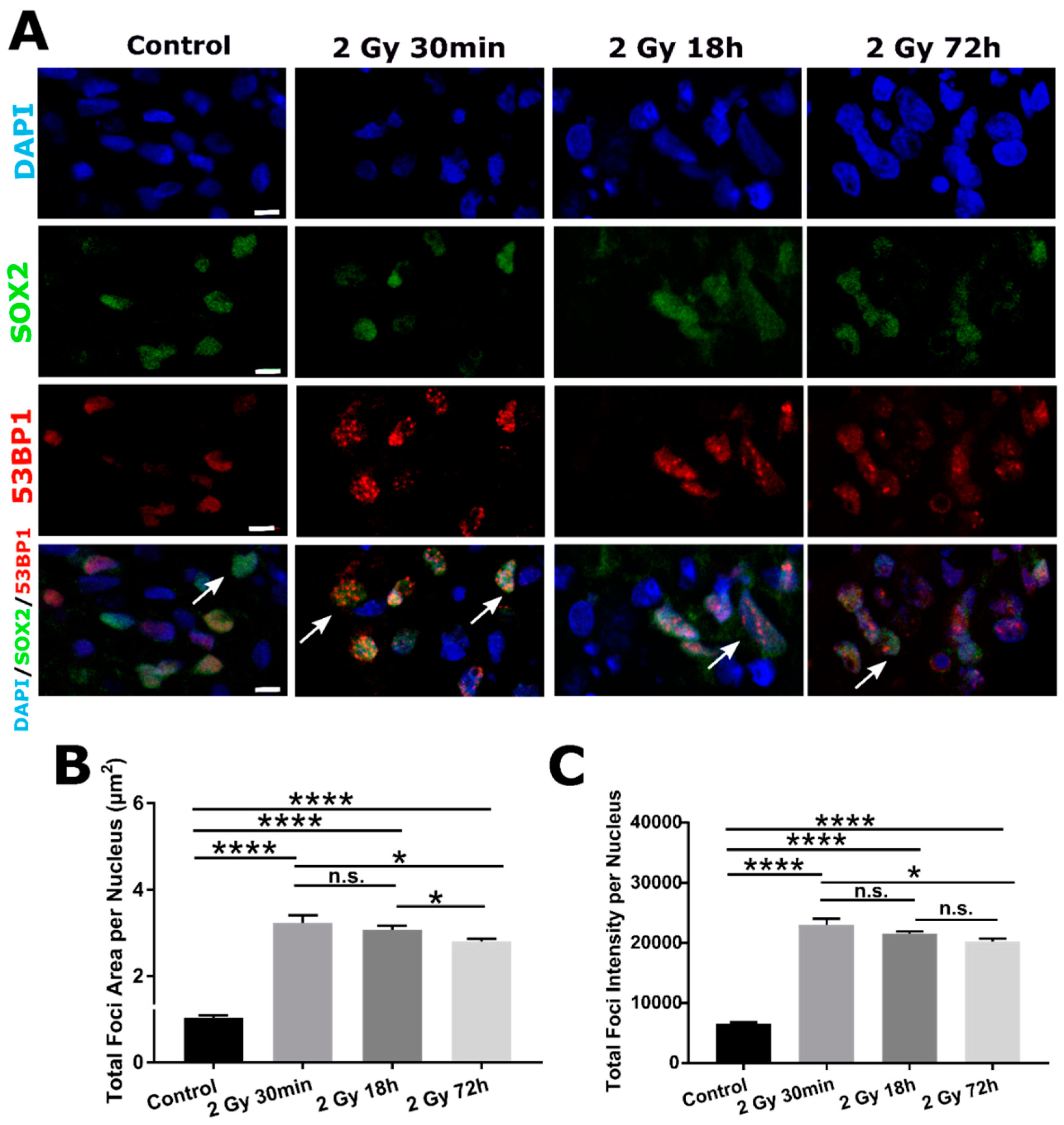
2.3. DSB DNA Damage Induced by 2 Gy Irradiation Is Most Persistent in Neuronal Progenitor Cells
3. Discussion
4. Materials and Methods
4.1. Cerebral Organoids Cultivation
4.2. Preparation of Frozen Tissue Sections
4.3. X-ray Irradiation
4.4. Immunofluorescent Staining
4.5. Microscopy and Image Analysis
4.6. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawada, J.; Kaneda, S.; Kirihara, T.; Maroof, A.; Levi, T.; Eggan, K.; Fujii, T.; Ikeuchi, Y. Generation of a Motor Nerve Organoid with Human Stem Cell-Derived Neurons. Stem. Cell Rep. 2017, 9, 1441–1449. [Google Scholar] [CrossRef] [Green Version]
- Hor, J.H.; Soh, E.S.; Tan, L.Y.; Lim, V.J.W.; Santosa, M.M.; Winanto; Ho, B.X.; Fan, Y.; Soh, B.S.; Ng, S.Y. Cell cycle inhibitors protect motor neurons in an organoid model of Spinal Muscular Atrophy. Cell Death Dis. 2018, 9, 1100. [Google Scholar] [CrossRef]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.D.; Goke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.P.; Lokman, H.; et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 2016, 19, 248–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monzel, A.S.; Smits, L.M.; Hemmer, K.; Hachi, S.; Moreno, E.L.; van Wuellen, T.; Jarazo, J.; Walter, J.; Bruggemann, I.; Boussaad, I.; et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Rep. 2017, 8, 1144–1154. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Kadoshima, T.; Soen, M.; Narii, N.; Ishida, Y.; Ohgushi, M.; Takahashi, J.; Eiraku, M.; Sasai, Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015, 6, 8896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Pasca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Krefft, O.; Jabali, A.; Iefremova, V.; Koch, P.; Ladewig, J. Generation of Standardized and Reproducible Forebrain-type Cerebral Organoids from Human Induced Pluripotent Stem Cells. J. Vis. Exp. 2018, 56768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, B.; Mathew, R.K.; Polson, E.S.; Williams, J.; Wurdak, H. Spontaneous Glioblastoma Spheroid Infiltration of Early-Stage Cerebral Organoids Models Brain Tumor Invasion. SLAS Discov. 2018, 23, 862–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef] [Green Version]
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically engineered cerebral organoids model brain tumor formation. Nat. Methods 2018, 15, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hempen, C.; Weiss, E.; Hess, C.F. Dexamethasone treatment in patients with brain metastases and primary brain tumors: Do the benefits outweigh the side-effects? Support. Care Cancer 2002, 10, 322–328. [Google Scholar] [CrossRef]
- Shaw, M.G.; Ball, D.L. Treatment of brain metastases in lung cancer: Strategies to avoid/reduce late complications of whole brain radiation therapy. Curr. Treat. Opt. Oncol. 2013, 14, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.L.; Adileh, M.; Hsu, K.S.; Hua, G.; Lee, S.G.; Li, C.; Fuller, J.D.; Rotolo, J.A.; Bodo, S.; Klingler, S.; et al. Organoids Reveal That Inherent Radiosensitivity of Small and Large Intestinal Stem Cells Determines Organ Sensitivity. Cancer Res. 2020, 80, 1219–1227. [Google Scholar] [CrossRef] [Green Version]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.C.; Chan, A.S.; Law, S.C.; Chan, J.H.; Tse, V.K. Impact of radionecrosis on cognitive dysfunction in patients after radiotherapy for nasopharyngeal carcinoma. Cancer 2003, 97, 2019–2026. [Google Scholar] [CrossRef]
- Mineyeva, O.A.; Bezriadnov, D.V.; Kedrov, A.V.; Lazutkin, A.A.; Anokhin, K.V.; Enikolopov, G.N. Radiation Induces Distinct Changes in Defined Subpopulations of Neural Stem and Progenitor Cells in the Adult Hippocampus. Front. Neurosci. 2018, 12, 1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhle, A.; Huber, P.E. Normal tissue: Radiosensitivity, toxicity, consequences for planning. Radiologe 2018, 58, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Falcke, S.E.; Ruhle, P.F.; Deloch, L.; Fietkau, R.; Frey, B.; Gaipl, U.S. Clinically Relevant Radiation Exposure Differentially Impacts Forms of Cell Death in Human Cells of the Innate and Adaptive Immune System. Int. J. Mol. Sci. 2018, 19, 3574. [Google Scholar] [CrossRef] [Green Version]
- Goodarzi, A.A.; Jeggo, P.A. Irradiation induced foci (IRIF) as a biomarker for radiosensitivity. Mutat. Res. 2012, 736, 39–47. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Singh, K.; Almasan, A. Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol. Biol. 2012, 920, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Ward, I.M.; Minn, K.; Jorda, K.G.; Chen, J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003, 278, 19579–19582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Hurk, M.; Bardy, C. Single-cell multimodal transcriptomics to study neuronal diversity in human stem cell-derived brain tissue and organoid models. J. Neurosci. Methods 2019, 325, 108350. [Google Scholar] [CrossRef] [PubMed]
- Achanta, P.; Capilla-Gonzalez, V.; Purger, D.; Reyes, J.; Sailor, K.; Song, H.; Garcia-Verdugo, J.M.; Gonzalez-Perez, O.; Ford, E.; Quinones-Hinojosa, A. Subventricular zone localized irradiation affects the generation of proliferating neural precursor cells and the migration of neuroblasts. Stem Cells 2012, 30, 2548–2560. [Google Scholar] [CrossRef] [Green Version]
- Bowles, K.R.; Silva, M.C.; Whitney, K.; Bertucci, T.; Berlind, J.E.; Lai, J.D.; Garza, J.C.; Boles, N.C.; Mahali, S.; Strang, K.H.; et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 2021, 184, 4547–4563.e17. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Li, J.; Cheng, L.; Franco, S.; Mahairaki, V. Human Forebrain Organoids from Induced Pluripotent Stem Cells: A Novel Approach to Model Repair of Ionizing Radiation-Induced DNA Damage in Human Neurons. Radiat. Res. 2020, 194, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Dokic, I.; Mairani, A.; Niklas, M.; Zimmermann, F.; Chaudhri, N.; Krunic, D.; Tessonnier, T.; Ferrari, A.; Parodi, K.; Jakel, O.; et al. Next generation multi-scale biophysical characterization of high precision cancer particle radiotherapy using clinical proton, helium-, carbon- and oxygen ion beams. Oncotarget 2016, 7, 56676–56689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowrouzi, A.; Sertorio, M.G.; Akbarpour, M.; Knoll, M.; Krunic, D.; Kuhar, M.; Schwager, C.; Brons, S.; Debus, J.; Wells, S.I.; et al. Personalized Assessment of Normal Tissue Radiosensitivity via Transcriptome Response to Photon, Proton and Carbon Irradiation in Patient-Derived Human Intestinal Organoids. Cancers 2020, 12, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeler, C.R.; Mirkovic, D.; Titt, U.; Blanchard, P.; Gunther, J.R.; Mahajan, A.; Mohan, R.; Grosshans, D.R. Clinical evidence of variable proton biological effectiveness in pediatric patients treated for ependymoma. Radiother. Oncol. 2016, 121, 395–401. [Google Scholar] [CrossRef] [Green Version]
- Mayo, C.; Yorke, E.; Merchant, T.E. Radiation associated brainstem injury. Int. J. Radiat. Oncol. Biol. Phys. 2010, 76, S36–S41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Cerdeno, V.; Noctor, S.C. Neural Progenitor Cell Terminology. Front. Neuroanat. 2018, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keogh, M.C.; Kim, J.A.; Downey, M.; Fillingham, J.; Chowdhury, D.; Harrison, J.C.; Onishi, M.; Datta, N.; Galicia, S.; Emili, A.; et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 2006, 439, 497–501. [Google Scholar] [CrossRef]
- Nowak, E.; Etienne, O.; Millet, P.; Lages, C.S.; Mathieu, C.; Mouthon, M.A.; Boussin, F.D. Radiation-induced H2AX phosphorylation and neural precursor apoptosis in the developing brain of mice. Radiat. Res. 2006, 165, 155–164. [Google Scholar] [CrossRef]
- Liu, S.K.; Olive, P.L.; Bristow, R.G. Biomarkers for DNA DSB inhibitors and radiotherapy clinical trials. Cancer Metastasis Rev. 2008, 27, 445–458. [Google Scholar] [CrossRef]
- Jacobs, K.M.; Misri, S.; Meyer, B.; Raj, S.; Zobel, C.L.; Sleckman, B.P.; Hallahan, D.E.; Sharma, G.G. Unique epigenetic influence of H2AX phosphorylation and H3K56 acetylation on normal stem cell radioresponses. Mol. Biol. Cell 2016, 27, 1332–1345. [Google Scholar] [CrossRef]
- Lemke, D.; Weiler, M.; Blaes, J.; Wiestler, B.; Jestaedt, L.; Klein, A.C.; Low, S.; Eisele, G.; Radlwimmer, B.; Capper, D.; et al. Primary glioblastoma cultures: Can profiling of stem cell markers predict radiotherapy sensitivity? J. Neurochem. 2014, 131, 251–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourton, E.C.; Plowman, P.N.; Smith, D.; Arlett, C.F.; Parris, C.N. Prolonged expression of the gamma-H2AX DNA repair biomarker correlates with excess acute and chronic toxicity from radiotherapy treatment. Int. J. Cancer 2011, 129, 2928–2934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannesen, T.B.; Lien, H.H.; Hole, K.H.; Lote, K. Radiological and clinical assessment of long-term brain tumour survivors after radiotherapy. Radiother. Oncol. 2003, 69, 169–176. [Google Scholar] [CrossRef]
- Raber, J.; Rola, R.; LeFevour, A.; Morhardt, D.; Curley, J.; Mizumatsu, S.; VandenBerg, S.R.; Fike, J.R. Radiation-induced cognitive impairments are associated with changes in indicators of hippocampal neurogenesis. Radiat. Res. 2004, 162, 39–47. [Google Scholar] [CrossRef]
- Monzel, A.S.; Hemmer, K.; Kaoma, T.; Smits, L.M.; Bolognin, S.; Lucarelli, P.; Rosety, I.; Zagare, A.; Antony, P.; Nickels, S.L.; et al. Machine learning-assisted neurotoxicity prediction in human midbrain organoids. Parkinsonism Relat. Disord. 2020, 75, 105–109. [Google Scholar] [CrossRef]
- Alter, B.P. Radiosensitivity in Fanconi’s anemia patients. Radiother. Oncol. 2002, 62, 345–347. [Google Scholar] [CrossRef]
- Suri, J.S.; Rednam, S.; Teh, B.S.; Butler, E.; Paulino, A.C. Subsequent Malignancies in Patients With Li-Fraumeni Syndrome Treated With Radiation Therapy. Int. J. Radiat. Oncol. 2013, 87, S71–S72. [Google Scholar] [CrossRef]
- Sloan, S.A.; Darmanis, S.; Huber, N.; Khan, T.A.; Birey, F.; Caneda, C.; Reimer, R.; Quake, S.R.; Barres, B.A.; Pasca, S.P. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 2017, 95, 779–790.e6. [Google Scholar] [CrossRef] [PubMed]
- Ormel, P.R.; Vieira de Sa, R.; van Bodegraven, E.J.; Karst, H.; Harschnitz, O.; Sneeboer, M.A.M.; Johansen, L.E.; van Dijk, R.E.; Scheefhals, N.; Berdenis van Berlekom, A.; et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018, 9, 4167. [Google Scholar] [CrossRef]
- Yu, J. Vascularized Organoids: A More Complete Model. Int. J. Stem Cells 2021, 14, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Goncalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Antibody | Company | Subtype | Dilution |
|---|---|---|---|
| Nestin | Santa Cruz Biotechnology, TX, USA | Mouse monoclonal | 1:500 |
| Tuj1 (Anti-β -Tubulin III) | BioLegend, CA, USA | Mouse monoclonal | 1:500 |
| GFAP (Glial Fibrillary Acidic Protein) | Sigma-Aldrich (Merck), MO, USA | Rabbit polyclonal | 1:500 |
| CD11b (Integrin αM subunit) | Abcam, Cambridge, UK | Rabbit polyclonal | 1:500 |
| SOX2 | Merck Millipore, MA, USA | Rabbit polyclonal | 1:500 |
| γH2AX | Cell Biolabs, CA, USA | Mouse monoclonal | 1:100 |
| 53BP1 | NOVUS, CO, USA | Rabbit polyclonal | 1:500 |
| Antibody | Company | Subtype | Conjugate | Dilution |
|---|---|---|---|---|
| Goat anti-rabbit | Life Technologies, CA, USA | IgG | Alexa 647 | 1:400 |
| Goat anti-mouse | Life Technologies, CA, USA | IgG | Alexa 647 | 1:400 |
| Goat anti-rabbit | Life Technologies, CA, USA | IgG | Alexa 555 | 1:400 |
| Goat anti-mouse | Life Technologies, CA, USA | IgG | Alexa 555 | 1:400 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bojcevski, J.; Wang, C.; Liu, H.; Abdollahi, A.; Dokic, I. Assessment of Normal Tissue Radiosensitivity by Evaluating DNA Damage and Repair Kinetics in Human Brain Organoids. Int. J. Mol. Sci. 2021, 22, 13195. https://doi.org/10.3390/ijms222413195
Bojcevski J, Wang C, Liu H, Abdollahi A, Dokic I. Assessment of Normal Tissue Radiosensitivity by Evaluating DNA Damage and Repair Kinetics in Human Brain Organoids. International Journal of Molecular Sciences. 2021; 22(24):13195. https://doi.org/10.3390/ijms222413195
Chicago/Turabian StyleBojcevski, Jovana, Changwen Wang, Haikun Liu, Amir Abdollahi, and Ivana Dokic. 2021. "Assessment of Normal Tissue Radiosensitivity by Evaluating DNA Damage and Repair Kinetics in Human Brain Organoids" International Journal of Molecular Sciences 22, no. 24: 13195. https://doi.org/10.3390/ijms222413195






