Retinoic Acid Promotes the In Vitro Growth, Patterning and Improves the Cellular Composition of Human Pluripotent Stem-Cell-Derived Intestinal Organoids
Abstract
:1. Introduction
2. Results
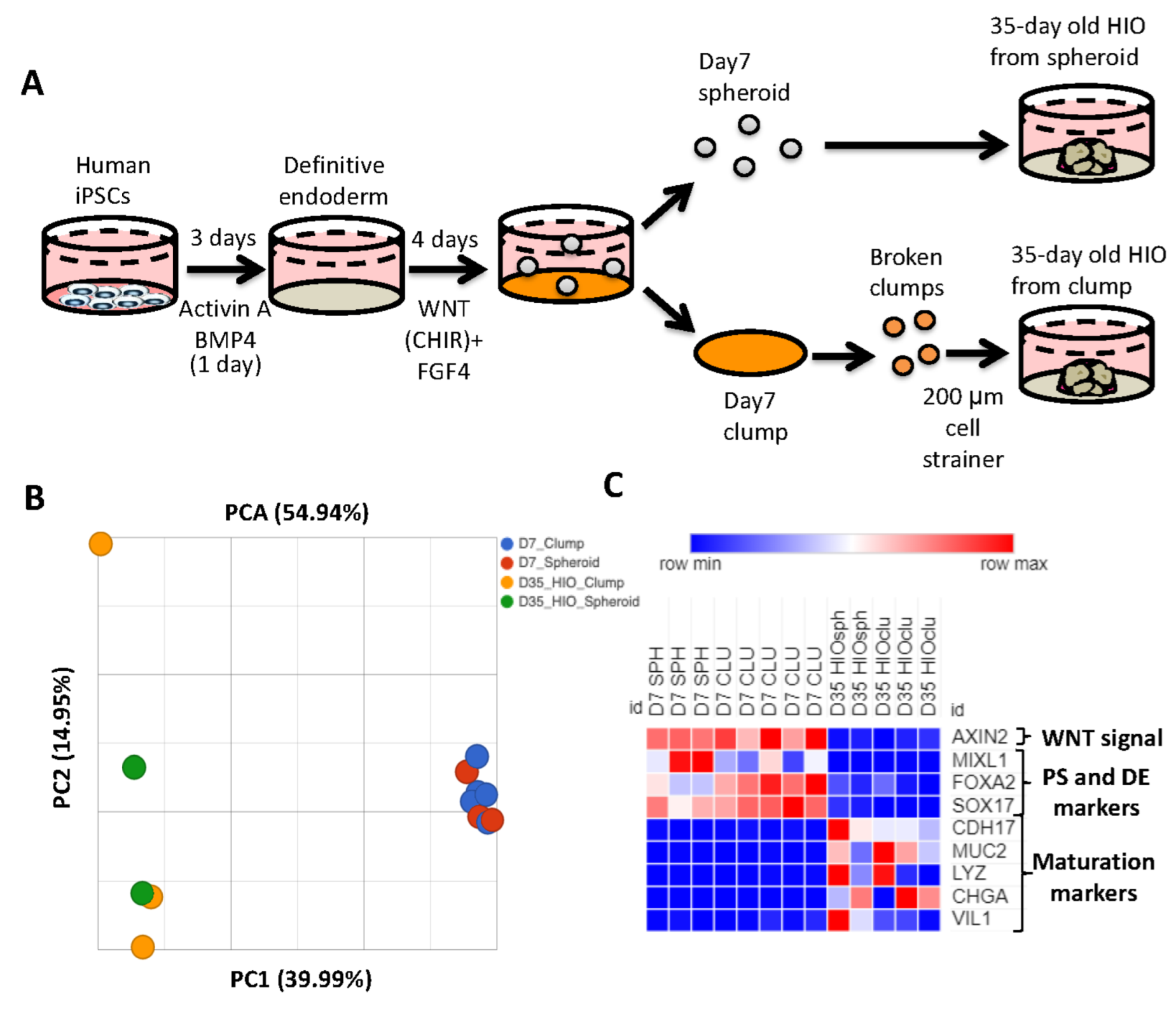
2.1. Spheroid- and Clump-Derived HIOs Display Similar Transcriptomic Profiles
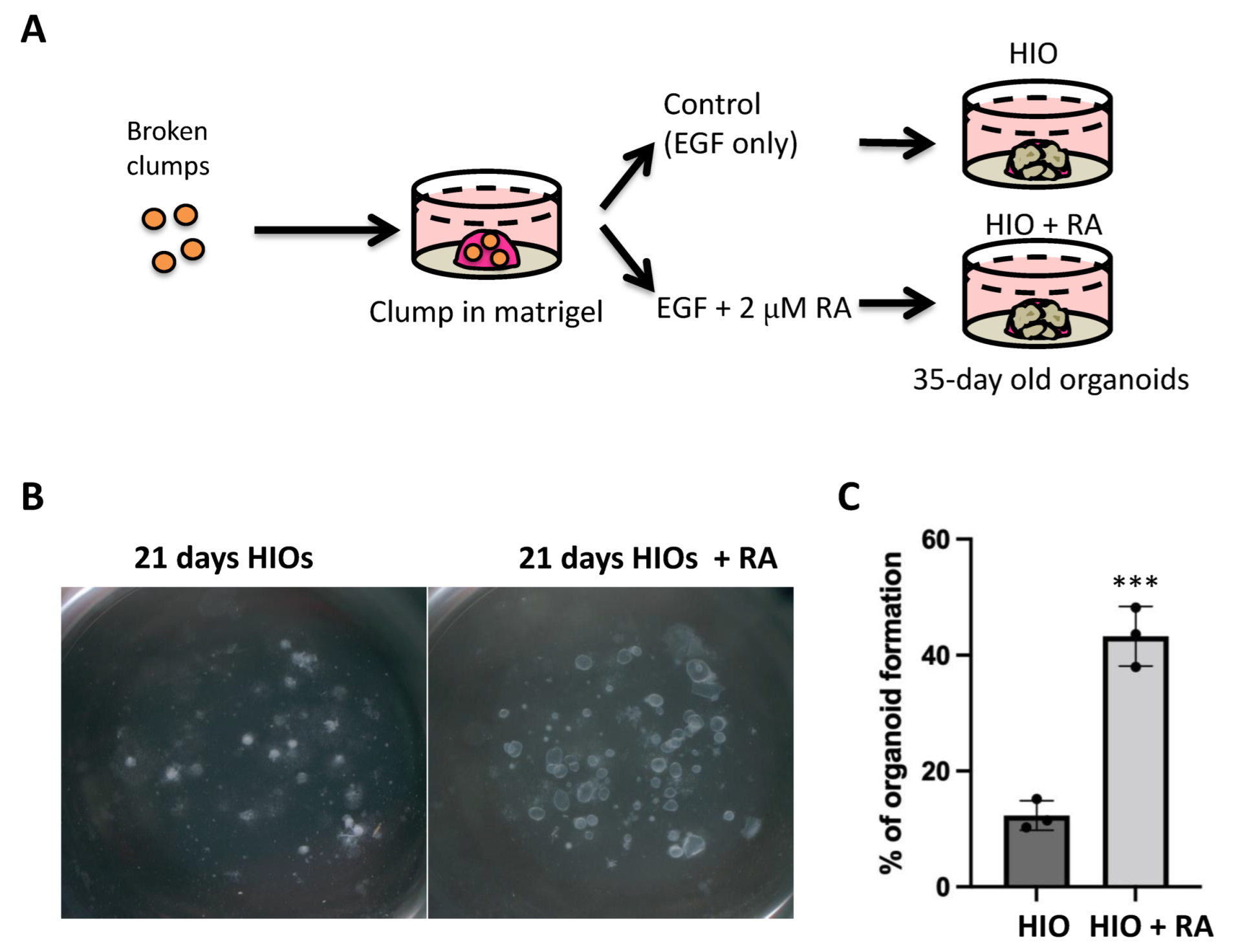
2.2. Retinoic Acid Improves HIO Outgrowth
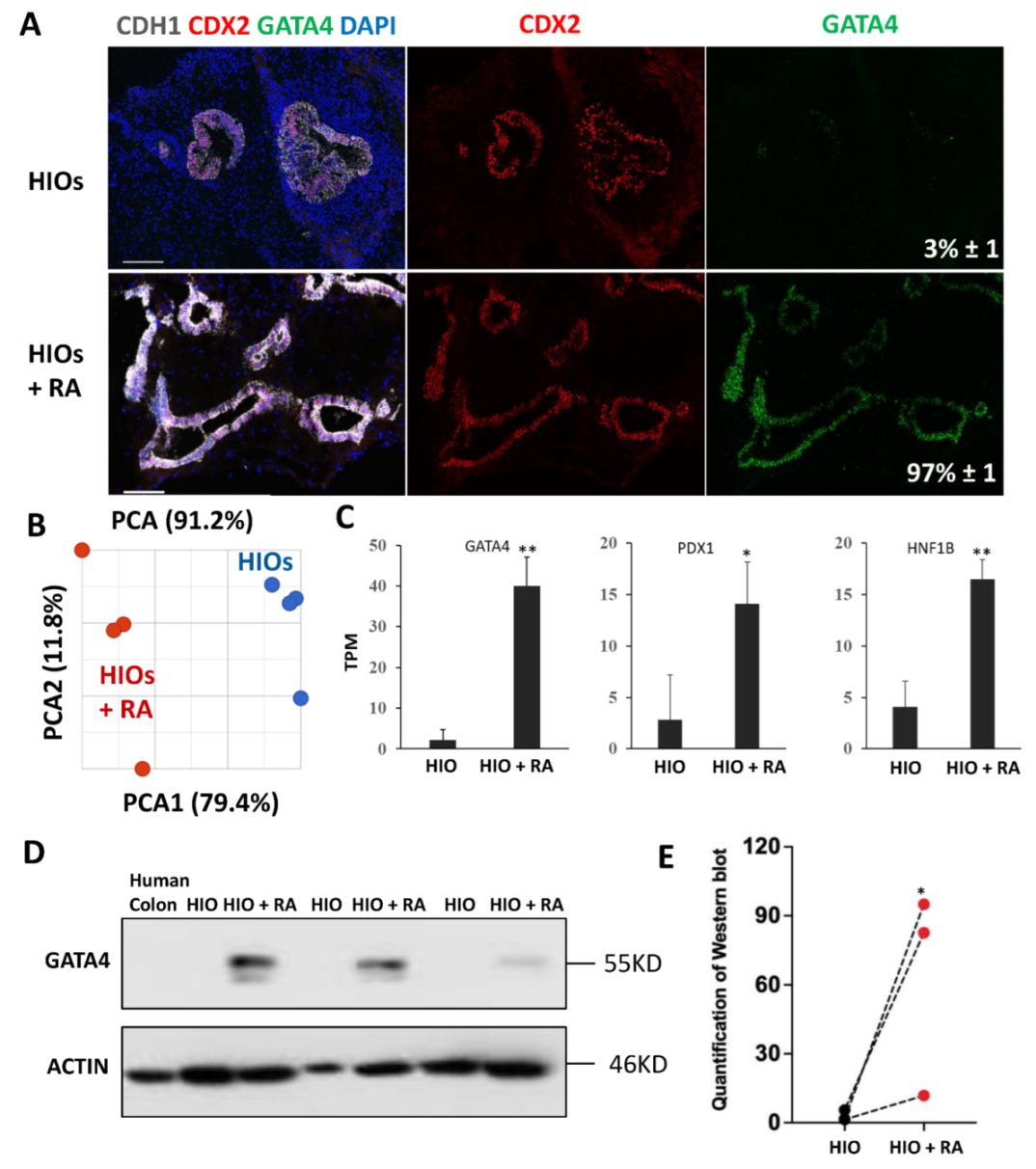
2.3. Retinoic Acid Improves the Patterning of HIOs
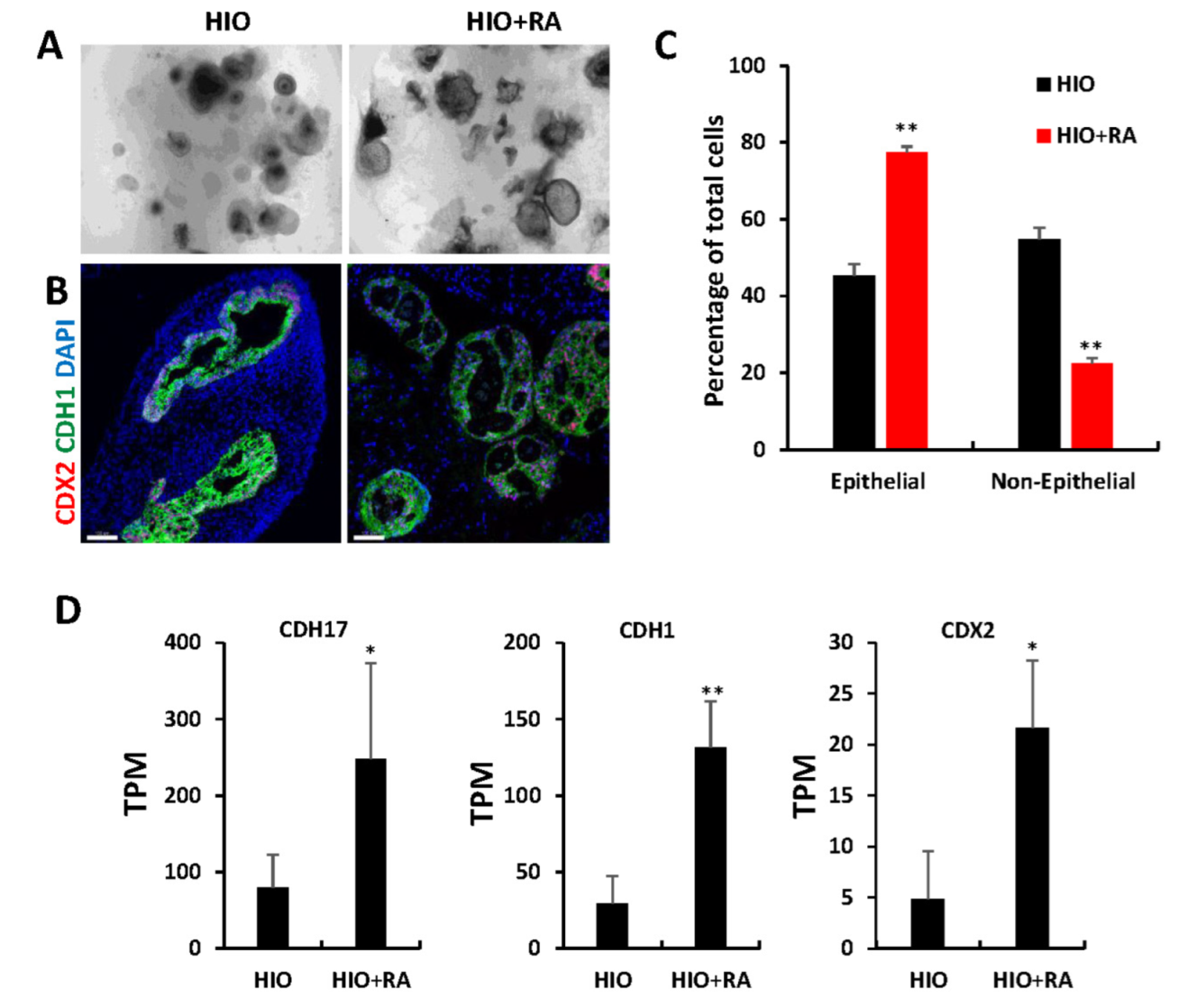
2.4. Retinoic Acid Promotes Epithelial Growth and Enriches ISEMFs in HIOs
2.5. Retinoic Acid Inhibits the Co-Development of Neurons in HIOs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Generation of Human Gut Tube Cultures
4.3. The Culture of Floating Spheroids and Clumps of Dissociated Mid/Hindgut Endoderm
4.4. Efficiency of Organoid Formation
4.5. Immunofluorescence Staining
4.6. Quantification of Immunofluorescence Images
4.7. Western Blot
4.8. RNA-Seq Processing
4.9. Quantification, Statistical Analysis and Data Availability
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Date, S.; Sato, T. Mini-Gut Organoids: Reconstitution of the Stem Cell Niche. Annu. Rev. Cell Dev. Biol. 2015, 31, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Clevers, H.; Sato, T. Modeling Human Digestive Diseases With CRISPR-Cas9–Modified Organoids. Gastroenterology 2019, 156, 562–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Múnera, J.O.; Sundaram, N.; Rankin, S.A.; Hill, D.; Watson, C.; Mahe, M.; Vallance, J.E.; Shroyer, N.F.; Sinagoga, K.L.; Zarzoso-Lacoste, A.; et al. Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling. Cell Stem Cell 2017, 21, 51–64.e6. [Google Scholar] [CrossRef] [Green Version]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.-H.; Nattiv, R.; Dedhia, P.H.; Nagy, M.S.; Chin, A.M.; Thomson, M.; Klein, O.D.; Spence, J.R. In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development. Development 2017, 144, 1045–1055. [Google Scholar] [CrossRef] [Green Version]
- Sinagoga, K.L.; McCauley, H.A.; Múnera, J.O.; Reynolds, N.A.; Enriquez, J.R.; Watson, C.; Yang, H.-C.; Helmrath, M.A.; Wells, J.M. Deriving functional human enteroendocrine cells from pluripotent stem cells. Development 2018, 145, dev165795. [Google Scholar] [CrossRef] [Green Version]
- Battle, M.A.; Bondow, B.J.; Iverson, M.A.; Adams, S.J.; Jandacek, R.J.; Tso, P.; Duncan, S.A. GATA4 is Essential for Jejunal Function in Mice. Gastroenterology 2008, 135, 1676–1686.e1. [Google Scholar] [CrossRef] [Green Version]
- Bosse, T.; Piaseckyj, C.M.; Burghard, E.; Fialkovich, J.J.; Rajagopal, S.; Pu, W.T.; Krasinski, S.D. Gata4 is Essential for the Maintenance of Jejunal-Ileal Identities in the Adult Mouse Small Intestine. Mol. Cell. Biol. 2006, 26, 9060–9070. [Google Scholar] [CrossRef] [Green Version]
- Holloway, E.M.; Wu, J.H.; Czerwinski, M.; Sweet, C.W.; Wu, A.; Tsai, Y.-H.; Huang, S.; Stoddard, A.E.; Capeling, M.M.; Glass, I.; et al. Differentiation of Human Intestinal Organoids with Endogenous Vascular Endothelial Cells. Dev. Cell 2020, 54, 516–528.e517. [Google Scholar] [CrossRef] [PubMed]
- Sarvestani, S.K.; Signs, S.; Hu, B.; Yeu, Y.; Feng, H.; Ni, Y.; Hill, D.R.; Fisher, R.C.; Ferrandon, S.; DeHaan, R.K.; et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 2021, 12, 262. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, T.J.; Duester, G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.; Wei, W.; Saund, R.S.; Xiang, P.; Cunningham, T.J.; Yi, Y.; Alder, O.; Lu, D.Y.; Savory, J.G.; Krentz, N.A.; et al. A regulatory network controls nephrocan expression and midgut patterning. Development 2014, 141, 3772–3781. [Google Scholar] [CrossRef] [Green Version]
- Bayha, E.; Jørgensen, M.C.; Serup, P.; Grapin-Botton, A. Retinoic Acid Signaling Organizes Endodermal Organ Specification along the Entire Antero-Posterior Axis. PLoS ONE 2009, 4, e5845. [Google Scholar] [CrossRef] [Green Version]
- Rankin, S.A.; McCracken, K.W.; Luedeke, D.M.; Han, L.; Wells, J.M.; Shannon, J.M.; Zorn, A.M. Timing is everything: Reiterative Wnt, BMP and RA signaling regulate developmental competence during endoderm organogenesis. Dev. Biol. 2018, 434, 121–132. [Google Scholar] [CrossRef]
- Plateroti, M.; Freund, J.-N.; Leberquier, C.; Kedinger, M. Mesenchyme-mediated effects of retinoic acid during rat intestinal development. J. Cell Sci. 1997, 110, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Swartz-Basile, D.A.; Rubin, D.C.; Levin, M.S. Retinoic acid stimulates early cellular proliferation in the adapting remnant rat small intestine after partial resection. J. Nutr. 1997, 127, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukonin, I.; Serra, D.; Challet Meylan, L.; Volkmann, K.; Baaten, J.; Zhao, R.; Meeusen, S.; Colman, K.; Maurer, F.; Stadler, M.B.; et al. Phenotypic landscape of intestinal organoid regeneration. Nature 2020, 586, 275–280. [Google Scholar] [CrossRef]
- Ding, S.; Song, Y.; Brulois, K.F.; Pan, J.; Co, J.Y.; Ren, L.; Feng, N.; Yasukawa, L.L.; Sánchez-Tacuba, L.; Wosen, J.E.; et al. Retinoic Acid and Lymphotoxin Signaling Promote Differentiation of Human Intestinal M Cells. Gastroenterology 2020, 159, 214–226.e1. [Google Scholar] [CrossRef] [PubMed]
- Fordham, R.P.; Yui, S.; Hannan, N.R.; Soendergaard, C.; Madgwick, A.; Schweiger, P.J.; Nielsen, O.H.; Vallier, L.; Pedersen, R.A.; Nakamura, T.; et al. Transplantation of Expanded Fetal Intestinal Progenitors Contributes to Colon Regeneration after Injury. Cell Stem Cell 2013, 13, 734–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamminen, K.; Balboa, D.; Toivonen, S.; Pakarinen, M.P.; Wiener, Z.; Alitalo, K.; Otonkoski, T. Intestinal Commitment and Maturation of Human Pluripotent Stem Cells Is Independent of Exogenous FGF4 and R-spondin1. PLoS ONE 2015, 10, e0134551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crespo, M.; Vilar, E.; Tsai, S.-Y.; Chang, K.; Amin, S.; Srinivasan, T.; Zhang, T.; Pipalia, N.H.; Chen, H.J.; Witherspoon, M.; et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 2017, 23, 878–884. [Google Scholar] [CrossRef]
- Arceci, R.J.; King, A.A.; Simon, M.C.; Orkin, S.H.; Wilson, D.B. Mouse GATA-4: A retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol. Cell. Biol. 1993, 13, 2235–2246. [Google Scholar] [CrossRef]
- Lahar, N.; Lei, N.Y.; Wang, J.; Jabaji, Z.; Tung, S.C.; Joshi, V.; Lewis, M.; Stelzner, M.; Martín, M.G.; Dunn, J.C. Intestinal Subepithelial Myofibroblasts Support in vitro and in vivo Growth of Human Small Intestinal Epithelium. PLoS ONE 2011, 6, e26898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLin, V.A.; Henning, S.J.; Jamrich, M. The Role of the Visceral Mesoderm in the Development of the Gastrointestinal Tract. Gastroenterology 2009, 136, 2074–2091. [Google Scholar] [CrossRef]
- Gracz, A.D.; Fuller, M.K.; Wang, F.; Li, L.; Stelzner, M.; Dunn, J.C.; Martin, M.G.; Magness, S.T. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells 2013, 31, 2024–2030. [Google Scholar] [CrossRef] [Green Version]
- Shoshkes-Carmel, M.; Wang, Y.J.; Wangensteen, K.J.; Tóth, B.; Kondo, A.; Massasa, E.E.; Itzkovitz, S.; Kaestner, K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 2018, 557, 242–246. [Google Scholar] [CrossRef]
- McCarthy, N.; Manieri, E.; Storm, E.E.; Saadatpour, A.; Luoma, A.M.; Kapoor, V.N.; Madha, S.; Gaynor, L.T.; Cox, C.; Keerthivasan, S.; et al. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 2020, 26, 391–402.e5. [Google Scholar] [CrossRef]
- Fukuda, M.; Mizutani, T.; Mochizuki, W.; Matsumoto, T.; Nozaki, K.; Sakamaki, Y.; Ichinose, S.; Okada, Y.; Tanaka, T.; Watanabe, M.; et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes Dev. 2014, 28, 1752–1757. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Acuña, R.; Quirós, M.; Farkas, A.E.; Dedhia, P.H.; Huang, S.; Siuda, D.; García-Hernández, V.; Miller, A.J.; Spence, J.R.; Nusrat, A.; et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017, 19, 1326–1335. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.; Imran Alsous, J.; Guggenheim, J.W.; Mak, M.; Munera, J.; Wells, J.M.; Kamm, R.D.; Asada, H.H.; Shvartsman, S.Y.; Griffith, L.G. A process engineering approach to increase organoid yield. Development 2017, 144, 1128–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlnhofer, B.M.; Thompson, C.A.; Walker, E.M.; Battle, M.A. GATA4 Regulates Epithelial Cell Proliferation to Control Intestinal Growth and Development in Mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 189–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.-H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling Human Development and Disease in Pluripotent Stem-Cell-Derived Gastric Organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noto, F.K.; Determan, M.R.; Cai, J.; Cayo, M.A.; Mallanna, S.K.; Duncan, S.A. Aneuploidy is permissive for hepatocyte-like cell differentiation from human induced pluripotent stem cells. BMC Res. Notes 2014, 7, 437. [Google Scholar] [CrossRef] [Green Version]
- Múnera, J.O.; Wells, J.M. Generation of Gastrointestinal Organoids from Human Pluripotent Stem Cells. Methods Mol. Biol. 2017, 1597, 167–177. [Google Scholar] [CrossRef]
- Watson, C.L.; Mahe, M.M.; Munera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, N.; Jeffcoat, B.; Maity, P.; Christensen, R.K.; Múnera, J.O. Retinoic Acid Promotes the In Vitro Growth, Patterning and Improves the Cellular Composition of Human Pluripotent Stem-Cell-Derived Intestinal Organoids. Int. J. Mol. Sci. 2022, 23, 8624. https://doi.org/10.3390/ijms23158624
Qu N, Jeffcoat B, Maity P, Christensen RK, Múnera JO. Retinoic Acid Promotes the In Vitro Growth, Patterning and Improves the Cellular Composition of Human Pluripotent Stem-Cell-Derived Intestinal Organoids. International Journal of Molecular Sciences. 2022; 23(15):8624. https://doi.org/10.3390/ijms23158624
Chicago/Turabian StyleQu, Na, Braxton Jeffcoat, Pritiprasanna Maity, Rachael K. Christensen, and Jorge O. Múnera. 2022. "Retinoic Acid Promotes the In Vitro Growth, Patterning and Improves the Cellular Composition of Human Pluripotent Stem-Cell-Derived Intestinal Organoids" International Journal of Molecular Sciences 23, no. 15: 8624. https://doi.org/10.3390/ijms23158624
APA StyleQu, N., Jeffcoat, B., Maity, P., Christensen, R. K., & Múnera, J. O. (2022). Retinoic Acid Promotes the In Vitro Growth, Patterning and Improves the Cellular Composition of Human Pluripotent Stem-Cell-Derived Intestinal Organoids. International Journal of Molecular Sciences, 23(15), 8624. https://doi.org/10.3390/ijms23158624






