A Comparative Study on Nickel Binding to Hpn-like Polypeptides from Two Helicobacter pylori Strains
Abstract
:1. Introduction
2. Results and Discussion
- -
- for Ni(II)-TRIS interaction: 3.2 kcal/mol;
- -
- for Ni(II)-HEPES interaction: 0.54 kcal/mol;
- -
- for Ni(II)-MOPS interaction: 2.07 kcal/mol.
3. Materials and Methods
3.1. Isothermal Titration Calorimetry
3.2. Potentiometric Titration
3.3. Mass Spectrometry
3.4. UV-Vis and CD Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seshadri, S.; Benoit, S.L.; Maier, R.J. Roles of His-rich hpn and hpn-like proteins in Helicobacter pylori nickel physiology. J. Bacteriol. 2007, 189, 4120–4126. [Google Scholar] [CrossRef] [PubMed]
- Witkowska, D.; Rowińska-Żyrek, M.; Valensin, G.; Kozłowski, H. Specific poly-histydyl and poly-cysteil protein sites involved in Ni2+ homeostasis in Helicobacter pylori. Impact of Bi3+ ions on Ni2+ binding to proteins. Structural and thermodynamic aspects. Coord. Chem. Rev. 2012, 256, 133–148. [Google Scholar] [CrossRef]
- Kusters, J.G.; van Vliet, A.H.; Kuipers, E.J. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006, 19, 449–490. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, S.; Lewkowicz, A.; Kędziora-Kornatowska, K. Association between infection of Helicobacter pylori and iron deficiency anemia of unknown origin: A systematic review. Med. Sci. Pulse 2021, 15, 60–65. [Google Scholar] [CrossRef]
- Fazoli, K.G.Z.; dos Santos, I.C.; de Silva Caetano, I.C.; Cervantes, F.H.C.; Pacheco, F.C.; Branco, L.A.; de Padua Pereira, U.; Barbosa, L.N.; Goncalves, D.D. Antibiotic Resistance in Enterobacteriaceae Family Members Isolated from Horses Used for Animal Traction. J. Pure Appl. Microbiol. 2020, 14, 1149–1156. [Google Scholar] [CrossRef]
- Benoit, S.L.; Schmalstig, A.A.; Glushka, J.; Maier, S.E.; Edison, A.S.; Maier, R.J. Nickel chelation therapy as an approach to combat multi-drug resistant enteric pathogens. Sci. Rep. 2019, 9, 13851. [Google Scholar] [CrossRef] [PubMed]
- Boyanova, L.; Hadzhiyski, P.; Kandilarov, N.; Markovska, R.; Mitov, I. Multidrug resistance in Helicobacter pylori: Current state and future directions. Expert Rev. Clin. Pharmacol. 2019, 12, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.J.; Benoit, S.L.; Seshadri, S. Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals 2007, 20, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Zhang, D.M.; Li, H.; Sun, H. Binding of Ni2+ to a histidine- and glutamine-rich protein, Hpn-like. J. Biol. Inorg. Chem. 2008, 13, 1121–1131. [Google Scholar] [CrossRef]
- Rowińska-Żyrek, M.; Witkowska, D.; Bielińska, S.; Kamysz, W.; Kozłowski, H. The –Cys-Cys- motif in Helicobacter pylori’s Hpn i HspA proteins is the essential anchoring site for metal ions. Dalton Trans. 2011, 40, 5604–5610. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Yang, N.; Sun, H. Metal-Binding Properties of an Hpn-Like Histidine-Rich Protein. Chem. A Eur. J. 2011, 17, 5852–5860. [Google Scholar] [CrossRef]
- Fischer, B.E.; Haring, U.K.; Tribolet, R.; Sigel, H. Stability of binary and ternary complexes containing 2-amino-2(hydroxymethyl)-1,3-propanediol (Tris) and adenosine 5′-triphosphate (ATP). Eur. J. Biochem. 1979, 94, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Grossoehme, N.E.; Spuches, A.M.; Wilcox, D.E. Application of isothermal titration calorimetry in bioinorganic chemistry. J. Biol. Inorg. Chem. 2010, 15, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Makowska, J.; Wyrzykowski, D.; Pilarski, B.; Neubauer, D.; Kamysz, E.; Tesmar, A.; Chmurzyński, L. Copper(II) coordination properties of GxG peptides: Key role of side chains of central residues on coordination of formed systems; combined potentiometric and ITC studies. J. Chem. Thermodyn. 2019, 128, 336–343. [Google Scholar] [CrossRef]
- Witkowska, D.; Bielinska, S.; Kamysz, W.; Kozlowski, H. Cu2+ and Ni2+ interactions with N-terminal fragments of Hpn and Hpn-like proteins from Helicobacter pylori: Unusual impact of poly-Gln sequence on the complex stability. J. Inorg. Biochem. 2011, 105, 208–214. [Google Scholar] [CrossRef]
- Bal, W.; Sokołowska, M.; Kurowska, E.; Faller, P. Binding of transition metal ions to albumin: Sites, affinities and rates. Biochim. Biophys. Acta 2013, 1830, 5444–5455. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.Q.; Huang, Q.; Zhang, Y.; Zhang, H.Q.; Lai, L. Binding thermodynamics of divalent metal ions to several biological buffers. Thermochim. Acta 2020, 691, 178721. [Google Scholar] [CrossRef]
- Ferreira, C.M.H.; Pinto, I.S.S.; Soares, E.V.; Soares, H.M.V.M. (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions—A review. RSC Adv. 2015, 5, 30989–31003. [Google Scholar] [CrossRef]
- Johnson, D.K.; Stevenson, M.J.; Almadidy, Z.A.; Jenkins, S.E.; Wilcox, D.E.; Grossoehme, N.E. Stabilization of Cu(I) for binding and calorimetric measurements in aqueous solution. Dalton Trans. 2015, 44, 16494–16505. [Google Scholar] [CrossRef]
- Migliorini, C.; Witkowska, D.; Valensin, D.; Kamysz, W.; Kozlowski, H. Competition between histamine-like and poly-imidazole coordination sites for Cu2+ and Zn2+ ions in zebra-fish peptide of prion-like protein. Dalton Trans. 2010, 39, 8663–8670. [Google Scholar] [CrossRef] [PubMed]
- Wątły, J.; Hecel, A.; Rowińska-Żyrek, M.; Kozłowski, H. Impact of histidine spacing on modified polyhistidine tag—Metal ion interactions. Inorg. Chim. Acta 2018, 472, 119–126. [Google Scholar] [CrossRef]
- Gonzalez, P.; Bossak, K.; Stefaniak, E.; Hureau, C.; Raibaut, L.; Bal, W.; Faller, P. N-Terminal Cu-Binding Motifs (Xxx-Zzz-His, Xxx-His) and Their Derivatives: Chemistry, Biology and Medicinal Applications. Chemistry 2018, 24, 8029–8041. [Google Scholar] [CrossRef]
- Johnson, R.A.; Manley, O.M.; Spuches, A.M.; Grossoehme, N.E. Dissecting ITC data of metal ions binding to ligands and proteins. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Chiera, N.M.; Rowinska-Zyrek, M.; Wieczorek, R.; Guerrini, R.; Witkowska, D.; Remelii, M.; Kozłowski, H. Unexpected impact of the number of glutamine residues on metal complex stability. Metallomics 2013, 5, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.; Matera-Witkiewicz, A.; Mikołajczyk, A.; Wątły, J.; Wilcox, D.; Witkowska, D.; Rowińska-Żyrek, M. Zn-Enhanced Asp-Rich Antimicrobial Peptides: N-Terminal Coordination by Zn(II) and Cu(II), Which Distinguishes Cu(II) Binding to Different Peptides. Int. J. Mol. Sci. 2021, 22, 6971. [Google Scholar] [CrossRef] [PubMed]
- Wyrzykowski, D.; Pilarski, B.; Jacewicz, D.; Chmurzyński, L. Investigation of metal–buffer interactions using isothermal titration calorimetry. J. Therm. Anal. Calorim. 2013, 111, 1829–1836. [Google Scholar] [CrossRef]
- Bianconi, M.L. Avoiding Buffer Interference in ITC Experiments: A Case Study from the Analysis of Entropy-Driven Reactions of Glucose-6-Phosphate Dehydrogenase. Methods Enzymol. 2016, 567, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Wezynfeld, N.E.; Bossak, K.; Goch, W.; Bonna, A.; Bal, W.; Frączyk, T. Human Annexins A1, A2, and A8 as Potential Molecular Targets for Ni(II) Ions. Chem. Res. Toxicol. 2014, 27, 1996–2009. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Prot. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Olson, J.W.; Maier, R.J. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc. Natl. Acad. Sci. USA 1995, 92, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Rey, L.; Imperial, J.; Palacios, J.M.; Ruiz-ArguÈeso, T. Purification of Rhizobium leguminosarum HypB, a nickel-binding protein required for hydrogenase synthesis. J. Bacteriol. 1994, 176, 6066–6073. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Pankratz, H.S.; Wang, S.; Scott, R.A.; Finnegan, M.G.; Johnson, M.K.; Ippolito, J.A.; Christianson, D.W.; Hausinger, R.P. Purification and characterization of Klebsiella aerogenes UreE protein: A nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993, 2, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Brayman, T.G.; Hausinger, R.P. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J. Bacteriol. 1996, 178, 5410–5416. [Google Scholar] [CrossRef] [PubMed]
- Watt, R.K.; Ludden, P.W. The identification, purification, and characterization of CooJ. A nickel-binding protein that is co-regulated with the Ni-containing CO dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 1998, 273, 10019–10025. [Google Scholar] [CrossRef] [PubMed]
- Hottenrott, S.; Schumann, T.; PluÈckthun, A.; Fischer, G.; Rahfeld, J.U. The Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 1997, 272, 15697–15701. [Google Scholar] [CrossRef] [PubMed]
- Kaluarachchi, H.; Sutherland, D.E.; Young, A.; Pickering, I.J.; Stillman, M.J.; Zamble, D.B. The Ni(II)-binding properties of the metallochaperone SlyD. J. Am. Chem. Soc. 2009, 131, 18489–18500. [Google Scholar] [CrossRef]
- Kansau, I.; Guillain, F.; Thiberge, J.M.; Labigne, A. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA). Mol. Microbiol. 1996, 22, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Cun, S.; Li, H.; Ge, R.; Lin, M.C.M.; Sun, H. A Histidine-rich and Cysteine-rich Metal-binding Domain at the C Terminus of Heat Shock Protein A from Helicobacter pylori: Implication for nickel homeostasis and bismuth susceptibility. J. Biol. Chem. 2008, 283, 15142–15151. [Google Scholar] [CrossRef]
- Loguercio, S.; Dian, C.; Flagiello, A.; Scannella, A.; Pucci, P.; Terradot, L.; Zagari, A. In HspA from Helicobacter pylori vicinal disulfide bridges are a key determinant of domain B structure. FEBS Lett. 2008, 582, 3537–3541. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.; Watt, R.M.; Sun, X.; Tanner, J.A.; He, Q.Y.; Huang, J.D.; Sun, H. Expression and characterization of a histidine- rich protein, Hpn: Potential for Ni2+ storage in Helicobacter pylori. Biochem. J. 2006, 393, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Shelake, R.M.; Ito, Y.; Masumoto, J.; Morita, E.H.; Hayashi, H. A novel mechanism of “metal gel-shift” by histidine-rich Ni2+-binding Hpn protein from Helicobacter pylori strain SS1. PLoS ONE 2017, 12, e0172182. [Google Scholar] [CrossRef] [PubMed]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 10, 1739–1753. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility programfor the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]


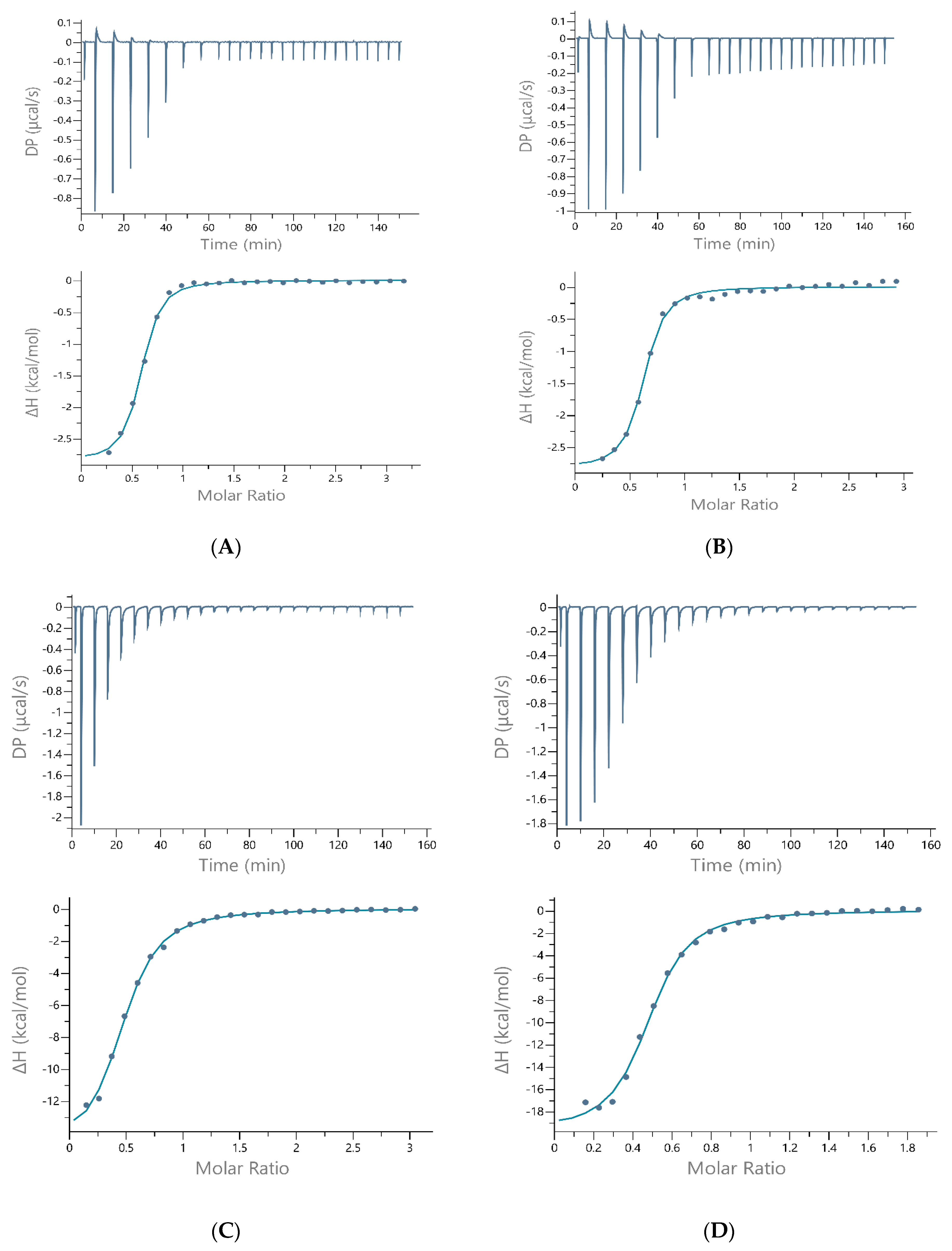

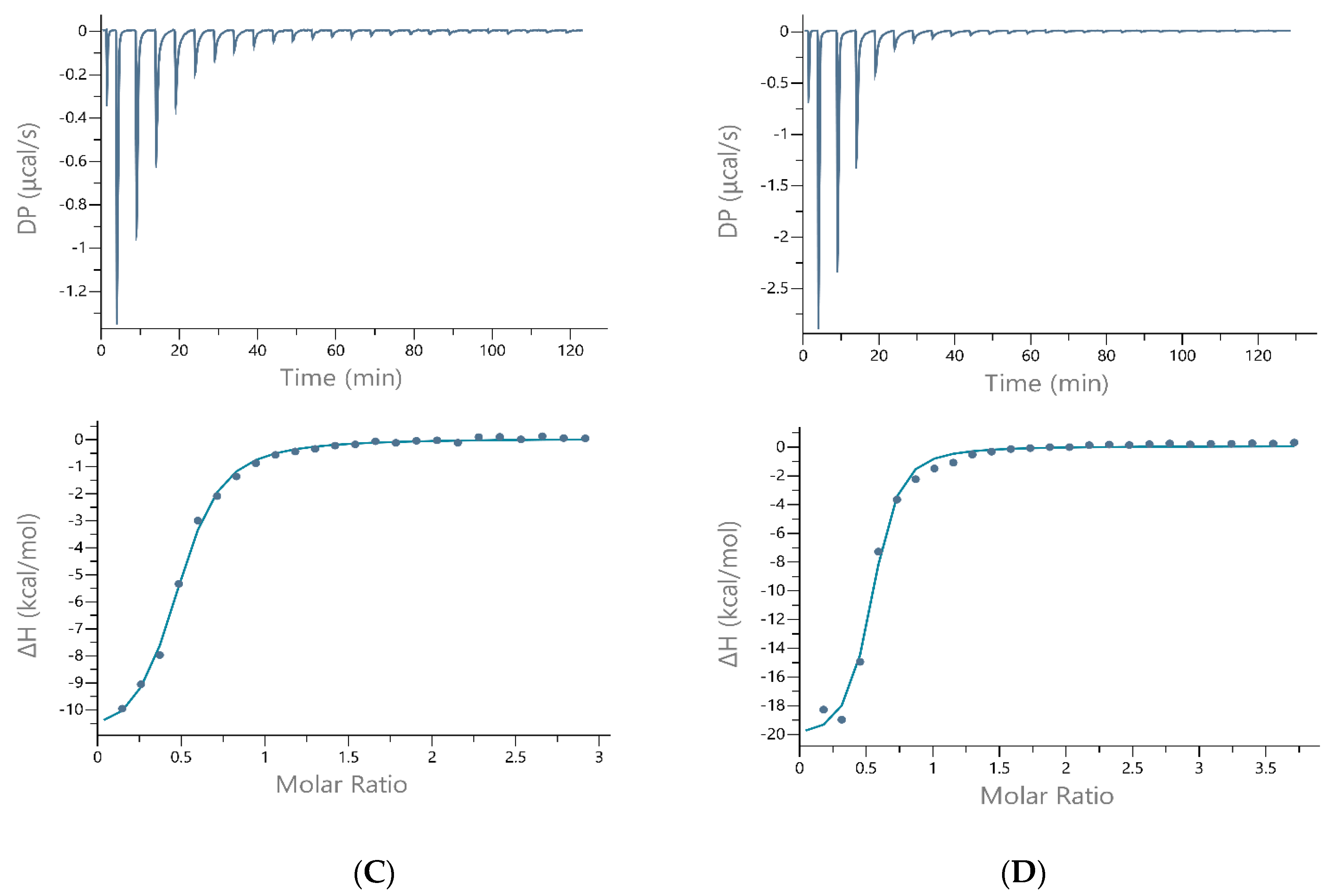


| Ligand | Data (Units) | TRIS | HEPES | MOPS |
|---|---|---|---|---|
| Hpnl1 | NITC | 0.51 ± 0.01 | 0.43 ± 0.01 | 0.54 ± 0.01 |
| KdITC (μM) | 3.24 ± 0.37 | 3.16 ± 0.66 | 2.01 ± 0.24 | |
| ΔHITC (kcal/mol) | −8.51 ± 0.18 | −3.31 ± 0.12 | −2.86 ± 0.06 | |
| −TΔSITC (kcal/mol) | 0.70 | −4.50 | −5.23 | |
| Hpnl2 | NITC | 0.46 ± 0.003 | 0.43 ± 0.01 | 0.59 ± 0.01 |
| KdITC (μM) | 2.44 ± 0.17 | 2.45 ± 0.37 | 2.12 ± 0.33 | |
| ΔHITC (kcal/mol) | −13.2 ± 0.14 | −3.1 ± 0.08 | −2.83 ± 0.07 | |
| −TΔSITC (kcal/mol) | 5.28 | −4.86 | −5.22 | |
| Hpnl3 | NITC | 0.51 ± 0.01 | 0.45 ± 0.01 | 0.46 ± 0.01 |
| KdITC (μM) | 4.64 ± 0.68 | 7.8 ± 0.54 | 8.3 ± 0.85 | |
| ΔHITC (kcal/mol) | −29.4 ± 0.94 | −19.4 ± 0.4 | −15.2 ± 0.44 | |
| −TΔSITC (kcal/mol) | 21.80 | 12.10 | 8.00 | |
| Hpnl3a | NITC | 0.43 ± 0.01 | 0.55 ± 0.01 | 0.47 ± 0.01 |
| KdITC (μM) | 1.03 ± 0.2 | 4.9 ± 0.06 | 2.53 ± 0.34 | |
| ΔHITC (kcal/mol) | −32.4 ± 0.75 | −23.9 ± 0.77 | −19.8 ± 0.53 | |
| −TΔSITC (kcal/mol) | 23.90 | 16.40 | 11.90 |
| Ligand | Data (Units) | TRIS | HEPES | MOPS |
|---|---|---|---|---|
| Hpnl1 | NITC | 0.45 ± 0.01 | 0.45 ± 0.02 | 0.54 ± 0.01 |
| KdITC (μM) | 11.4 ± 2.33 | 3.79 ± 0.57 | 5.09 ± 0.66 | |
| ΔHITC (kcal/mol) | −12.1 ± 0.7 | −3.96 ± 0.13 | −4.26 ± 0.11 | |
| −TΔSITC (kcal/mol) | 5.39 | −3.44 | −2.97 | |
| Hpnl2 | NITC | 0.48 ± 0.01 | 0.59 ± 0.03 | 0.51 ± 0.01 |
| KdITC (μM) | 7.98 ± 1.22 | 5.74 ± 1.06 | 4.12 ± 0.52 | |
| ΔHITC (kcal/mol) | −11.70 ± 0.46 | −1.86 ± 0.17 | −4.44 ± 0.12 | |
| −TΔSITC (kcal/mol) | 4.74 | −5.29 | −2.92 | |
| Hpnl3 | NITC | 0.94 ± 0.01 | 0.41 ± 0.01 | 0.45 ± 0.01 |
| KdITC (μM) | 9.47 ± 1.22 | 6.41 ± 0.55 | 5.27 ± 0.55 | |
| ΔHITC (kcal/mol) | −27.5 ± 0.98 | −15.6 ± 0.38 | −11.4 ± 0.27 | |
| −TΔSITC (kcal/mol) | 20.70 | 8.53 | 4.22 | |
| Hpnl3a | NITC | 0.93 ± 0.01 | 0.44 ± 0.01 | 0.49 ± 0.01 |
| KdITC (μM) | 1.06 ± 0.25 | 4.03 ± 0.86 | 2.11 ± 0.43 | |
| ΔHITC (kcal/mol) | −31.3 ± 0.99 | −22.8 ± 1.01 | −20.7 ± 0.65 | |
| −TΔSITC (kcal/mol) | 23.10 | 15.40 | 12.90 |
| Species | MAHHEQQQQQQA-NH2 (Hpnl1) | MAHHEQQHQA-NH2 (Hpnl2) | ||
|---|---|---|---|---|
| logβ | logK | logβ | logK | |
| LH | 7.53 (1) | 7.53 | 7.58 (1) | 7.58 |
| LH2 | 14.07 (1) | 6.54 | 14.35 (1) | 6.77 |
| LH3 | 20.04 (1) | 5.97 | 20.63 (1) | 6.28 |
| LH4 | 24.11 (2) | 4.07 | 26.36 (1) | 5.73 |
| LH5 | - | - | 30.31 (1) | 3.95 |
| Protein | Bacterium Name | No. of Histidine (H) Residues Per Histidine-Rich Domain | No. of Glutamine (Q) Per Protein Monomer | No.of Ni(II) Ions Per Monomer | Kd (µM) | Formula MW (kDa) | Apparent MW (kDa) | References |
|---|---|---|---|---|---|---|---|---|
| HypB | Bradyrhizobium japonicum | 24/39 | 5 | 9 | 2.3 | 32.5 | 38, 78(dimer) | [30] |
| Hyp B | Rhizobium leguminosarum | 17/32 | 6 | 4 | 2.5 | 32 | 39 | [1,31] |
| UreE | Klebsiella aerogenes | 10/15 | 5 | 3 | 9.6 | 17.5 | 35 (dimer) | [32,33] |
| CooJ | Rhodospirillum rubrum | 16/34 | 1 | 4 | 4.3 | 19 | 39 (dimer) | [34] |
| SlyD | Escherichia coli | 15/50 | 6 | 3–7 | 2 | 20.8 | 25 | [35,36] |
| HspA | Helicobacter pylori | 8/27 | 1 | 2 | 1.8 | 13 | 13 | [37,38,39] |
| Hpn | Helicobacter pylori | 28/60 | 2 | 5–6 | 7.1 | 7 | 7, 14, 20, 70, 136, 230… | [40,41] |
| Hpn-like (Hpnl) strain 11637 | Helicobacter pylori | 14/23 | 31 | 2 | 3.8 | 9 | 18 (dimer), 28, 36, 47, 201 | [1,11] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witkowska, D.; Szebesczyk, A.; Wątły, J.; Braczkowski, M.; Rowińska-Żyrek, M. A Comparative Study on Nickel Binding to Hpn-like Polypeptides from Two Helicobacter pylori Strains. Int. J. Mol. Sci. 2021, 22, 13210. https://doi.org/10.3390/ijms222413210
Witkowska D, Szebesczyk A, Wątły J, Braczkowski M, Rowińska-Żyrek M. A Comparative Study on Nickel Binding to Hpn-like Polypeptides from Two Helicobacter pylori Strains. International Journal of Molecular Sciences. 2021; 22(24):13210. https://doi.org/10.3390/ijms222413210
Chicago/Turabian StyleWitkowska, Danuta, Agnieszka Szebesczyk, Joanna Wątły, Michał Braczkowski, and Magdalena Rowińska-Żyrek. 2021. "A Comparative Study on Nickel Binding to Hpn-like Polypeptides from Two Helicobacter pylori Strains" International Journal of Molecular Sciences 22, no. 24: 13210. https://doi.org/10.3390/ijms222413210
APA StyleWitkowska, D., Szebesczyk, A., Wątły, J., Braczkowski, M., & Rowińska-Żyrek, M. (2021). A Comparative Study on Nickel Binding to Hpn-like Polypeptides from Two Helicobacter pylori Strains. International Journal of Molecular Sciences, 22(24), 13210. https://doi.org/10.3390/ijms222413210






