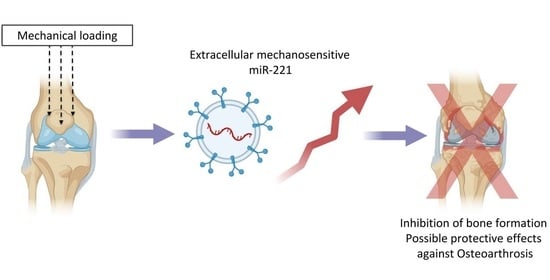
Extracellular Vesicles Allow Epigenetic Mechanotransduction between Chondrocytes and Osteoblasts
Abstract
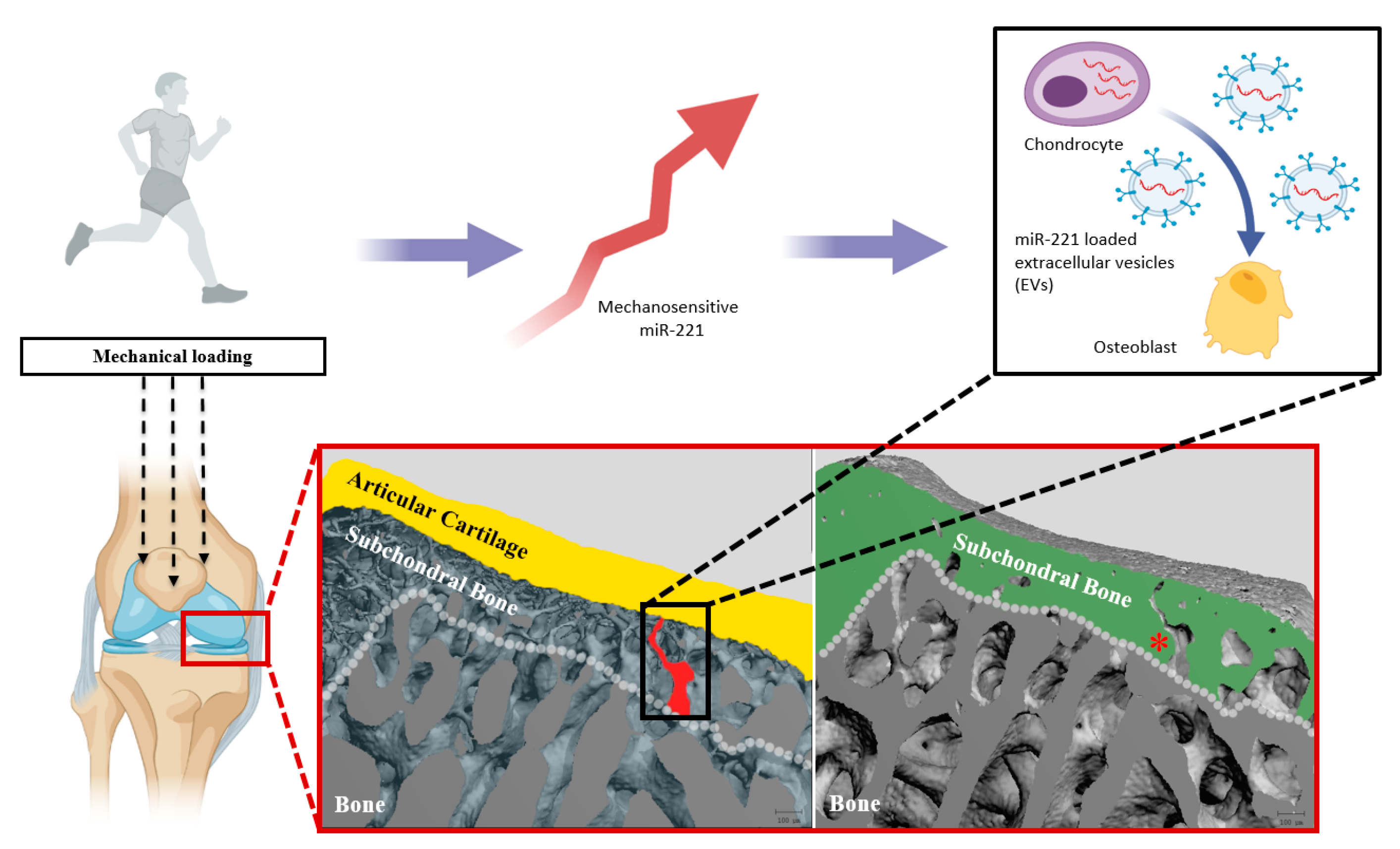
:1. Introduction
2. Results
2.1. Identification of Chondrocyte Secreted EVs
2.2. MiR-221-3p Expression in OA
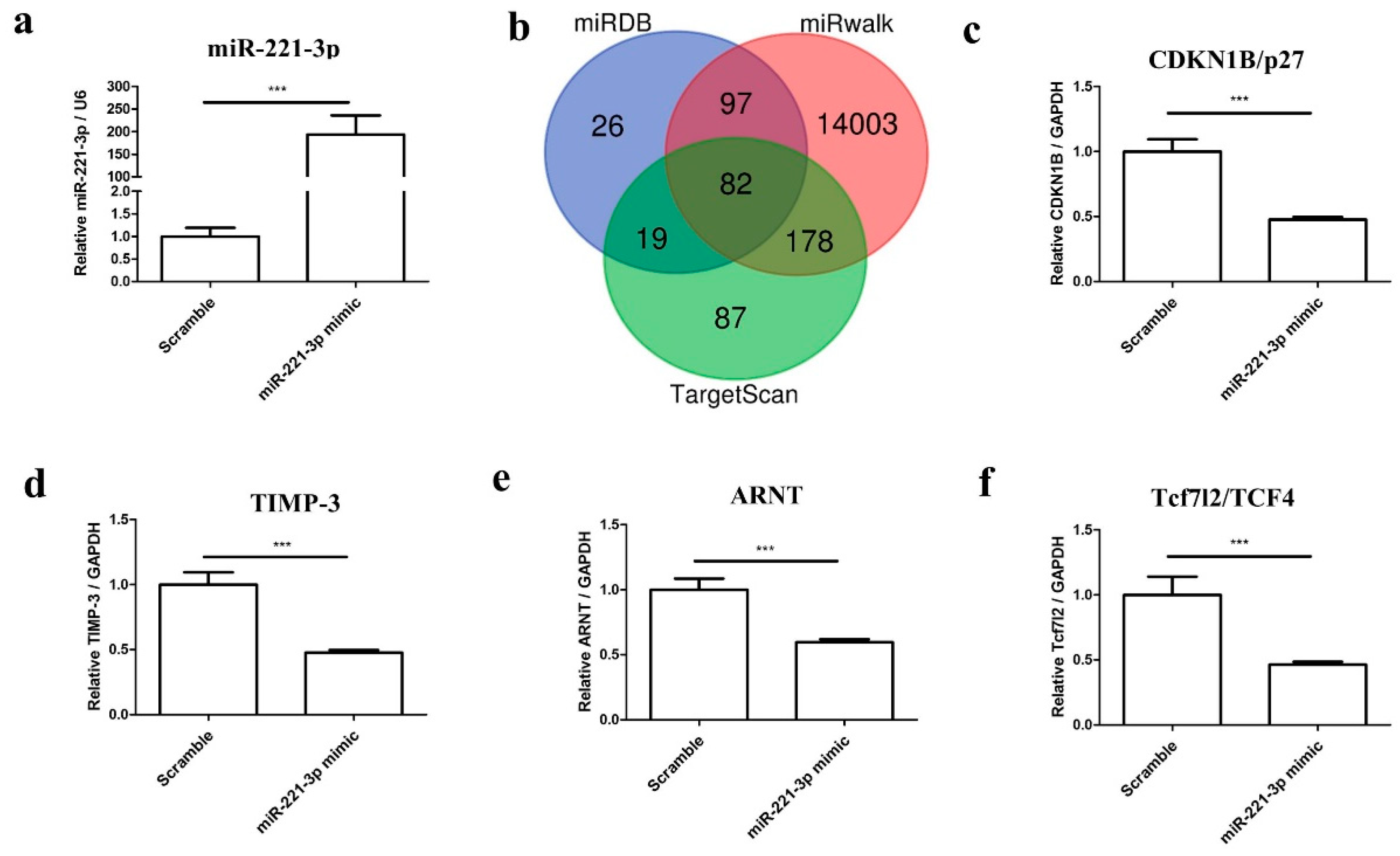
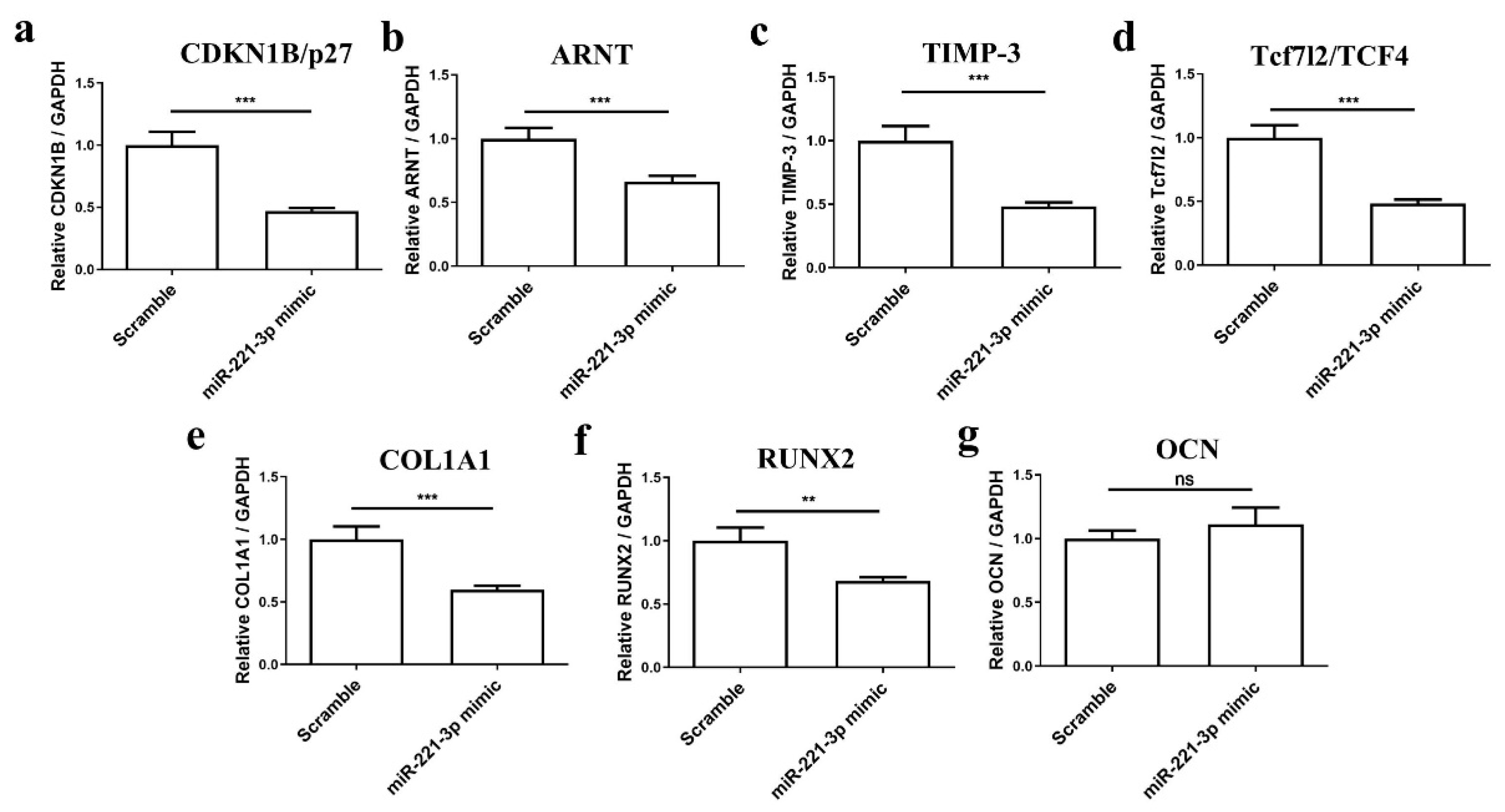
2.3. Mir-221-3p and Target Gene Analysis in Chondrocytes and Osteoblasts
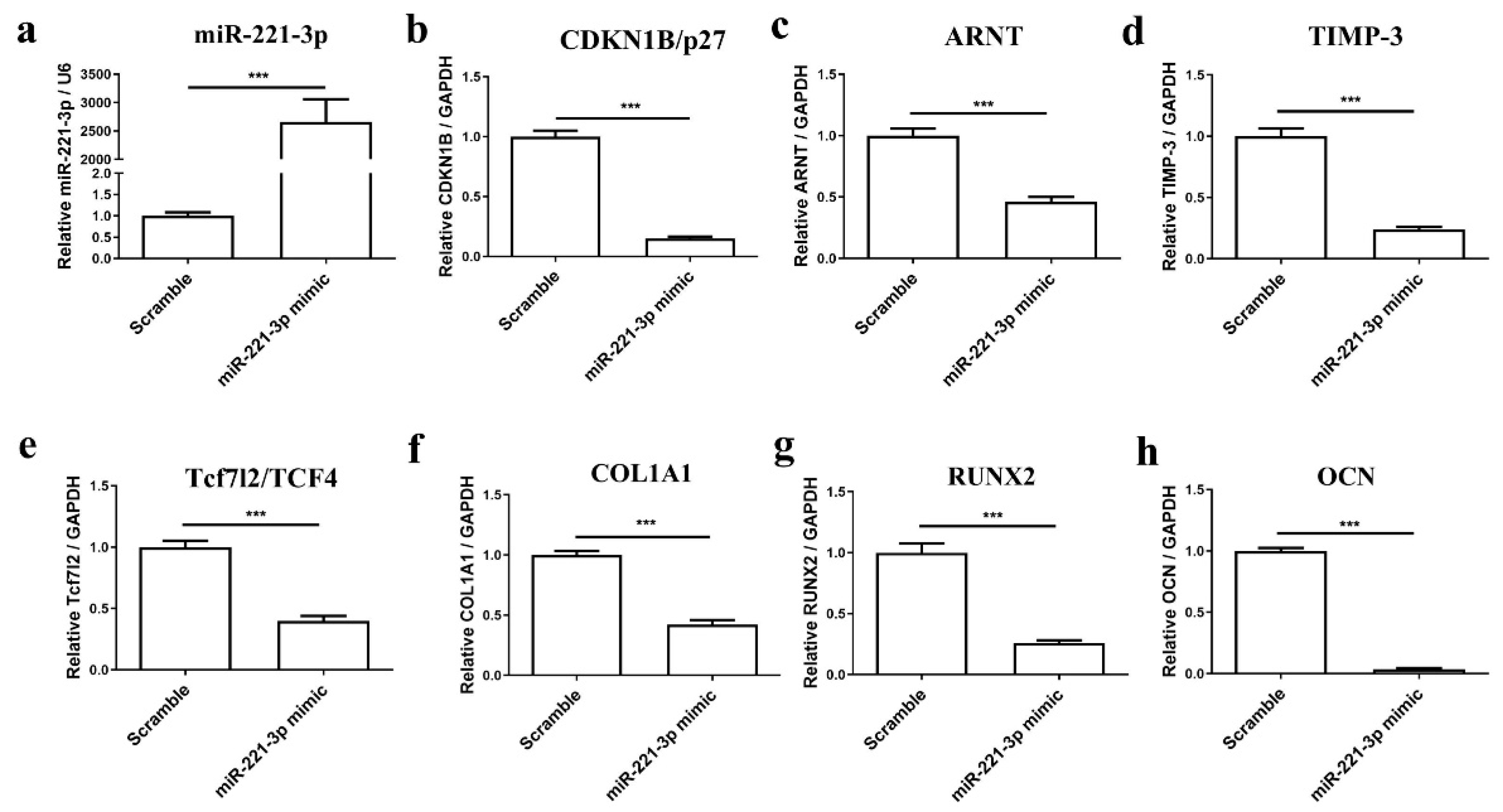
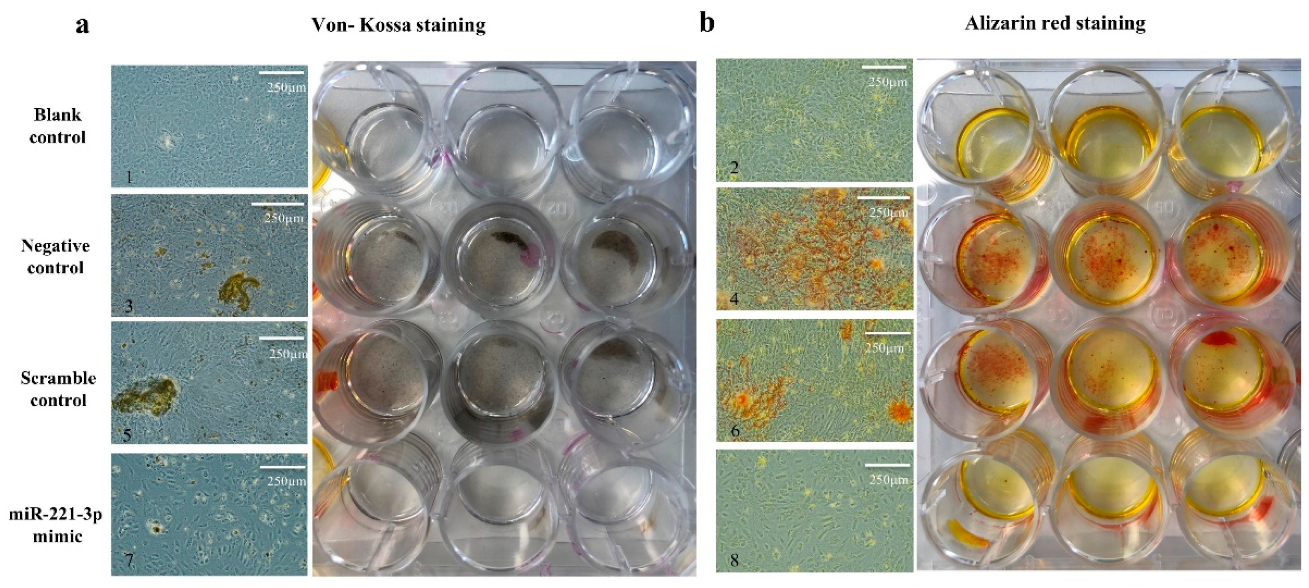
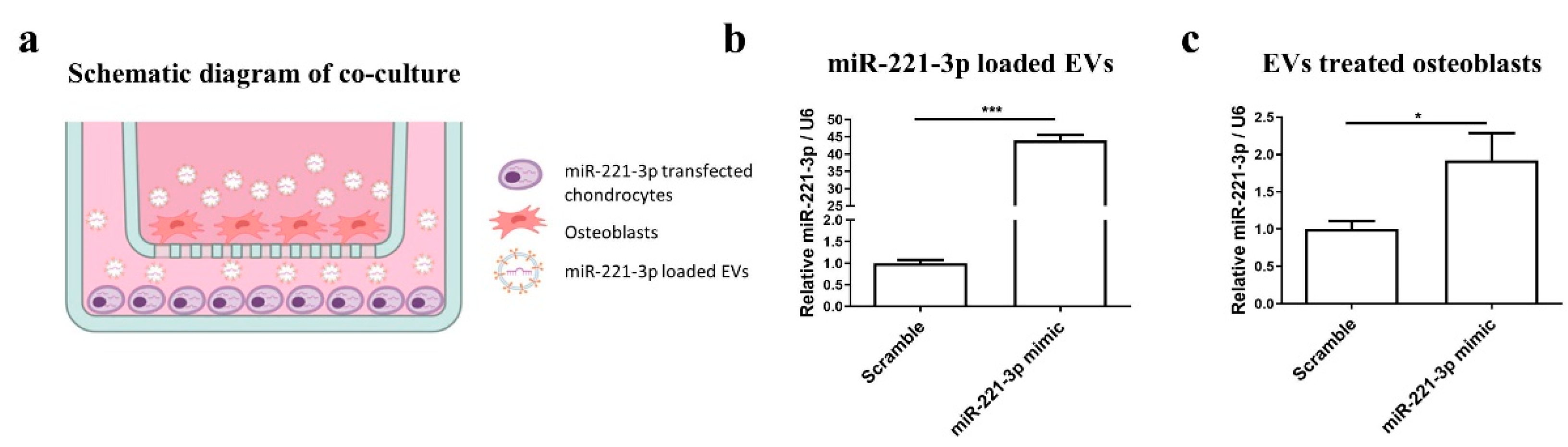
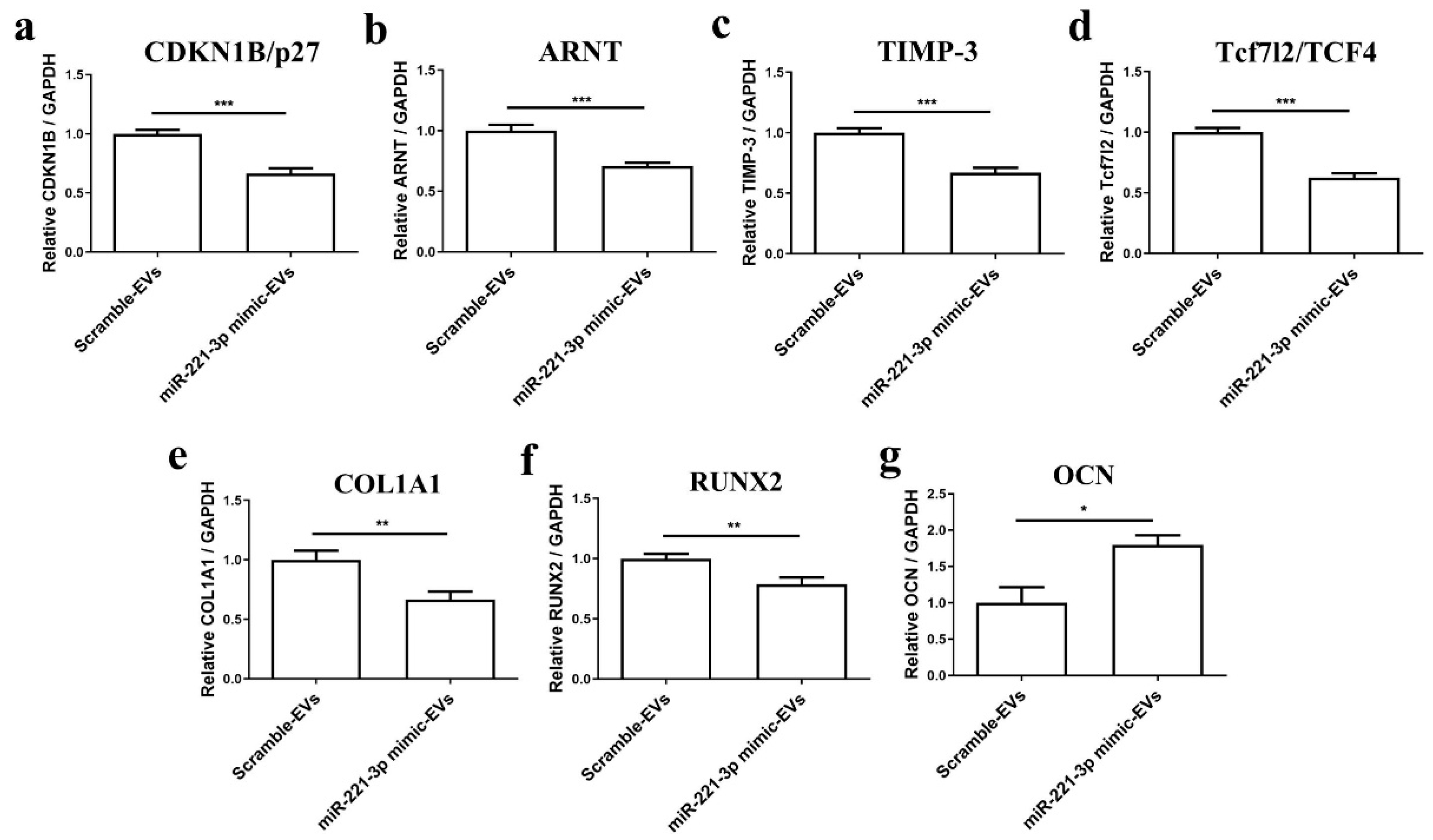
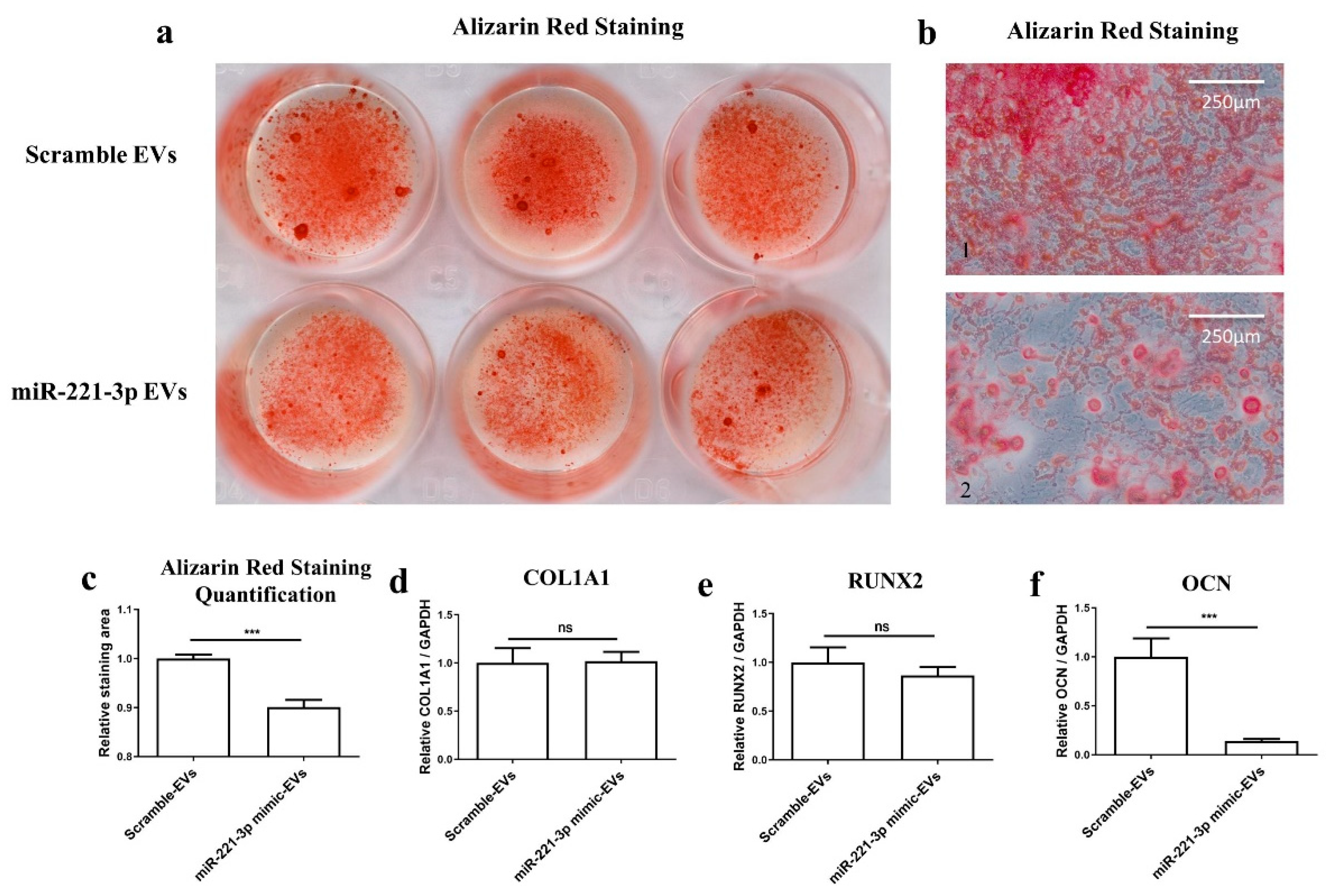
2.4. Effect of Mir-221-3p Loaded EVs on Osteoblasts
3. Discussion
4. Materials and Methods
4.1. Cell Isolation and Identification
4.2. Establishment of OA Model In Vitro
4.3. Transfection
4.4. Coculture of Chondrocytes and Osteoblasts
4.5. EVs Isolation and Identification
4.6. Target-Gene Prediction and Data Analysis
4.7. Western Blot Analysis
4.8. RNA Isolation and Real-Time qRT-PCR
4.9. Software and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hügle, T. Update Osteoarthritis. Rev. Med. Suisse 2020, 16, 500–502. [Google Scholar] [PubMed]
- Hunt, M.A.; Charlton, J.M.; Esculier, J.-F. Osteoarthritis Year in Review 2019: Mechanics. Osteoarthr. Cartil. 2020, 28, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A Disease of the Joint as an Organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [Green Version]
- Kloppenburg, M.; Berenbaum, F. Osteoarthritis Year in Review 2019: Epidemiology and Therapy. Osteoarthr. Cartil. 2020, 28, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Kolasinski, S.L.; Neogi, T.; Hochberg, M.C.; Oatis, C.; Guyatt, G.; Block, J.; Callahan, L.; Copenhaver, C.; Dodge, C.; Felson, D.; et al. 2019 American College of Rheumatology/Arthritis Foundation Guideline for the Management of Osteoarthritis of the Hand, Hip, and Knee. Arthritis Care Res. 2020, 72, 149–162. [Google Scholar] [CrossRef]
- Cui, A.; Li, H.; Wang, D.; Zhong, J.; Chen, Y.; Lu, H. Global, Regional Prevalence, Incidence and Risk Factors of Knee Osteoarthritis in Population-Based Studies. EClinicalMedicine 2020, 29, 1–13. [Google Scholar] [CrossRef]
- Zhang, W.; Ouyang, H.; Dass, C.R.; Xu, J. Current Research on Pharmacologic and Regenerative Therapies for Osteoarthritis. Bone Res. 2016, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shortt, C.; Zhang, F.; Bater, M.Q.; Cowman, M.K.; Kirsch, T. Extracellular Vesicles Released From Articular Chondrocytes Play a Major Role in Cell-Cell Communication. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2020, 38, 731–739. [Google Scholar] [CrossRef]
- Reynard, L.N.; Barter, M.J. Osteoarthritis Year in Review 2019: Genetics, Genomics and Epigenetics. Osteoarthr. Cartil. 2020, 28, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray Analysis Shows That Some MicroRNAs Downregulate Large Numbers of Target MRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Yu, X.-M.; Meng, H.-Y.; Yuan, X.-L.; Wang, Y.; Guo, Q.-Y.; Peng, J.; Wang, A.-Y.; Lu, S.-B. MicroRNAs’ Involvement in Osteoarthritis and the Prospects for Treatments. Evid. Based Complement. Alternat. Med. 2015, 2015, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, X.; Böker, K.O.; Taheri, S.; Hawellek, T.; Lehmann, W.; Schilling, A.F. The Interaction between MicroRNAs and the Wnt/β-Catenin Signaling Pathway in Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 9887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhao, F.-C.; Pang, Y.; Li, D.-Y.; Yao, S.-C.; Sun, S.-S.; Guo, K.-J. Downregulation of MiR-221-3p Contributes to IL-1β-Induced Cartilage Degradation by Directly Targeting the SDF1/CXCR4 Signaling Pathway. J. Mol. Med. Berl. Ger. 2017, 95, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, L.; Gibson, G.J. Chondrocyte MiRNAs 221 and 483-5p Respond to Loss of Matrix Interaction by Modulating Proliferation and Matrix Synthesis. Connect. Tissue Res. 2015, 56, 236–243. [Google Scholar] [CrossRef]
- Lolli, A.; Narcisi, R.; Lambertini, E.; Penolazzi, L.; Angelozzi, M.; Kops, N.; Gasparini, S.; van Osch, G.J.V.M.; Piva, R. Silencing of Antichondrogenic MicroRNA-221 in Human Mesenchymal Stem Cells Promotes Cartilage Repair In Vivo. Stem Cells Dayt. Ohio 2016, 34, 1801–1811. [Google Scholar] [CrossRef] [Green Version]
- Hecht, N.; Johnstone, B.; Angele, P.; Walker, T.; Richter, W. Mechanosensitive MiRs Regulated by Anabolic and Catabolic Loading of Human Cartilage. Osteoarthr. Cartil. 2019, 27, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Grad, S.; Lee, C.R.; Gorna, K.; Gogolewski, S.; Wimmer, M.A.; Alini, M. Surface Motion Upregulates Superficial Zone Protein and Hyaluronan Production in Chondrocyte-Seeded Three-Dimensional Scaffolds. Tissue Eng. 2005, 11, 249–256. [Google Scholar] [CrossRef]
- Musumeci, G. The Effect of Mechanical Loading on Articular Cartilage. J. Funct. Morphol. Kinesiol. 2016, 1, 154. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.-S.; Chen, W.-C.; Huang, C.-H.; Cheng, C.-K.; Chan, K.-K.; Chang, T.-K. The Effect of Graft Strength on Knee Laxity and Graft In-Situ Forces after Posterior Cruciate Ligament Reconstruction. PLoS ONE 2015, 10, e0168280. [Google Scholar] [CrossRef] [Green Version]
- Findlay, D.M.; Kuliwaba, J.S. Bone-Cartilage Crosstalk: A Conversation for Understanding Osteoarthritis. Bone Res. 2016, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.L.; Meng, H.Y.; Wang, Y.C.; Peng, J.; Guo, Q.Y.; Wang, A.Y.; Lu, S.B. Bone-Cartilage Interface Crosstalk in Osteoarthritis: Potential Pathways and Future Therapeutic Strategies. Osteoarthr. Cartil. 2014, 22, 1077–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taheri, S.; Winkler, T.; Schenk, L.S.; Neuerburg, C.; Baumbach, S.F.; Zustin, J.; Lehmann, W.; Schilling, A.F. Developmental Transformation and Reduction of Connective Cavities within the Subchondral Bone. Int. J. Mol. Sci. 2019, 20, 770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taheri, S.; Yoshida, T.; Böker, K.O.; Foerster, R.H.; Jochim, L.; Flux, A.L.; Grosskopf, B.; Lehmann, W.; Schilling, A.F. Investigating the Microchannel Architectures Inside the Subchondral Bone in Relation to Estimated Hip Reaction Forces on the Human Femoral Head. Calcif. Tissue Int. 2021, 1, 1–15. [Google Scholar] [CrossRef]
- Pan, J.; Zhou, X.; Li, W.; Novotny, J.E.; Doty, S.B.; Wang, L. In Situ Measurement of Transport between Subchondral Bone and Articular Cartilage. J. Orthop. Res. 2009, 27, 1347–1352. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yu, D.; Liu, Z.; Zhou, F.; Dai, J.; Wu, B.; Zhou, J.; Heng, B.C.; Zou, X.H.; Ouyang, H.; et al. Exosomes from Embryonic Mesenchymal Stem Cells Alleviate Osteoarthritis through Balancing Synthesis and Degradation of Cartilage Extracellular Matrix. Stem. Cell Res. Ther. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolhe, R.; Hunter, M.; Liu, S.; Jadeja, R.N.; Pundkar, C.; Mondal, A.K.; Mendhe, B.; Drewry, M.; Rojiani, M.V.; Liu, Y.; et al. Gender-Specific Differential Expression of Exosomal MiRNA in Synovial Fluid of Patients with Osteoarthritis. Sci. Rep. 2017, 7, 2029. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Hu, S.; Zhang, Z.; Wu, P.; Zhao, X.; Lin, R.; Liao, W.; Kang, Y. Exosomal MiR-95-5p Regulates Chondrogenesis and Cartilage Degradation via Histone Deacetylase 2/8. J. Cell. Mol. Med. 2018, 22, 5354–5366. [Google Scholar] [CrossRef]
- Wang, R.; Jiang, W.; Zhang, L.; Xie, S.; Zhang, S.; Yuan, S.; Jin, Y.; Zhou, G. Intra-Articular Delivery of Extracellular Vesicles Secreted by Chondrogenic Progenitor Cells from MRL/MpJ Superhealer Mice Enhances Articular Cartilage Repair in a Mouse Injury Model. Stem Cell Res. Ther. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Dunn, W.; DuRaine, G.; Reddi, A.H. Profiling MicroRNA Expression in Bovine Articular Cartilage and Implications for Mechanotransduction. Arthritis Rheum. 2009, 60, 2333–2339. [Google Scholar] [CrossRef]
- Stadnik, P.S.; Gilbert, S.J.; Tarn, J.; Charlton, S.; Skelton, A.J.; Barter, M.J.; Duance, V.C.; Young, D.A.; Blain, E.J. Regulation of MicroRNA-221, -222, -21 and -27 in Articular Cartilage Subjected to Abnormal Compressive Forces. J. Physiol. 2021, 599, 143–155. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Gee, M.M.M. A Comparison of Methods for the Isolation and Separation of Extracellular Vesicles from Protein and Lipid Particles in Human Serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakase, I.; Noguchi, K.; Fujii, I.; Futaki, S. Vectorization of Biomacromolecules into Cells Using Extracellular Vesicles with Enhanced Internalization Induced by Macropinocytosis. Sci. Rep. 2016, 6, 34937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eguchi, T.; Sogawa, C.; Ono, K.; Matsumoto, M.; Tran, M.T.; Okusha, Y.; Lang, B.J.; Okamoto, K.; Calderwood, S.K. Cell Stress Induced Stressome Release Including Damaged Membrane Vesicles and Extracellular HSP90 by Prostate Cancer Cells. Cells 2020, 9, 755. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.-G.; Kong, M.-Q.; Zhou, S.; Sheng, Y.-F.; Wang, P.; Yu, T.; Inci, F.; Kuo, W.P.; Li, L.-J.; Demirci, U.; et al. An Integrated Double-Filtration Microfluidic Device for Isolation, Enrichment and Quantification of Urinary Extracellular Vesicles for Detection of Bladder Cancer. Sci. Rep. 2017, 7, 46224. [Google Scholar] [CrossRef] [Green Version]
- Pérez-González, R.; Gauthier, S.A.; Sharma, A.; Miller, C.; Pawlik, M.; Kaur, G.; Kim, Y.; Levy, E. A Pleiotropic Role for Exosomes Loaded with the Amyloid β Precursor Protein Carboxyl-Terminal Fragments in the Brain of Down Syndrome Patients. Neurobiol. Aging 2019, 84, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1–47. [Google Scholar] [CrossRef] [Green Version]
- Mohd Noor, N.A.; Abdullah Nurul, A.; Ahmad Mohd Zain, M.R.; Wan Nor Aduni, W.K.; Azlan, M. Extracellular Vesicles from Mesenchymal Stem Cells as Potential Treatments for Osteoarthritis. Cells 2021, 10, 1287. [Google Scholar] [CrossRef]
- Tan, S.S.H.; Tjio, C.K.E.; Wong, J.R.Y.; Wong, K.L.; Chew, J.R.J.; Hui, J.H.P.; Toh, W.S. Mesenchymal Stem Cell Exosomes for Cartilage Regeneration: A Systematic Review of Preclinical In Vivo Studies. Tissue Eng. Part B Rev. 2021, 27, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Taghiyar, L.; Jahangir, S.; Khozaei Ravari, M.; Shamekhi, M.A.; Eslaminejad, M.B. Cartilage Repair by Mesenchymal Stem Cell-Derived Exosomes: Preclinical and Clinical Trial Update and Perspectives. Adv. Exp. Med. Biol. 2021, 12, 73–93. [Google Scholar] [CrossRef]
- Hu, X.-H.; Zhao, Z.-X.; Dai, J.; Geng, D.-C.; Xu, Y.-Z. MicroRNA-221 Regulates Osteosarcoma Cell Proliferation, Apoptosis, Migration, and Invasion by Targeting CDKN1B/P27. J. Cell. Biochem. 2019, 120, 4665–4674. [Google Scholar] [CrossRef]
- Poulet, B.; Liu, K.; Plumb, D.; Vo, P.; Shah, M.; Staines, K.; Sampson, A.; Nakamura, H.; Nagase, H.; Carriero, A.; et al. Overexpression of TIMP-3 in Chondrocytes Produces Transient Reduction in Growth Plate Length but Permanently Reduces Adult Bone Quality and Quantity. PLoS ONE 2016, 11, e0167971. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, X.; Xing, L.; Tian, F. Wnt Signaling: A Promising Target for Osteoarthritis Therapy. Cell Commun. Signal. CCS 2019, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Li, J.; Lu, C.; Xie, D.; Liu, J.; Zhong, C.; Wu, X.; Dai, R.; Zhang, H.; Guan, D.; et al. HIF1α Inhibition Facilitates Leflunomide-AHR-CRP Signaling to Attenuate Bone Erosion in CRP-Aberrant Rheumatoid Arthritis. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Garofalo, M.; Quintavalle, C.; Romano, G.; Croce, C.M.; Condorelli, G. MiR221/222 in Cancer: Their Role in Tumor Progression and Response to Therapy. Curr. Mol. Med. 2012, 12, 27–33. [Google Scholar] [CrossRef]
- Glass, D.A.; Bialek, P.; Ahn, J.D.; Starbuck, M.; Patel, M.S.; Clevers, H.; Taketo, M.M.; Long, F.; McMahon, A.P.; Lang, R.A.; et al. Canonical Wnt Signaling in Differentiated Osteoblasts Controls Osteoclast Differentiation. Dev. Cell 2005, 8, 751–764. [Google Scholar] [CrossRef] [Green Version]
- Weivoda, M.M.; Ruan, M.; Hachfeld, C.M.; Pederson, L.; Howe, A.; Davey, R.A.; Zajac, J.D.; Kobayashi, Y.; Williams, B.O.; Westendorf, J.J.; et al. Wnt Signaling Inhibits Osteoclast Differentiation by Activating Canonical and Noncanonical CAMP/PKA Pathways. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2016, 31, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gao, Y.; Cai, L.; Li, F.; Lou, Y.; Xu, N.; Kang, Y.; Yang, H. MicroRNA-221 Is Involved in the Regulation of Osteoporosis through Regulates RUNX2 Protein Expression and Osteoblast Differentiation. Am. J. Transl. Res. 2017, 9, 126–135. [Google Scholar]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Boskey, A.L.; Wians, F.H.; Hauschka, P.V. The Effect of Osteocalcin on in Vitro Lipid-Induced Hydroxyapatite Formation and Seeded Hydroxyapatite Growth. Calcif. Tissue Int. 1985, 37, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.; Ghosh, J.; Dhara, S.K. Osteogenic Differentiation Potential of Porcine Bone Marrow Mesenchymal Stem Cell Subpopulations Selected in Different Basal Media. Biol. Open 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Qin, X.; Jiang, Q.; Komori, H.; Sakane, C.; Fukuyama, R.; Matsuo, Y.; Ito, K.; Miyazaki, T.; Komori, T. Runt-Related Transcription Factor-2 (Runx2) Is Required for Bone Matrix Protein Gene Expression in Committed Osteoblasts in Mice. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2021, 36, 1–15. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y.; Liang, C.; Chen, L.; Zhang, G.; Qian, A. Mechanosensitive MiRNAs and Bone Formation. Int. J. Mol. Sci. 2017, 18, 1684. [Google Scholar] [CrossRef] [Green Version]
- Kuang, Y.; Zheng, X.; Zhang, L.; Ai, X.; Venkataramani, V.; Kilic, E.; Hermann, D.M.; Majid, A.; Bähr, M.; Doeppner, T.R. Adipose-Derived Mesenchymal Stem Cells Reduce Autophagy in Stroke Mice by Extracellular Vesicle Transfer of MiR-25. J. Extracell. Vesicles 2020, 10, 1–20. [Google Scholar] [CrossRef]
- Burr, D.B.; Gallant, M.A. Bone Remodelling in Osteoarthritis. Nat. Rev. Rheumatol. 2012, 8, 665–673. [Google Scholar] [CrossRef]
- Goldring, M.B.; Goldring, S.R. Osteoarthritis. J. Cell. Physiol. 2007, 213, 626–634. [Google Scholar] [CrossRef]
- Stucker, S.; Chen, J.; Watt, F.E.; Kusumbe, A.P. Bone Angiogenesis and Vascular Niche Remodeling in Stress, Aging, and Diseases. Front. Cell Dev. Biol. 2020, 8, 1–20. [Google Scholar] [CrossRef]
- Bouland, C.; Philippart, P.; Dequanter, D.; Corrillon, F.; Loeb, I.; Bron, D.; Lagneaux, L.; Meuleman, N. Cross-Talk Between Mesenchymal Stromal Cells (MSCs) and Endothelial Progenitor Cells (EPCs) in Bone Regeneration. Front. Cell Dev. Biol. 2021, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Takawale, A.; Zhang, P.; Azad, A.; Wang, W.; Wang, X.; Murray, A.G.; Kassiri, Z. Myocardial Overexpression of TIMP3 after Myocardial Infarction Exerts Beneficial Effects by Promoting Angiogenesis and Suppressing Early Proteolysis. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H224–H236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.-L.; Liu, Y.-M.; Sung, T.-Y.; Huang, T.-C.; Cheng, Y.-W.; Liou, J.-P.; Pan, S.-L. TIMP3 Expression Associates with Prognosis in Colorectal Cancer and Its Novel Arylsulfonamide Inducer, MPT0B390, Inhibits Tumor Growth, Metastasis and Angiogenesis. Theranostics 2019, 9, 6676–6689. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.-L.; Li, H.-T.; Liu, S.-H. TIMP3/TGF-β1 Axis Regulates Mechanical Loading-induced Chondrocyte Degeneration and Angiogenesis. Mol. Med. Rep. 2020, 22, 2637–2644. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.R.; Chen, Q.; Takahashi, Y.; Zhou, K.K.; Park, K.; Ma, J. MicroRNA-200b Downregulates Oxidation Resistance 1 (Oxr1) Expression in the Retina of Type 1 Diabetes Model. Investig. Ophthalmol. Vis. Sci. 2013, 54, 1689–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genz, B.; Coleman, M.A.; Irvine, K.M.; Kutasovic, J.R.; Miranda, M.; Gratte, F.D.; Tirnitz-Parker, J.E.E.; Olynyk, J.K.; Calvopina, D.A.; Weis, A.; et al. Overexpression of MiRNA-25-3p Inhibits Notch1 Signaling and TGF-β-Induced Collagen Expression in Hepatic Stellate Cells. Sci. Rep. 2019, 9, 8541. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, A.; Hosseini, S.; Baghaban Eslaminejad, M. Engineered-Extracellular Vesicles as an Optimistic Tool for MicroRNA Delivery for Osteoarthritis Treatment. Cell. Mol. Life Sci. CMLS 2021, 78, 79–91. [Google Scholar] [CrossRef]
- Webber, J.; Clayton, A. How Pure Are Your Vesicles? J. Extracell. Vesicles 2013, 2, 823–831. [Google Scholar] [CrossRef]
- Wang, P.; Mao, Z.; Pan, Q.; Lu, R.; Huang, X.; Shang, X.; Zhang, R.; You, H. Histone Deacetylase-4 and Histone Deacetylase-8 Regulate Interleukin-1β-Induced Cartilage Catabolic Degradation through MAPK/JNK and ERK Pathways. Int. J. Mol. Med. 2018, 41, 2117–2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momen-Heravi, F. Isolation of Extracellular Vesicles by Ultracentrifugation. Methods Mol. Biol. Clifton NJ 2017, 1660, 25–32. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Res. Int. 2018, 2018, 1–27. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, X.; Böker, K.O.; Taheri, S.; Lehmann, W.; Schilling, A.F. Extracellular Vesicles Allow Epigenetic Mechanotransduction between Chondrocytes and Osteoblasts. Int. J. Mol. Sci. 2021, 22, 13282. https://doi.org/10.3390/ijms222413282
Shang X, Böker KO, Taheri S, Lehmann W, Schilling AF. Extracellular Vesicles Allow Epigenetic Mechanotransduction between Chondrocytes and Osteoblasts. International Journal of Molecular Sciences. 2021; 22(24):13282. https://doi.org/10.3390/ijms222413282
Chicago/Turabian StyleShang, Xiaobin, Kai Oliver Böker, Shahed Taheri, Wolfgang Lehmann, and Arndt F. Schilling. 2021. "Extracellular Vesicles Allow Epigenetic Mechanotransduction between Chondrocytes and Osteoblasts" International Journal of Molecular Sciences 22, no. 24: 13282. https://doi.org/10.3390/ijms222413282
APA StyleShang, X., Böker, K. O., Taheri, S., Lehmann, W., & Schilling, A. F. (2021). Extracellular Vesicles Allow Epigenetic Mechanotransduction between Chondrocytes and Osteoblasts. International Journal of Molecular Sciences, 22(24), 13282. https://doi.org/10.3390/ijms222413282