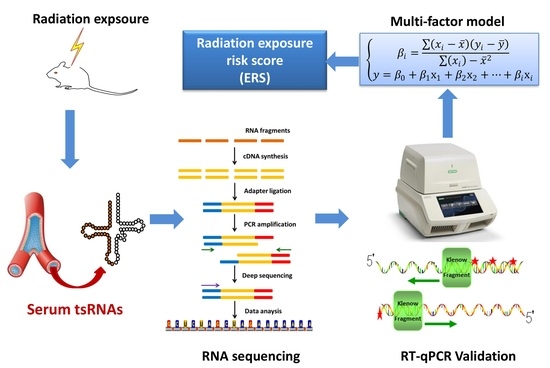
Circulating tRNA-Derived Small RNAs as Novel Radiation Biomarkers of Heavy Ion, Proton and X-ray Exposure
Abstract
1. Introduction
2. Results
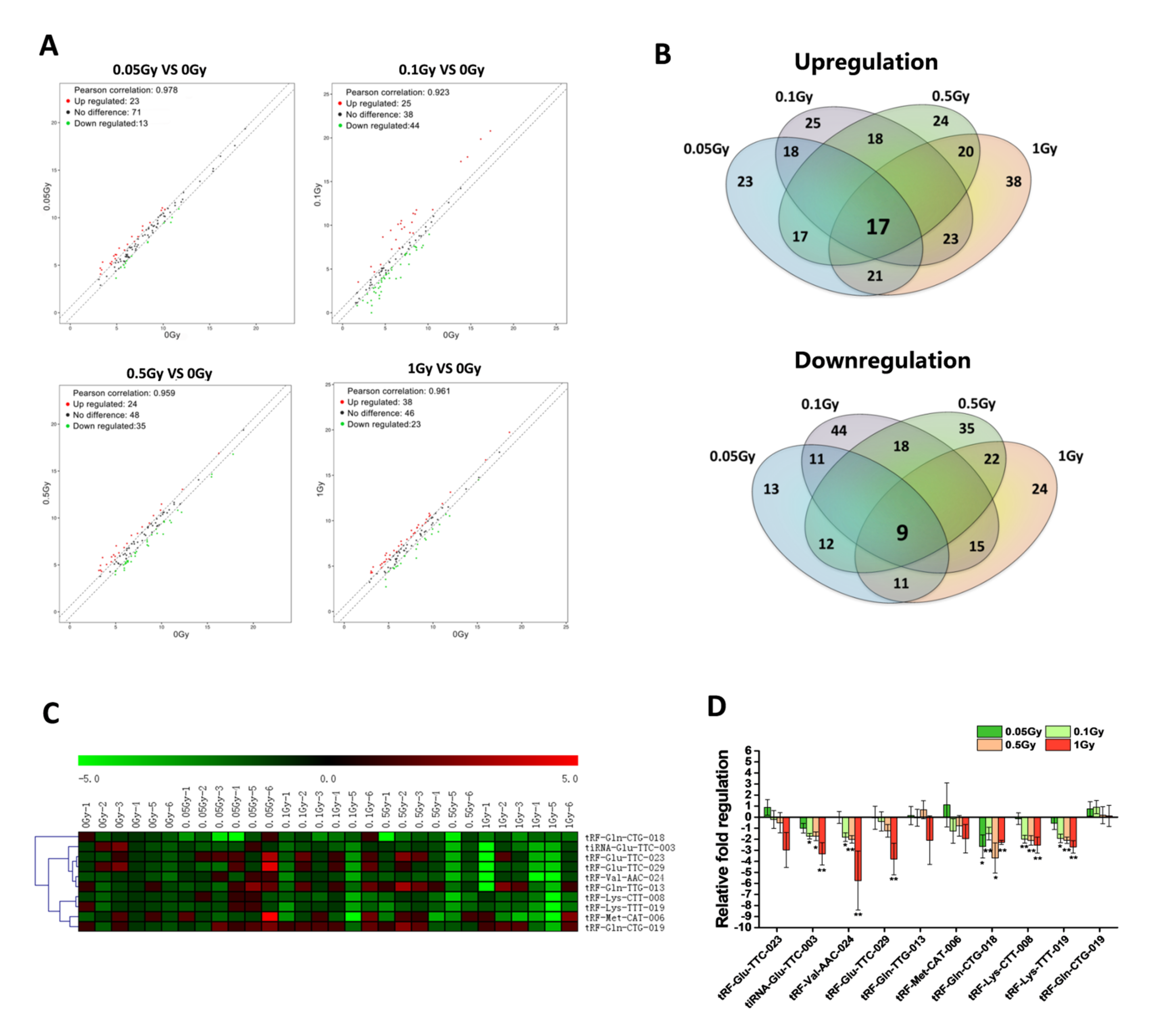
2.1. Profiling of tsRNA in Serum after Total-Body Exposure of Mice to Carbon Ions
2.2. Screening and Validation of Differentially Expressed Serum tsRNAs after Carbon Ion Irradiation
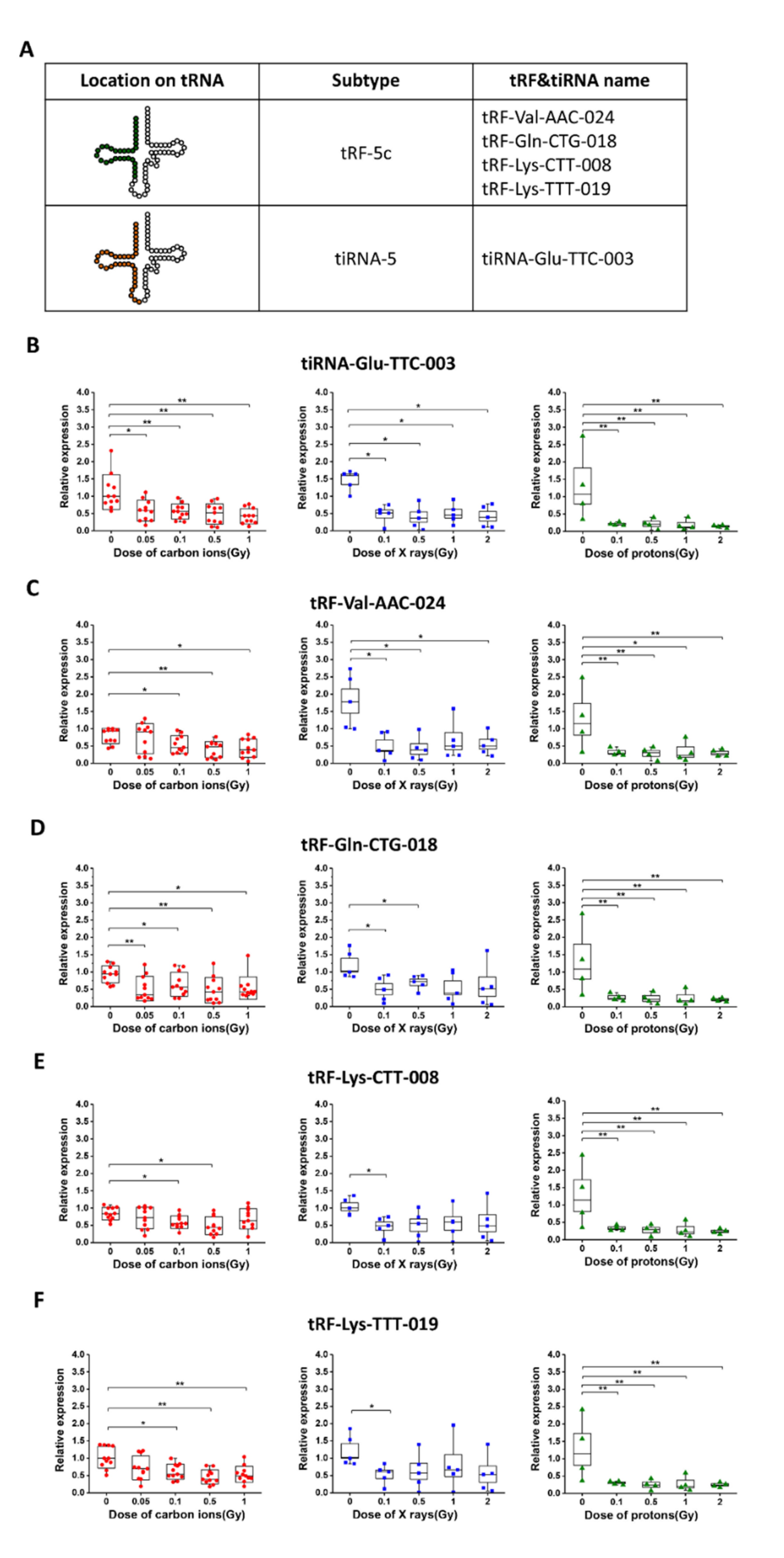
2.3. The Dose Responses of Five Selected tsRNAs after Exposure of Mice to Carbon Ions, X-rays, and Protons
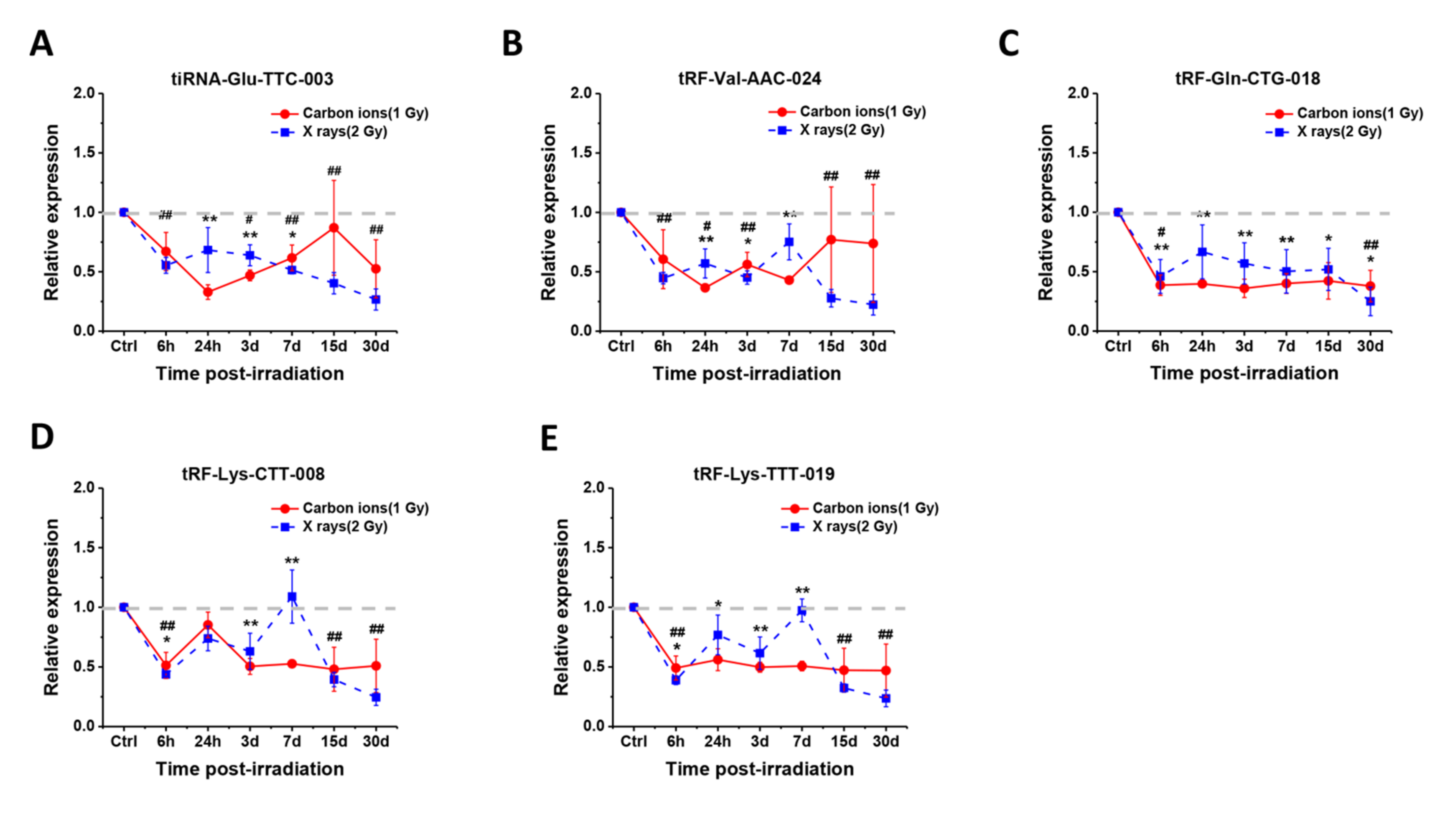
2.4. Temporal Changes of the Five Selected tsRNAs in Serum of Mice after Radiation Exposure
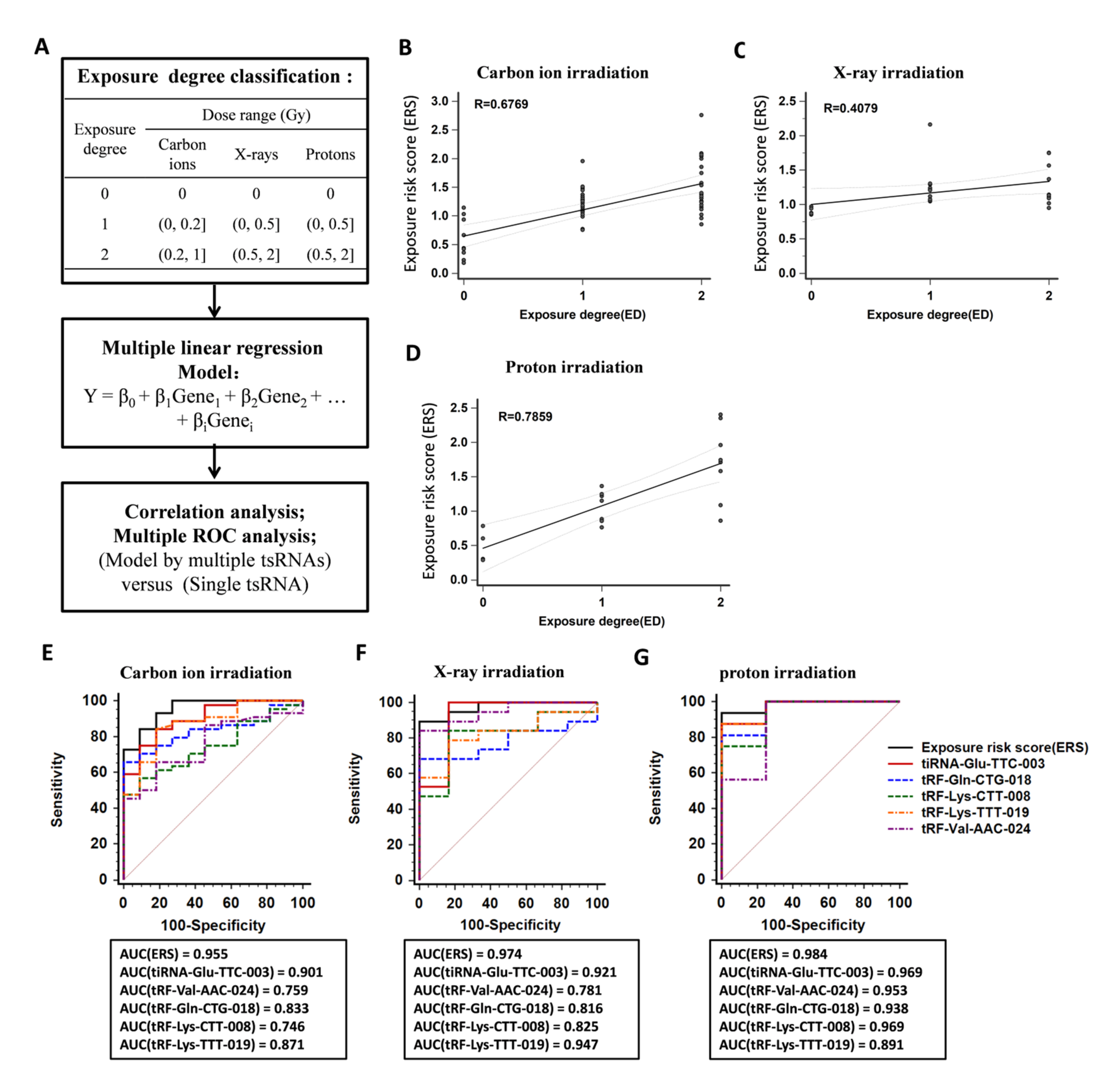
2.5. Establishment of a Model Based on Multiple tsRNA Biomarkers to Indicate Radiation Exposure
0.646(XtRF-Lys-CTT-008) − 0.077(XtRF-Lys-TTT-019)
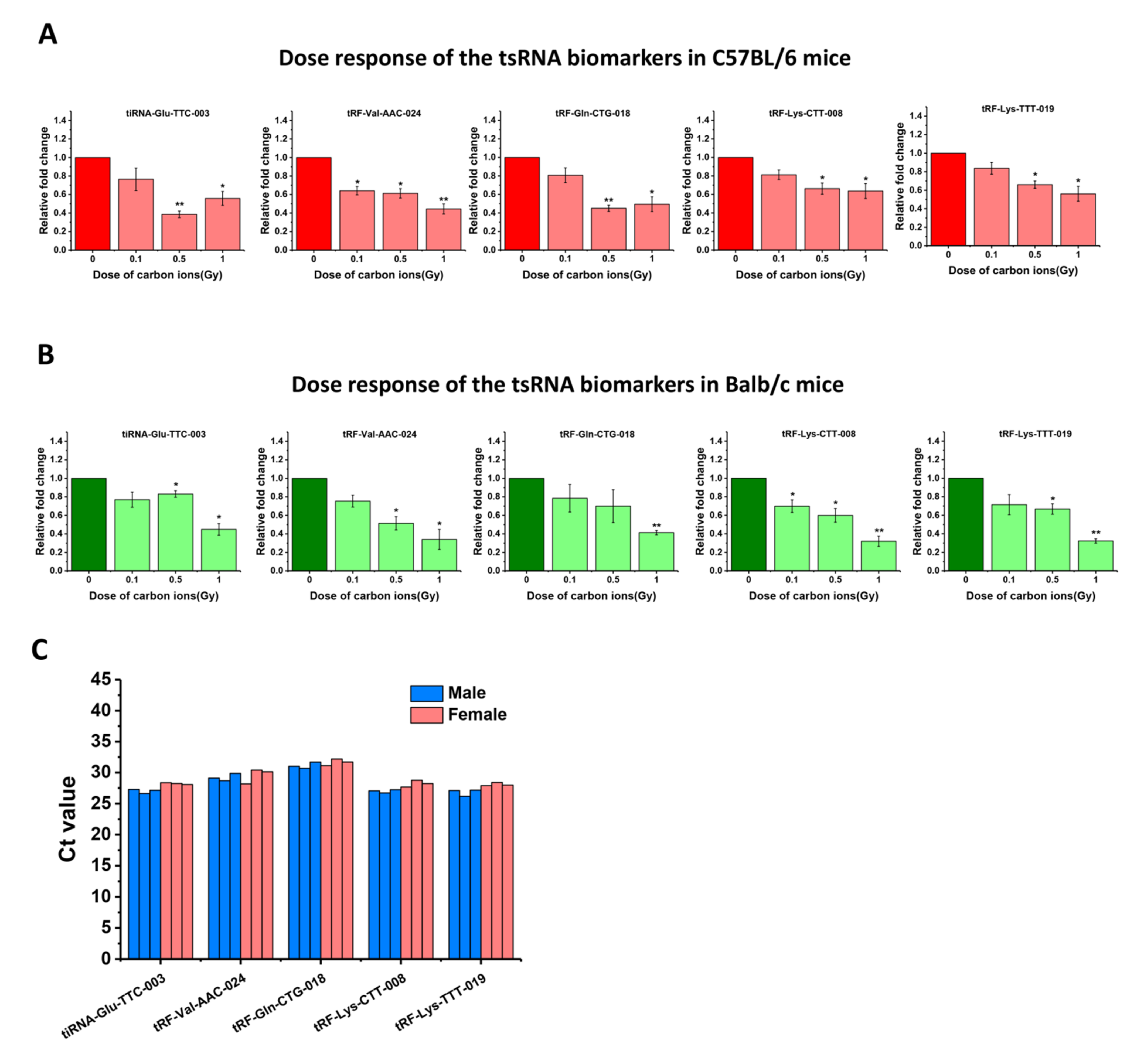
2.6. The Radiation Responses of the Five tsRNA Biomarkers in Different Mice Species and Their Expression Levels in Human Serum
3. Discussion
4. Materials and Methods
4.1. Mice and Irradiation
4.2. Mice Serum Extraction
4.3. Human Blood Sampling
4.4. Serum RNA Sequencing
4.5. Quantitative RT-PCR
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caffrey, J.A.; Hamby, D.M. A review of instruments and methods for dosimetry in space. Adv. Space Res. 2011, 47, 563–574. [Google Scholar] [CrossRef]
- Obe, G.; Johannes, I.; Johannes, C.; Hallman, K.; Reitz, G.; Facius, R. Chromosomal aberrations in blood lymphocytes of astronauts after long-term space flights. Int. J. Radiat. Biol. 1997, 72, 727–734. [Google Scholar] [CrossRef] [PubMed]
- George, K.A.; Rhone, J.; Chappell, L.J.; Cucinotta, F.A. Cytogenetic biodosimetry using the blood lymphocytes of astronauts. Acta Astronaut. 2013, 92, 97–102. [Google Scholar] [CrossRef][Green Version]
- Rothkamm, K.; Barnard, S.; Ainsbury, E.A.; Al-hafidh, J.; Barquinero, J.F.; Lindholm, C.; Moquet, J.; Perala, M.; Roch-Lefevre, S.; Scherthan, H.; et al. Manual versus automated gamma-H2AX foci analysis across five European laboratories: Can this assay be used for rapid biodosimetry in a large scale radiation accident? Mutat. Res./Genet. Toxicol. Environ. Mutagenesis 2013, 756, 170–173. [Google Scholar] [CrossRef]
- Andrievski, A.; Wilkins, R.C. The response of-H2AX in human lymphocytes and lymphocytes subsets measured in whole blood cultures. Int. J. Radiat. Biol. 2009, 85, 369–376. [Google Scholar] [CrossRef]
- Pinto, M.M.P.D.; Santos, N.F.G.; Amaral, A. Current status of biodosimetry based on standard cytogenetic methods. Radiat. Environ. Biophys. 2010, 49, 567–581. [Google Scholar] [CrossRef]
- Ioanna, S.; Sourvinou, A.M. Quantification of Circulating miRNAs in Plasma Effect of Preanalytical and Analytical Parameters on Their Isolation and Stability. J. Mol. Diagn. 2013, 15, 827–834. [Google Scholar] [CrossRef]
- Roya, T.; De Almeida, S.R.M.; Gooding, J.J. Toward biosensors for the detection of circulating microRNA as a cancer biomarker: An overview of the challenges and successes. WIREs Nanomed. Nanobiotechnol. 2014, 7, 580–592. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.Y.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Umu, S.U.; Langseth, H.; Bucherjohannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2018, 15, 242–250. [Google Scholar] [CrossRef]
- Yadav, M.; Bhayana, S.; Liu, J.; Lu, L.C.; Huang, J.S.; Ma, Y.; Qamri, Z.; Mo, X.K.; Jacob, D.S.; Parasa, S.T.; et al. Two-miRNA-based finger-stick assay for estimation of absorbed ionizing radiation dose. Sci. Transl. Med. 2020, 12, eaaw5831. [Google Scholar] [CrossRef]
- Maachowska, B.; Tomasik, B.; Stawiski, K.; Kulkarni, S.; Guha, C.; Chowdhury, D.; Fendler, W. Circulating microRNAs as Biomarkers of Radiation Exposure: A Systematic Review and Meta-Analysis. Int. J. Radiat. Oncol. 2020, 106, 390–402. [Google Scholar] [CrossRef]
- Wei, W.; Jufang, W.; Jinpeng, H.; Xiaodong, X. Serum microRNA as noninvasive indicator for space radiation. Acta Astronaut. 2018, 152, 101–104. [Google Scholar] [CrossRef]
- Wei, Z.Y.; Batagov, A.O.; Schinelli, S.; Wang, J.T.; Wang, Y.; El Fatimy, R.; Rabinovsky, R.; Balaj, L.; Chen, C.C.; Hochberg, F.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145. [Google Scholar] [CrossRef]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef]
- Polacek, N.; Ivanov, P. The regulatory world of tRNA fragments beyond canonical tRNA biology. RNA Biol. 2020, 17, 1057–1059. [Google Scholar] [CrossRef]
- Guan, L.; Karaiskos, S.; Grigoriev, A. Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA Biol. 2020, 17, 1070–1080. [Google Scholar] [CrossRef]
- Gonskikh, Y.; Gerstl, M. Modulation of mammalian translation by a ribosome-associated tRNA half. RNA Biol. 2020, 17, 1125–1136. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef]
- Fricker, R.; Brogli, R.; Luidalepp, H.; Wyss, L.; Fasnacht, M.; Joss, O.; Zywicki, M.; Helm, M.; Schneider, A.; Cristodero, M. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 2019, 10, 118. [Google Scholar] [CrossRef]
- Goans, R.E.; Holloway, E.C.; Berger, M.E.; Ricks, R.C. Early dose assessment following severe radiation accidents. Health Phys. 1997, 72, 513–518. [Google Scholar] [CrossRef]
- Sproull, M.; Kramp, T.; Tandle, A.; Shankavaram, U.; Camphausen, K. Serum Amyloid A as a Biomarker for Radiation Exposure. Radiat. Res. 2015, 184, 14–23. [Google Scholar] [CrossRef]
- Tinganelli, W.; Durante, M. Carbon Ion Radiobiology. Cancers 2020, 12, 3022. [Google Scholar] [CrossRef]
- Guipaud, O. Serum and Plasma Proteomics and Its Possible Use as Detector and Predictor of Radiation Diseases. Adv. Exp. Med. Biol. 2013, 990, 61–86. [Google Scholar] [CrossRef]
- Balzano, F.; Deiana, M.; Dei Giudici, S.; Oggiano, A.; Baralla, A.; Pasella, S.; Mannu, A.; Pescatori, M.; Porcu, B.; Fanciulli, G.; et al. miRNA Stability in Frozen Plasma Samples. Molecules 2015, 20, 19030–19040. [Google Scholar] [CrossRef]
- Jacob, N.K.; Cooley, J.V.; Yee, T.N.; Jacob, J.; Alder, H.; Wickramasinghe, P.; Maclean, K.H.; Chakravarti, A. Identification of Sensitive Serum microRNA Biomarkers for Radiation Biodosimetry. PLoS ONE 2013, 8, e57603. [Google Scholar] [CrossRef]
- Wei, W.; He, J.; Wang, J.; Ding, N.; Wang, B.; Lin, S.; Zhang, X.; Hua, J.; Li, H.; Hu, B. Serum microRNAs as Early Indicators for Estimation of Exposure Degree in Response to Ionizing Irradiation. Radiat. Res. 2017, 188, 342–354. [Google Scholar] [CrossRef]
- Semkova, J.; Dachev, T.; Koleva, R.; Bankov, N.; Maltchev, S.; Benghin, V.; Shurshakov, V.; Petrov, V. Observation of radiation environment in the International Space Station in 2012-March 2013 by Liulin-5 particle telescope. J. Space Weather Space Clim. 2014, 4, 2–10. [Google Scholar] [CrossRef]
- Kinoshita, N.; Sueki, K.; Sasa, K.; Kitagawa, J.; Ikarashi, S.; Nishimura, T.; Wong, Y.S.; Satou, Y.; Handa, K.; Takahashi, T.; et al. Assessment of individual radionuclide distributions from the Fukushima nuclear accident covering central-east Japan. Proc. Natl. Acad. Sci. USA 2011, 108, 19526–19529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Lu, D.; Li, X.; Feng, Y.G.; Cui, Q.; Song, X.J. Heavy ion mutagenesis combined with triclosan screening provides a new strategy for improving the arachidonic acid yield in Mortierella alpina. BMC Biotechnol. 2018, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Yamakawa, A.; Boffelli, D.; Mote, P.; Martin, D.I. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013, 14, 298. [Google Scholar] [CrossRef]
- Acharya, S.S.; Fendler, W.; Watson, J.; Hamilton, A.; Pan, Y.F.; Gaudiano, E.; Moskwa, P.; Bhanja, P.; Saha, S.; Guha, C.; et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci. Transl. Med. 2015, 7, 287ra69. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, K.; Zhou, G.; Hu, W. MicroRNAs Responding to Space Radiation. Int. J. Mol. Sci. 2020, 21, 6603. [Google Scholar] [CrossRef]
- Wang, X.; Matuszek, Z.; Huang, Y.; Parisien, M.; Dai, Q.; Clark, W.; Schwartz, M.H.; Pan, T. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 2018, 24, 1305–1313. [Google Scholar] [CrossRef]
- Rashad, S.; Han, X.; Sato, K.; Mishima, E.; Abe, T.; Tominaga, T.; Niizuma, K. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020, 17, 1092–1103. [Google Scholar] [CrossRef]
- Francesca, P.; Franco, C.; Ambrogia, B.; Enrica, C.; Annalisa, D.S.; Carmine, T.; Rosanna, N. Radiation-induced circulating miRNA expression in blood of head and neck cancer patients. Radiat. Environ. Biophys. 2020, 59, 237–244. [Google Scholar] [CrossRef]
- Anderson, P.; Ivanov, P. tRNA fragments in human health and disease. FEBS Lett. 2016, 588, 4297–4304. [Google Scholar] [CrossRef]
- Sobala, A.; Hutvagner, G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef]
- Marabita, F.; de Candia, P.; Torri, A.; Tegner, J.; Abrignani, S.; Rossi, R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief. Bioinform. 2016, 17, 204–212. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, W.; Bai, H.; Chen, Y.; Zhang, T.; Zhang, Y.; Hua, J.; He, J.; Ding, N.; Zhou, H.; Wang, J. Circulating tRNA-Derived Small RNAs as Novel Radiation Biomarkers of Heavy Ion, Proton and X-ray Exposure. Int. J. Mol. Sci. 2021, 22, 13476. https://doi.org/10.3390/ijms222413476
Wei W, Bai H, Chen Y, Zhang T, Zhang Y, Hua J, He J, Ding N, Zhou H, Wang J. Circulating tRNA-Derived Small RNAs as Novel Radiation Biomarkers of Heavy Ion, Proton and X-ray Exposure. International Journal of Molecular Sciences. 2021; 22(24):13476. https://doi.org/10.3390/ijms222413476
Chicago/Turabian StyleWei, Wenjun, Hao Bai, Yaxiong Chen, Tongshan Zhang, Yanan Zhang, Junrui Hua, Jinpeng He, Nan Ding, Heng Zhou, and Jufang Wang. 2021. "Circulating tRNA-Derived Small RNAs as Novel Radiation Biomarkers of Heavy Ion, Proton and X-ray Exposure" International Journal of Molecular Sciences 22, no. 24: 13476. https://doi.org/10.3390/ijms222413476
APA StyleWei, W., Bai, H., Chen, Y., Zhang, T., Zhang, Y., Hua, J., He, J., Ding, N., Zhou, H., & Wang, J. (2021). Circulating tRNA-Derived Small RNAs as Novel Radiation Biomarkers of Heavy Ion, Proton and X-ray Exposure. International Journal of Molecular Sciences, 22(24), 13476. https://doi.org/10.3390/ijms222413476