Quality Traits, Volatile Organic Compounds, and Expression of Key Flavor Genes in Strawberry Genotypes over Harvest Period
Abstract
:1. Introduction
2. Results and Discussion
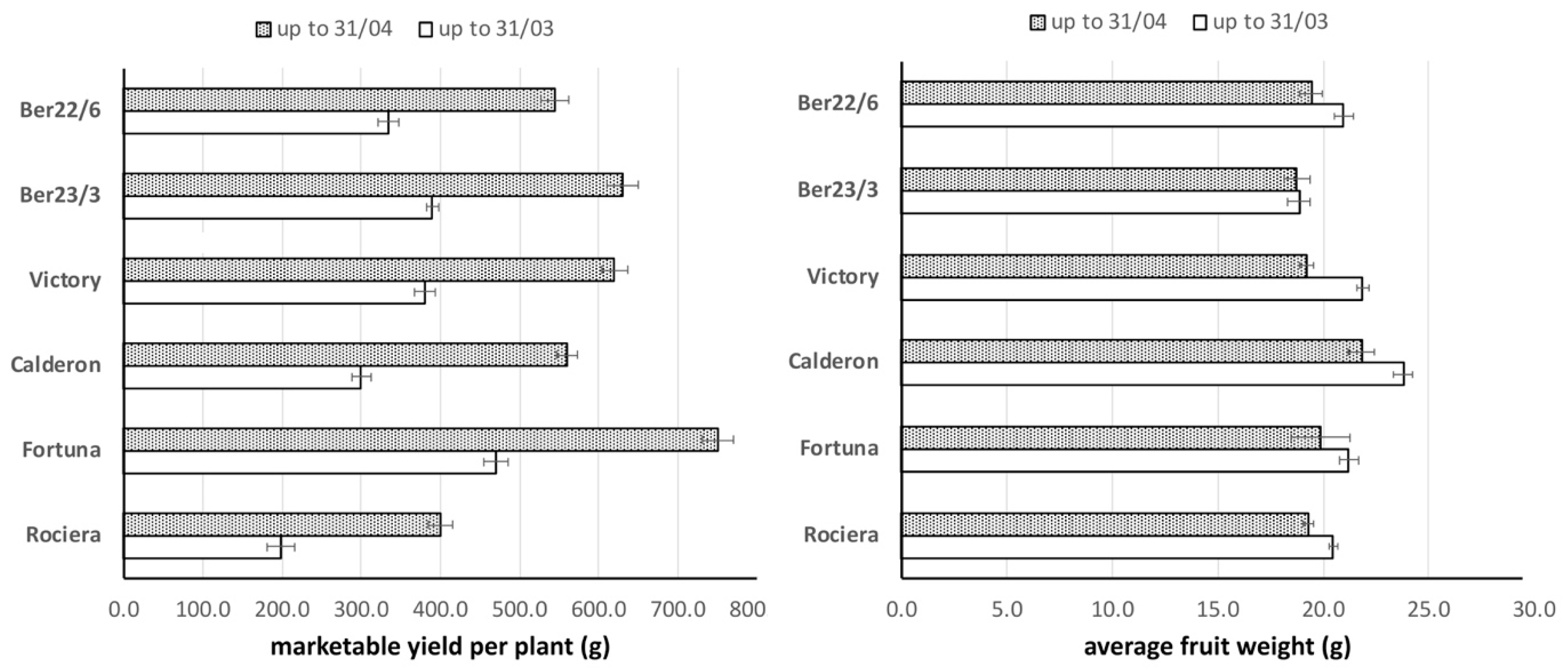
2.1. Fruit Yield and Weight
2.2. Fruit Quality Parameters
2.2.1. Sugars
2.2.2. Phenolics and Anthocyanins
2.2.3. Ascorbic Acid and Antioxidant Activity
2.3. Strawberry Volatiles
Effect of Genotype and Harvest Time on Volatiles
2.4. Expression Analysis of Key Flavor Genes and Transcription Factors (TFs)
2.5. Correlation of Chemical and Genetic Data
3. Materials and Methods
3.1. Plant Cultivation
3.2. Fruit Measurements and Experimental Design
3.3. Determination of Total Phenolics, Sugars, Anthocyanins and Antioxidant Capacity
3.4. Determination of Glucose and Ascorbic Acid Content
3.5. GC/MS Analysis of Strawberry Volatiles
3.6. RNA Isolation and Gene Expression Analysis by Quantitative Real-Time PCR (qRT-PCR)
3.7. Statistical Analysis of Chemical Traits and Gene Expression Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, Z.; Hasing, T.; Johnson, T.S.; Garner, D.M.; Schwieterman, M.L.; Barbey, C.R.; Colquhoun, T.A.; Sims, C.A.; Resende, M.F.R.; Whitaker, V.M. Strawberry sweetness and consumer preference are enhanced by specific volatile compounds. Hortic. Res. 2021, 8, 1–15. [Google Scholar] [CrossRef]
- Mezzetti, B.; Giampieri, F.; Zhang, Y.-T.; Zhong, C.-F. Status of strawberry breeding programs and cultivation systems in Europe and the rest of the world. J. Berry Res. 2018, 8, 205–221. [Google Scholar] [CrossRef]
- Prescott, J. Multisensory processes in flavour perception and their influence on food choice. Curr. Opin. Food Sci. 2015, 3, 47–52. [Google Scholar] [CrossRef]
- Yan, J.-W.; Ban, Z.-J.; Lu, H.-Y.; Li, D.; Poverenov, E.; Luo, Z.-S.; Li, L. The aroma volatile repertoire in strawberry fruit: A review. J. Sci. Food Agric. 2018, 98, 4395–4402. [Google Scholar] [CrossRef] [PubMed]
- Di Vittori, L.; Mazzoni, L.; Battino, M.; Mezzetti, B. Pre-harvest factors influencing the quality of berries. Sci. Hortic. 2018, 233, 310–322. [Google Scholar] [CrossRef]
- Ulrich, D.; Kecke, S.; Olbricht, K. What Do We Know about the Chemistry of Strawberry Aroma? J. Agric. Food Chem. 2018, 66, 3291–3301. [Google Scholar] [CrossRef]
- Zabetakis, I.; Holden, M.A. Strawberry Flavour: Analysis and Biosynthesis. J. Sci. Food Agric. 1997, 74, 421–434. [Google Scholar] [CrossRef]
- Cannon, R.J.; Agyemang, D.; Curto, N.L.; Yusuf, A.; Chen, M.Z.; Janczuk, A.J. In-depth analysis of Ciflorette strawberries (Fragaria × ananassa ‘Ciflorette’) by multidimensional gas chromatography and gas chromatography-olfactometry. Flavour Fragr. J. 2015, 30, 302–319. [Google Scholar] [CrossRef]
- Aharoni, A.; Giri, A.P.; Verstappen, F.W.; Bertea, C.M.; Sevenier, R.; Sun, Z.; Jongsma, M.; Schwab, W.; Bouwmeester, H. Gain and Loss of Fruit Flavor Compounds Produced by Wild and Cultivated Strawberry Species. Plant Cell 2004, 16, 3110–3131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koeduka, T.; Fridman, E.; Gang, D.R.; Vassão, D.G.; Jackson, B.L.; Kish, C.M.; Orlova, I.; Spassova, S.M.; Lewis, N.; Noel, J.P.; et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 2006, 103, 10128–10133. [Google Scholar] [CrossRef] [Green Version]
- Aragüez, I.; Osorio, S.; Hoffmann, T.; Rambla, J.L.; Medina-Escobar, N.; Granell, A.; Botella, M.Á.; Schwab, W.; Valpuesta, V. Eugenol Production in Achenes and Receptacles of Strawberry Fruits Is Catalyzed by Synthases Exhibiting Distinct Kinetics. Plant Physiol. 2013, 163, 946–958. [Google Scholar] [CrossRef] [Green Version]
- Medina-Puche, L.; Cumplido-Laso, G.; Amil-Ruiz, F.; Hoffmann, T.; Ring, L.; Franco, A.R.; Caballero, J.L.; Schwab, W.; Blanco, J.M.; Portales, R.B. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 2014, 65, 401–417. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Shi, Y.; Ma, Y.; Yang, X.; Yin, X.; Zhang, Y.; Xiao, Y.; Liu, W.; Li, Y.; Li, S.; et al. The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 2020, 18, 2267–2279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina-Puche, L.; Molina-Hidalgo, F.J.; Boersma, M.; Schuurink, R.C.; López-Vidriero, I.; Solano, R.; Franco-Zorrilla, J.M.; Caballero, J.L.; Portales, R.B.; Muñoz-Blanco, J. An R2R3-MYB Transcription Factor Regulates Eugenol Production in Ripe Strawberry Fruit Receptacles. Plant Physiol. 2015, 168, 598–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Hidalgo, F.J.; Medina-Puche, L.; Cañete-Gómez, C.; Franco-Zorrilla, J.M.; López-Vidriero, I.; Solano, R.; Caballero, J.L.; Rodríguez-Franco, A.; Blanco-Portales, R.; Muñoz-Blanco, J.; et al. The fruit-specific transcription factor FaDOF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 2017, 68, 4529–4543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afrin, S.; Gasparrini, M.; Forbes-Hernandez, T.Y.; Reboredo-Rodriguez, P.; Mezzetti, B.; Varela-López, A.; Giampieri, F.; Battino, M. Promising Health Benefits of the Strawberry: A Focus on Clinical Studies. J. Agric. Food Chem. 2016, 64, 4435–4449. [Google Scholar] [CrossRef] [PubMed]
- Forbes-Hernandez, T.Y.; Gasparrini, M.; Afrin, S.; Bompadre, S.; Mezzetti, B.; Quiles, J.L.; Giampieri, F.; Battino, M. The Healthy Effects of Strawberry Polyphenols: Which Strategy behind Antioxidant Capacity? Crit. Rev. Food Sci. Nutr. 2016, 56, S46–S59. [Google Scholar] [CrossRef]
- Li, D.; Wang, P.; Luo, Y.; Zhao, M.; Chen, F. Health benefits of anthocyanins and molecular mechanisms: Update from recent decade. Crit. Rev. Food Sci. Nutr. 2017, 57, 1729–1741. [Google Scholar] [CrossRef]
- Cervantes, L.; Martínez-Ferri, E.; Soria, C.; Ariza, M.T. Bioavailability of phenolic compounds in strawberry, raspberry and blueberry: Insights for breeding programs. Food Biosci. 2020, 37, 100680. [Google Scholar] [CrossRef]
- Diamanti, J.; Capocasa, F.; Battino, M.; Mezzetti, B. Evaluation of F. × ananassa intra-specific and inter-specific backcrosses to generate new genetic material with increased fruit nutritional quality. J. Berry Res. 2010, 1, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Fredericks, C.H.; Fanning, K.J.; Gidley, M.J.; Netzel, G.; Zabaras, D.; Herrington, M.; Netzel, M. High-anthocyanin strawberries through cultivar selection. J. Sci. Food Agric. 2013, 93, 846–852. [Google Scholar] [CrossRef]
- Mezzetti, B.; Balducci, F.; Capocasa, F.; Zhong, C.-F.; Cappelletti, R.; Di Vittori, L.; Mazzoni, L.; Giampieri, F.; Battino, M. Breeding Strawberry for Higher Phytochemicals Content and Claim It: Is It Possible? Int. J. Fruit Sci. 2016, 16, 194–206. [Google Scholar] [CrossRef]
- Correia, P.; Pestana, M.; Martínez, F.; Ribeiro, E.; Gama, F.; Saavedra, T.; Palencia, P. Relationships between strawberry fruit quality attributes and crop load. Sci. Hortic. 2011, 130, 398–403. [Google Scholar] [CrossRef]
- Zeliou, K.; Papasotiropoulos, V.; Manoussopoulos, Y.; Lamari, F.N. Physical and chemical quality characteristics and antioxidant properties of strawberry cultivars (Fragaria × ananassa Duch.) in Greece: Assessment of their sensory impact. J. Sci. Food Agric. 2018, 98, 4065–4073. [Google Scholar] [CrossRef]
- Davik, J.; Bakken, A.K.; Holte, K.; Blomhoff, R. Effects of genotype and environment on total anti-oxidant capacity and the content of sugars and acids in strawberries (Fragaria × ananassa Duch.). J. Hortic. Sci. Biotechnol. 2006, 81, 1057–1063. [Google Scholar] [CrossRef]
- Pangborn, R.M. Relative Taste Intensities of Selected Sugars and Organic Acids. J. Food Sci. 1963, 28, 726–733. [Google Scholar] [CrossRef]
- Doty, T.E. Fructose sweetness: A new dimension. Cereal Foods World 1976, 21, 62–63. [Google Scholar]
- Kawanobu, S.; Wajima, T.; Zushi, K.; Mori, T.; Matsuzoe, N. Seasonal Variations in the Maturation Period, Anthocyanin Content, and Ascorbic Acid Content in Strawberry Fruits. Environ. Control. Biol. 2010, 48, 175–184. [Google Scholar] [CrossRef]
- Tomić, J.; Pešaković, M.; Milivojevic, J.; Miletić, R.; Karaklajić-Stajić, Ž.; Paunović, S.; Milinković, M. Changes in anthocyanins and total phenols in fruit of three strawberry cultivars during five harvests. Acta Hortic. 2016, 1139, 633–638. [Google Scholar] [CrossRef]
- Jimenez-Garcia, S.N.; Guevara-Gonzalez, R.G.; Miranda-Lopez, R.; Feregrino-Perez, A.A.; Torres-Pacheco, I.; Vazquez-Cruz, M.A. Functional properties and quality characteristics of bioactive compounds in berries: Biochemistry, biotechnology, and genomics. Food Res. Int. 2013, 54, 1195–1207. [Google Scholar] [CrossRef]
- Giampieri, F.; Tulipani, S.; Alvarez-Suarez, J.M.; Quiles, J.L.; Mezzetti, B.; Battino, M. The strawberry: Composition, nutritional quality, and impact on human health. Nutrition 2012, 28, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Tulipani, S.; Mezzetti, B.; Capocasa, F.; Bompadre, S.; Beekwilder, J.; de Vos, C.H.R.; Capanoglu, E.; Bovy, A.; Battino, M. Antioxidants, Phenolic Compounds, and Nutritional Quality of Different Strawberry Genotypes. J. Agric. Food Chem. 2008, 56, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Kangina, I.B.; Bondarenko, N.F. Vliyanie meteorologicheskikh faktorov na kachestvo yagod zemlyaniki. The effect of meteorological factors on strawberry quality. Sadovodstvo 1987, 35, 65–69. [Google Scholar]
- Ariza, M.; Martínez-Ferri, E.; Domínguez, P.; Medina, J.; Miranda, L.; Soria, C. Effects of harvest time on functional compounds and fruit antioxidant capacity in ten strawberry cultivars. J. Berry Res. 2015, 5, 71–80. [Google Scholar] [CrossRef]
- National Institute of Standards and Technology. NIST WebBook. Available online: http://webbook.nist.gov/chemistry/ (accessed on 1 July 2021).
- Van Den Dool, H.; Kratz, P.D. A generalization of the retention index system including linear temperature programmed gas—liquid partition chromatography. J. Chromatogr. A 1963, 11, 463–471. [Google Scholar] [CrossRef]
- Acree, T.; Arn, H. Flavornet and Human Odor Space. Available online: http://www.flavornet.org/flavornet.html (accessed on 24 August 2021).
- Pérez, A.G.; Sanz, C. Strawberry Flavor. In Handbook of Fruit and Vegetable Flavors of Fruit and Vegetables Flavors; Hui, Y.H., Chen, F., Nollet, L.M.L., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; pp. 431–449. ISBN 978-047-062-283-4. [Google Scholar]
- Gaborieau, S.; Cendrès, A.; Page, D.; Ginies, C.; Renard, C.M. Variability of free and glycosylated volatiles from strawberries destined for the fresh market and for processing, assessed using direct enzymatic hydrolysis. LWT 2018, 98, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Zhang, Y.-J.; Liu, N.; Ye, D.-Q.; Gong, X.; Qin, Y.; Liu, Y.-L. Volatile compounds in wild strawberry and their odorants of wild strawberry wines: Effects of different stages of fermentation. Int. J. Food Prop. 2017, 20, S399–S415. [Google Scholar] [CrossRef] [Green Version]
- Hussein, B.; Aldhaher, Z.A.; Abdu-Allah, S.N.; Hamdan, A. Bioactivity of Phenolic Compounds Extracted from Strawberry against Formation of Pseudomonas aeruginosa Biofilm. Int. J. Pharm. Qual. Assur. 2019, 10, 160–167. [Google Scholar] [CrossRef]
- Wintoch, H.; Krammer, G.; Schreier, P. Glycosidically bound aroma compounds from two strawberry fruit species, Fragaria vesca f. semperflorens and Fragaria × ananassa, cv. Korona. Flavour Fragr. J. 1991, 6, 209–215. [Google Scholar] [CrossRef]
- Álvarez-Fernández, M.A.; Cerezo, A.B.; Rodríguez, A.M.C.; Troncoso, A.M.; García-Parrilla, M.C. Composition of Nonanthocyanin Polyphenols in Alcoholic-Fermented Strawberry Products Using LC–MS (QTRAP), High-Resolution MS (UHPLC-Orbitrap-MS), LC-DAD, and Antioxidant Activity. J. Agric. Food Chem. 2015, 63, 2041–2051. [Google Scholar] [CrossRef]
- Kosar, M.; Kafkas, E.; Paydas, S.; Baser, K.H.C. Phenolic Composition of Strawberry Genotypes at Different Maturation Stages. J. Agric. Food Chem. 2004, 52, 1586–1589. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; de Castro, M.D.L. Ultrasound-assisted extraction of phenolic compounds from strawberries prior to liquid chromatographic separation and photodiode array ultraviolet detection. J. Chromatogr. A 2005, 1100, 1–7. [Google Scholar] [CrossRef]
- Pérez, A.G.; Olías, R.; Sanz, C.; Olías, J.M. Furanones in Strawberries: Evolution during Ripening and Postharvest Shelf Life. J. Agric. Food Chem. 1996, 44, 3620–3624. [Google Scholar] [CrossRef]
- Jouquand, C.; Chandler, C.; Plotto, A.; Goodner, K. A Sensory and Chemical Analysis of Fresh Strawberries Over Harvest Dates and Seasons Reveals Factors That Affect Eating Quality. J. Am. Soc. Hortic. Sci. 2008, 133, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Salameh, D.; Brandam, C.; Medawar, W.; Lteif, R.; Strehaiano, P. Highlight on the problems generated by p-coumaric acid analysis in wine fermentations. Food Chem. 2008, 107, 1661–1667. [Google Scholar] [CrossRef] [Green Version]
- Samykanno, K.; Pang, E.; Marriott, P.J. Genotypic and environmental effects on flavor attributes of ‘Albion’ and ‘Juliette’ strawberry fruits. Sci. Hortic. 2013, 164, 633–642. [Google Scholar] [CrossRef]
- Pelayo-Zaldivar, C.; Ebeler, S.E.; Kader, A.A. Cultivar and harvest date effects on flavor and other quality attributes of California strawberries. J. Food Qual. 2005, 28, 78–97. [Google Scholar] [CrossRef]
- Parra-Palma, C.; Úbeda, C.; Gil, M.; Ramos, P.; Castro, R.I.; Morales-Quintana, L. Comparative study of the volatile organic compounds of four strawberry cultivars and it relation to alcohol acyltransferase enzymatic activity. Sci. Hortic. 2019, 251, 65–72. [Google Scholar] [CrossRef]
- Beekwilder, J.; Alvarez-Huerta, M.; Neef, E.; Verstappen, F.W.; Bouwmeester, H.; Aharoni, A. Functional Characterization of Enzymes Forming Volatile Esters from Strawberry and Banana. Plant Physiol. 2004, 135, 1865–1878. [Google Scholar] [CrossRef] [Green Version]
- Yauk, Y.-K.; Souleyre, E.J.; Matich, A.J.; Chen, X.; Wang, M.Y.; Plunkett, B.; Dare, A.P.; Espley, R.; Tomes, S.; Chagné, D.; et al. Alcohol acyl transferase 1 links two distinct volatile pathways that produce esters and phenylpropenes in apple fruit. Plant J. 2017, 91, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Yu, M.; Zhang, B.; Xu, J.; Ma, R. Differences in PpAAT1 Activity in High- and Low-Aroma Peach Varieties Affect γ-Decalactone Production. Plant Physiol. 2020, 182, 2065–2080. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.H.; Pillet, J.; Plotto, A.; Bai, J.; Whitaker, V.M.; Folta, K.M. Identification of a strawberry flavor gene candidate using an integrated genetic-genomic-analytical chemistry approach. BMC Genom. 2014, 15, 217. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Sevilla, J.F.; Cruz-Rus, E.; Valpuesta, V.; A Botella, M.; Amaya, I. Deciphering gamma-decalactone biosynthesis in strawberry fruit using a combination of genetic mapping, RNA-Seq and eQTL analyses. BMC Genom. 2014, 15, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Rus, E.; Sesmero, R.; Ángel-Pérez, J.A.; Sevilla, J.F.S.; Ulrich, D.; Amaya, I. Validation of a PCR test to predict the presence of flavor volatiles mesifurane and γ-decalactone in fruits of cultivated strawberry (Fragaria × ananassa). Mol. Breed. 2017, 37, 1–15. [Google Scholar] [CrossRef]
- Oh, Y.; Barbey, C.R.; Chandra, S.; Bai, J.; Fan, Z.; Plotto, A.; Pillet, J.; Folta, K.M.; Whitaker, V.M.; Lee, S. Genomic Characterization of the Fruity Aroma Gene, FaFAD1, Reveals a Gene Dosage Effect on γ-Decalactone Production in Strawberry (Fragaria × ananassa). Front. Plant Sci. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Singh, P.; Kalunke, R.M.; Giri, A.P. Towards comprehension of complex chemical evolution and diversification of terpene and phenylpropanoid pathways in Ocimum species. RSC Adv. 2015, 5, 106886–106904. [Google Scholar] [CrossRef]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.Y.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Aharoni, A.; De Vos, C.H.R.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.M.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, H.; Wang, X.; Chen, Q.; Jiang, L.; Lin, Y. Virus-induced gene silencing as a tool for FaMYB5 gene functional studies in strawberry. Acta Hortic. Sin. 2017, 44, 33–42. [Google Scholar]
- Parra-Palma, C.; Morales-Quintana, L.; Ramos, P. Phenolic Content, Color Development, and Pigment−Related Gene Expression: A Comparative Analysis in Different Cultivars of Strawberry during the Ripening Process. Agronomy 2020, 10, 588. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Mao, W.; Jia, M.; Xing, S.; Ali, U.; Zhao, Y.; Chen, Y.; Cao, M.; Dai, Z.; Zhang, K.; et al. FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB. J. Exp. Bot. 2018, 69, 4805–4820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, R.; Imran, M.; Amanullah, S.; Rothenberg, D.O.; Wang, Q.; Wang, L.; Fan, Y. Functional Characterization of Hedychium coronarium J. Koenig MYB132 Confers the Potential Role in Floral Aroma Synthesis. Plants 2021, 10, 2014. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Abbas, F.; Zhou, Y.; Yu, R.; Fan, Y. Auxin-Responsive R2R3-MYB Transcription Factors HcMYB1 and HcMYB2 Activate Volatile Biosynthesis in Hedychium coronarium Flowers. Front. Plant Sci. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A., Jr. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Laurentin, A.; Edwards, C. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 2003, 315, 143–145. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Giusti, M.M.; Wrolstad, R.E. Characterization and measurement of anthocyanins by UV-visible spectroscopy. In Current Protocols in Food Analytical Chemistry; Wrolstad, R.E., Acree, T.E., An, H., Decker, E.A., Penner, M.H., Reid, D.S., Schwartz, S.J., Shoemaker, C.F., Sporns, P., Eds.; John Wiley & Sons: New York, NY, USA, 2001; pp. F1.2.1–F1.2.13. [Google Scholar] [CrossRef]
- López, R.; Aznar, M.; Cacho, J.; Ferreira, V. Determination of minor and trace volatile compounds in wine by solid-phase extraction and gas chromatography with mass spectrometric detection. J. Chromatogr. A 2002, 966, 167–177. [Google Scholar] [CrossRef]
- Zavala, J.F.A.; Wang, S.Y.; Wang, C.Y.; A González-Aguilar, G. Effect of storage temperatures on antioxidant capacity and aroma compounds in strawberry fruit. LWT 2004, 37, 687–695. [Google Scholar] [CrossRef]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry, 4th ed.; Allured Publishing Corporation: Carol Stream, IL, USA, 2007; ISBN 978-1-932633-21-4. [Google Scholar]
- Negri, A.S.; Allegra, D.; Simoni, L.; Rusconi, F.; Tonelli, C.; Espen, L.; Galbiati, M. Comparative analysis of fruit aroma patterns in the domesticated wild strawberries “Profumata di Tortona” (F. moschata) and “Regina delle Valli” (F. vesca). Front. Plant Sci. 2015, 6, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Cheng, S.; Zhang, Y.; Du, B.; Feng, C.; Zhou, Y.; Mei, X.; Jiang, Y.; Duan, X.; Yang, Z. Differential responses of four biosynthetic pathways of aroma compounds in postharvest strawberry (Fragaria × ananassa Duch.) under interaction of light and temperature. Food Chem. 2017, 221, 356–364. [Google Scholar] [CrossRef]
- Baldi, P.; Orsucci, S.; Moser, M.; Brilli, M.; Giongo, L.; Ammour, A.S. Gene expression and metabolite accumulation during strawberry (Fragaria × ananassa) fruit development and ripening. Planta 2018, 248, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Salvatierra, A.; Pimentel, P.; Moya-Leon, M.A.; Caligari, P.D.; Herrera, R. Comparison of transcriptional profiles of flavonoid genes and anthocyanin contents during fruit development of two botanical forms of Fragaria chiloensis ssp. chiloensis. Phytochemistry 2010, 71, 1839–1847. [Google Scholar] [CrossRef] [PubMed]









| No | RIexp | RIlit | Compound | Compound Category | Flavor Impact | Range of Peak Area Percentages | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | 777 | 785 | 2-Methylpropanoic acid (isobutyric acid) | Acids | a: rancid, butter, cheese | 0.08–0.94 | b, c, d |
| 2 | 799 | 802 | 2,3-Butanediol | Alcohols | a: fruit, onion; f: creamy | <0.31 | e, f |
| 3 | 803 | 802 | Ethyl butanoate | Esters | a: apple; b: estery, fruity, sweet; g | <0.33 | b, c, d, g |
| 4 | 805 | 790 | Butanoic acid (butyric acid) | Acids | a: rancid, cheese, sweat; b: cheesy, fruity; g | tr.<4.23 | b, c, d, g |
| 5 | 831 | 811b | Methyl 2-hydroxybutanoate (methyl 2-hydroxy butyrate) | Esters | <0.21 | b | |
| 6 | 834 | 830/832/839 | Furfural | Furans | a: bread, almond, sweet | <0.22 | b, c, d |
| 7 | 856 | 827 (Kovat’s index) | Maleic anhydridestd (cis-butenedioic anhydride; 2,5-furandione) | Furans/Anhydrides | 0.59–8.83 | ||
| 8 | 871 | 846/886 | 2-Methylbutanoic acid | Acids | b: cheesy, stinky; g | 0.03–4.86 | b, c, d, g |
| 9 | 890 | 890 | Styrene (vinylbenzene) | Aromatic compounds | a: balsamic, gasoline | <1.44 | b |
| 10 | 915 | 915/916/922 | Butyrolactone [dihydrofuran-2(3H)-one] | Furans/Lactones | a: caramel-sweet | 0.12–0.66 | c |
| 11 | 925 | 924 | Methyl n-hexanoate (methyl caproate) | Esters | a: fruit, fresh, sweet | <0.12 | b, c, d, g |
| 12 | 947 | 949 (ZB-5) | Citraconic anhydride (3-methyl-2,5-furandione) | Furans/Anhydrides | 0.73–9.00 | b | |
| 13 | 979 | 977 (VF-5MS) | 3-Hydroxybutanoic acid | Acids | <6.49 | ||
| 14 | 990 | 981 | Phenol | Aromatic compounds | a: phenol | <0.51 | b,f |
| 15 | 1001 | 996 | Ethyl hexanoate (ethyl caproate) | Esters | a: apple, peel, fruit; b: fruity, sweet; g | <0.15 | b, c, d, g |
| 16 | 992 | 996/1001/1003/1005/1006 | α-Phellandrene * | Terpenes | a: dill | <0.17 * | d |
| 17 | 992 | 987/994/970/988/992/998/990/993 | 2-Pentylfuran * | Furans | a: green bean, butter | b, d | |
| 18 | 1006 | 977/981 | Hexanoic acid (n-caproic acid) | Acids | b: sour, cheesy | 0.44–12.84 | b, c, d, g |
| 19 | 1016 | 995/1008/1013-1018 | Terpilene (α-terpinene) | Terpenes | a: lemon | <0.11 | b, d |
| 20 | 1026 | 1022 (non-polar) | Succinic anhydride std (dihydro-2,5-furandione) | Furans/Anhydrides | 0.59–3.15 | e | |
| 21 | 1028 | 1007/1020/1022/1025–1033 | Limonene | Terpenes | a: citrus, mint | <0.30 | b, c, d |
| 22 | 1038 | 1020/1033–1036/1042 | Benzyl Alcohol | Aromatic compounds | a: sweet, flower; g | 0.43–1.39 | b, c, d, g |
| 23 | 1040 | 967 (DB-1) | Itaconic anhydride * (dihydro-3-methylene-2,5-furandione) | Furans/Anhydrides | 0.90–2.75 * | e | |
| 24 | 1040 | 1014b | Pantolactone * [dihydro-3-hydroxy-4,4-dimethyl-2(3H)-furanone; 2-hydroxy-3,3-dimethyl-γ-butyrolactone] | Furans/Lactones | a: cotton candy | b | |
| 25 | 1049 | 1017/1020/1029/1033/1037–1040 | cis-Ocimene (cis-3,7-dimethyl-1,3,6-octatriene) | Terpenes | a: citrus, herb, flower | <0.16 | b |
| 26 | 1057 | 1056 | γ-Hexalactone [γ-ethyl-γ-butyrolactone, dihydro-5-ethyl-2(3H)-furanone, γ-caprolactone] | Furans/Lactones | <0.27 | b, c, d | |
| 27 | 1063 | 1065 | Mesifurane [2,5-dimethyl-4-methoxy-3(2H)-furanone; DMMF] | Furans | b: toffee, sugary, sweet; g | 0.24–1.29 | b, c, d, g |
| 28 | 1076 | 1055/1072/1097 | Furaneol [2,5-dimethyl-4-hydroxy-3(2H)-furanone; DMHF] | Furans | a: caramel; b: sweet, candy, caramellic; g | 0.42–4.55 | b, c, d, g |
| 29 | 1088 | 1067/1071/1076/1080/1085–1089 | Terpinolene [p-mentha-1,4(8)-diene] | Terpenes | <0.16 | b, d | |
| 30 | 1089 | 1065/1069/1087–1088/1091 | trans-Linalool oxide (furanoid) [trans-5-ethenyltetrahydro-α,α,5-trimethyl-2-furanmethanol] | Terpenes | a: flower | <0.18 | b, c |
| 31 | 1098 | 1084 (DB-5) | δ-Hexalactone (tetrahydro-6-methyl-2H-pyran-2-one; δ-caprolactone) | Lactones | <0.26 | b, c, d | |
| 32 | 1102 | 1079/1082/1092/1094/1097–1105 | Linalool (3,7-dimethyl-1,6-octadien-3-ol) | Terpenes | a: flower, lavender; b: floral | 0.06–0.63 | b, c, d |
| 33 | 1115 | - | Levoglucosenone * | Ketones | 0.20–1.03 * | h | |
| 34 | 1115 | 1109/1114–1119/1139 | 2-Phenylethyl Alcohol * (benzeneethanol) | Aromatic compounds | a: honey, spice, rose, lilac | b, c, d | |
| 35 | 1173 | 1163 (DB-5MS) | 4-Ethylphenol | Aromatic compounds | a: must | 0.14–0.87 | |
| 36 | 1180 | 1159/1178 | Benzoic Acid | Aromatic compounds | a: urine | 0.09–0.45 | b, c, d |
| 37 | 1192 | 1187/1192 | 1-Dodecene | Alkenes | <0.20 | ||
| 38 | 1216 | 1197 (DB-5) | 1,2-Benzenediol (pyrocatechol; 2-hydroxyphenol; catechol) | Aromatic compounds | <0.52 | i | |
| 39 | 1228 | 1223–1224 (SPB-5) | Coumaran (2,3-dihydrobenzofuran) | Aromatic compounds | e: green tea | 1.35–5.62 | e |
| 40 | 1319 | 1326 (VF-5MS) | Salicylic acid std * (o-hydroxybenzoic acid; phenol-2-carboxylic acid) | Aromatic compounds | 0.13–1.30 * | c, d | |
| 41 | 1319 | 1314–1318 | 2-Methoxy-4-vinylphenol * | Aromatic compounds | b, c, d | ||
| 42 | 1386 | 1343b | cis-Cinnamic acid [(Z)-3-phenyl-2-propenoic acid) | Aromatic compounds/Phenylpropanoids | 0.07–0.47 | b, d, i | |
| 43 | 1442 | 1432 | Tyrosol (4-hydroxyphenylethyl alcohol) | Aromatic compounds | <0.88 | b, c, i | |
| 44 | 1471 | 1450/1462 (DB-1) | trans-Cinnamic acid std [(E)-3-phenyl-2-propenoic acid] | Aromatic compounds/Phenylpropanoids | a: honey | 19.41–37.28 | b, c, d, i |
| 45 | 1472 | 1470 | γ-Decalactone [5-hexyldihydro-2(3H)-furanone] | Furans/Lactones | a: peach, fat; b: sweet, peach, lactonic; g | <2.49 | b, c, d, g |
| 46 | 1553 | 1491 (LM-5) | Levoglucosan * (1,6-anhydro-β-D-glucopyranose) | Others | 0.30–7.88 * | ||
| 47 | 1553 | 1538 (VF-5MS) | p-Salicylic acid * std (4-hydroxybenzoic acid; paraben-acid; 4-carboxyphenol ) | Aromatic compounds | |||
| 48 | 1566 | 1544/1561–1563/1568/1569 | trans-Nerolidol [(E)-3,7,11-trimethyldodeca-1,6,10-trien-3-ol] | Terpenes | a: wax; g | <0.57 | b, c, d, g |
| 49 | 1571 | 1559/1566/−1568/1570/1573/1576 | Dodecanoic acid (lauric acid) | Acids/Fatty acids | <0.23 | b, c, d | |
| 50 | 1659 | 1659 | Homovanilic acid * (4-hydroxy-3-methoxybenzeneacetic acid; vanilacetic acid) | Aromatic compounds | 1.08–2.69 * | j | |
| 51 | 1659 | 1658 | Bisabolol oxide II * | Terpenes | |||
| 52 | 1683 | 1675 | γ-Dodecalactone [dihydro-5-octyl-2(3H)-furanone] | Furans/Lactones | a: sweet, flower, fruit; g | <0.24 | b, c, d, g |
| 53 | 1765 | 1759/1767/1769/1770/1777/1780/1787/1790 | Tetradecanoic acid (myristic acid) | Acids/Fatty acids | <0.53 | b, c, d | |
| 54 | 1864 | 1869/1878 | Pentadecanoic acid | Acids/Fatty acids | <0.44 | c | |
| 55 | 1881 | 1871/1879/1882 | 1-Hexadecanol (cetyl alcohol) | Alcohols | a: flower, wax | <0.33 | |
| 56 | 1888 | 1881 (DB-1) | Ferulic acid [(E)-4-hydroxy-3-methoxycinnamic acid] | Aromatic compounds/Phenylpropanoids | <0.31 | j, l | |
| 57 | 1944 | 1953 (HP-5) | Z-11-Hexadecenoic acid | Acids/Fatty acids | <0.89 | ||
| 58 | 1968 | 1962/1963/1969/1971/1972/1975/1977/1978/1991/1995/2003 | n-Hexadecanoic acid (palmitic acid) | Acids/Fatty acids | g | 0.66–3.70 | b, c, d, g |
| 59 | 2086 (HP-5) | Heptadecanoic acid (margaric acid) | Acids/Fatty acids | <0.11 | c, d | ||
| 60 | 2095/2104/2130/2140/2144/2170 | Linoleic acid [(9Z,12Z)-octadeca-9,12-dienoic acid] | Acids/Fatty acids | <0.32 | b, c, d | ||
| 61 | 2102/2141/2152/2175 | Oleic Acid [(Z)-octadec-9-enoic acid] | Acids/Fatty acids | a: fat | <1.65 | b, c, d | |
| 62 | 2172/2178/2180/2188 | Stearic acid (Octadecanoic acid) | Acids/Fatty acids | 0.16–1.60 | c |
| No | Compound | F Value (Significance) | ||
|---|---|---|---|---|
| Genotype (G) | Harvest Time (T) | G × T | ||
| 4 | Butanoic acid | 4.528 * | 3.204 | 1.238 |
| 7 | Maleic anhydride | 5.622 ** | 96.337 *** | 3.126 * |
| 8 | 2-Methylbutanoic acid | 27.251 *** | 3.782 * | 4.455 ** |
| 9 | Styrene | 0.761 | 10.253 ** | 0.772 |
| 12 | Citraconic anhydride | 0.511 | 41.542 *** | 1.421 |
| 13 | 3-Hydroxybutanoic acid | 14.102 *** | 3.226 | 1.885 |
| 18 | Hexanoic acid | 6.905 ** | 2.516 | 1.162 |
| 20 | Succinic anhydride | 18.202 *** | 71.895 *** | 1.754 |
| 22 | Benzyl Alcohol | 2.106 | 2.725 | 1.245 |
| 27 | Mesifurane | 4.238 * | 9.043 ** | 2.504 * |
| 28 | Furaneol | 10.051 *** | 0.678 | 2.087 |
| 32 | Linalool | 35.484 *** | 5.703 ** | 4.911 ** |
| 39 | Coumaran | 2.337 | 36.473 *** | 1.749 |
| 44 | trans-Cinnamic acid | 1.845 | 7.947 ** | 0.774 |
| 45 | γ-Decalactone | 15.220 *** | 0.088 | 2.019 |
| 48 | trans-Nerolidol | 10.032 *** | 21.210 *** | 8.688 *** |
| 58 | n-Hexadecanoic acid | 1.814 | 13.736 *** | 3.986 ** |
| 61 | Oleic Acid | 1.830 | 11.371 *** | 2.079 |
| 62 | Stearic acid | 0.815 | 5.865 * | 1.503 |
| I | Esters | 9.828 *** | 28.988 *** | 9.912 *** |
| II | Terpenes | 57.438 *** | 33.892 *** | 26.161 *** |
| III | Phenylpropanes | 2.761 | 15.185 *** | 1.226 |
| IV | Aromatics | 1.411 | 17.622 *** | 1.183 |
| V | Short-chain Acids | 6.215 ** | 8.228 ** | 2.515 * |
| VI | Fatty Acids | 2.079 | 16.449 *** | 4.047 ** |
| VII | Alcohols | 13.772 *** | 8.754 ** | 2.426 |
| VIII | Alkenes | 3.289 * | 78.197 *** | 4.270 ** |
| IX | Furans/Lactones | 0.450 | 62.117 *** | 1.601 |
| X | Lactones | 7.724 ** | 1.354 | 0.650 |
| XI | Anhydrides | 1.558 | 79.254 *** | 1.484 |
| No | Gene | F Value (Significance) | ||
|---|---|---|---|---|
| Genotype (G) | Harvest Time (T) | G × T | ||
| 1 | SAAT | 86.308 ** | 46.455 ** | 7.211 ** |
| 2 | FaFAD1 | 440.261 ** | 164.941 ** | 32.417 ** |
| 3 | FaNES1 | 6.338 * | 39.625 ** | 8.138 ** |
| 4 | FaEGS2 | 162.835 ** | 0.280 | 8.875 ** |
| 5 | FaMYB10 | 19.579 ** | 119.515 ** | 5.430 ** |
| 6 | FaEOBII | 107.590 ** | 102.887 ** | 202.098 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonardou, V.K.; Doudoumis, E.; Tsormpatsidis, E.; Vysini, E.; Papanikolopoulos, T.; Papasotiropoulos, V.; Lamari, F.N. Quality Traits, Volatile Organic Compounds, and Expression of Key Flavor Genes in Strawberry Genotypes over Harvest Period. Int. J. Mol. Sci. 2021, 22, 13499. https://doi.org/10.3390/ijms222413499
Leonardou VK, Doudoumis E, Tsormpatsidis E, Vysini E, Papanikolopoulos T, Papasotiropoulos V, Lamari FN. Quality Traits, Volatile Organic Compounds, and Expression of Key Flavor Genes in Strawberry Genotypes over Harvest Period. International Journal of Molecular Sciences. 2021; 22(24):13499. https://doi.org/10.3390/ijms222413499
Chicago/Turabian StyleLeonardou, Varvara K., Evangelos Doudoumis, Evangelos Tsormpatsidis, Eleni Vysini, Theofanis Papanikolopoulos, Vasileios Papasotiropoulos, and Fotini N. Lamari. 2021. "Quality Traits, Volatile Organic Compounds, and Expression of Key Flavor Genes in Strawberry Genotypes over Harvest Period" International Journal of Molecular Sciences 22, no. 24: 13499. https://doi.org/10.3390/ijms222413499
APA StyleLeonardou, V. K., Doudoumis, E., Tsormpatsidis, E., Vysini, E., Papanikolopoulos, T., Papasotiropoulos, V., & Lamari, F. N. (2021). Quality Traits, Volatile Organic Compounds, and Expression of Key Flavor Genes in Strawberry Genotypes over Harvest Period. International Journal of Molecular Sciences, 22(24), 13499. https://doi.org/10.3390/ijms222413499








