Studying the Function of Phytoplasma Effector Proteins Using a Chemical-Inducible Expression System in Transgenic Plants
Abstract
:1. Introduction
2. Results
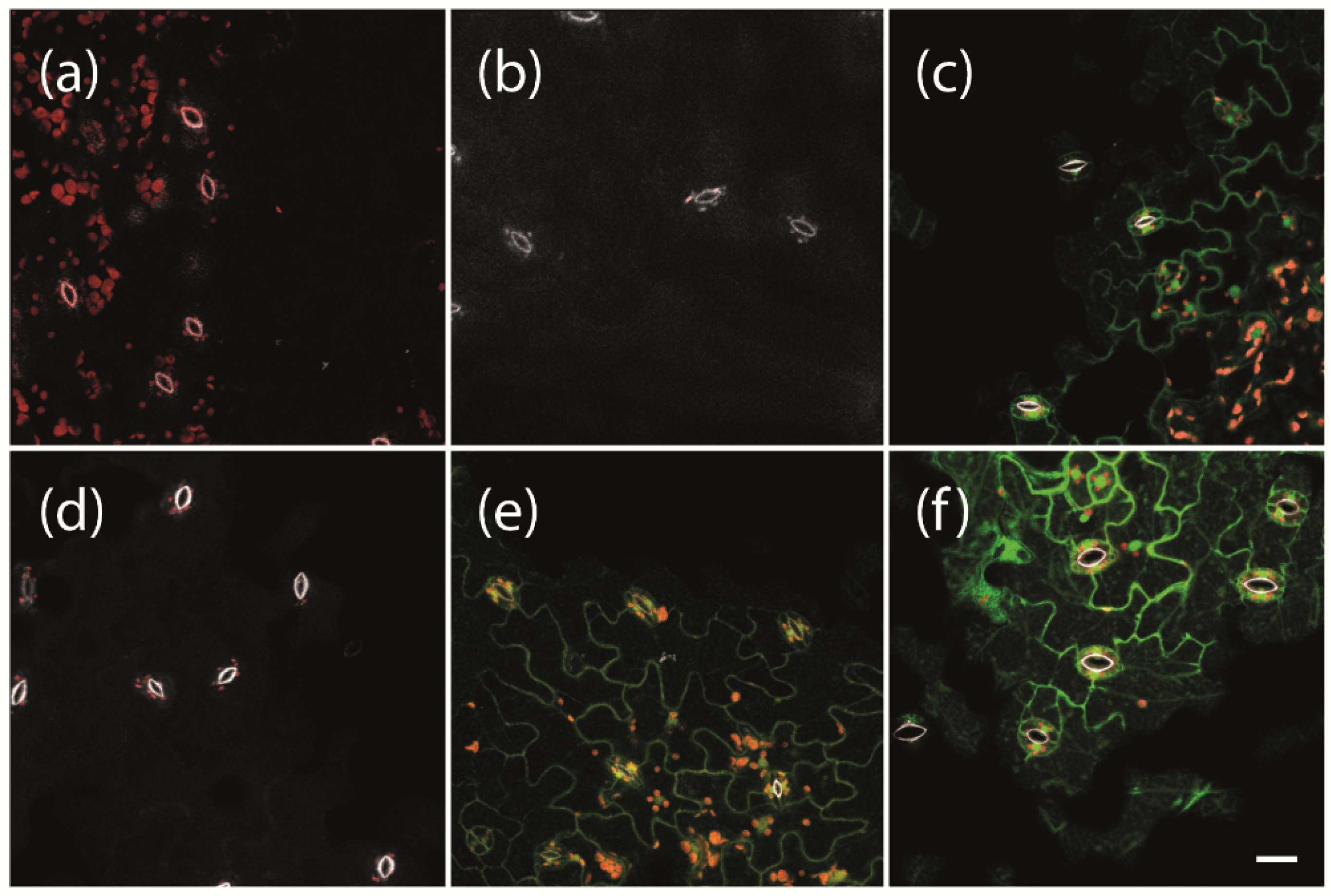
2.1. Establishment of an Inducible Expression System for Phytoplasma Effector Proteins
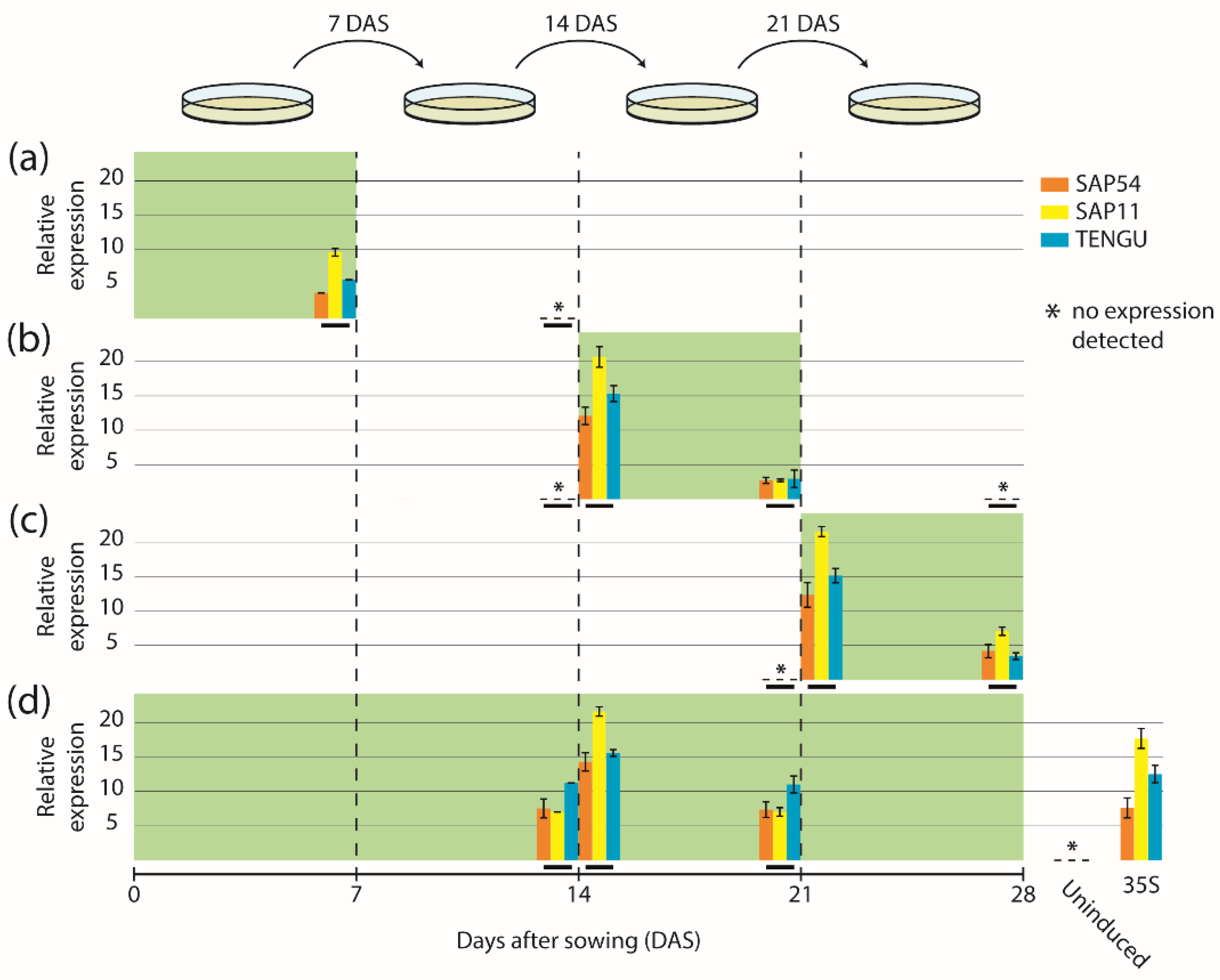
2.2. Comparative Time-Course Analyses of SAP54, SAP11, and TENGU Induction
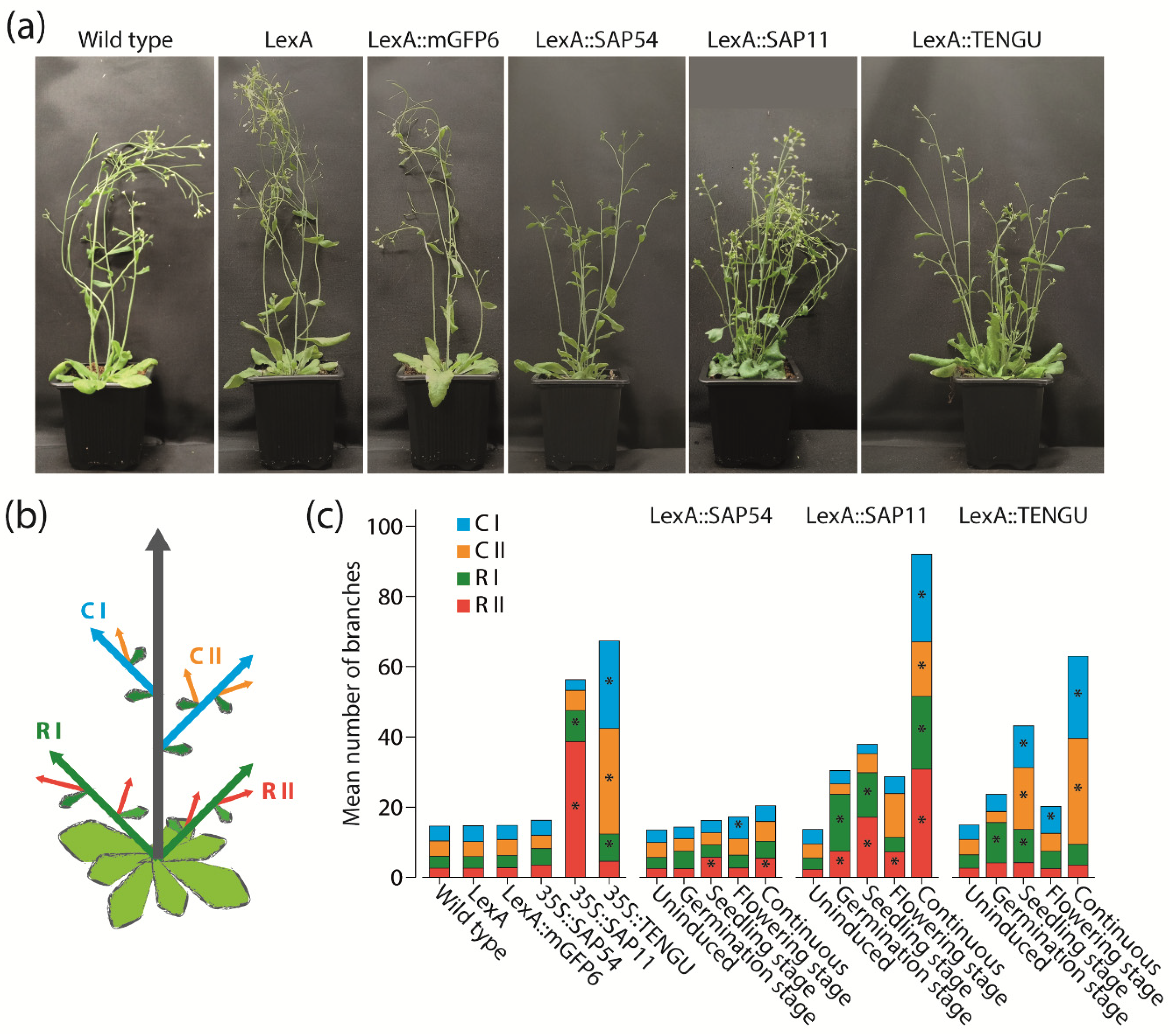
2.3. Timely Induction of Effector Proteins Affects Plant Habit of Arabidopsis
2.4. Timely Induction of Effector Protein Expression Alters Arabidopsis Flower Development
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Generation of Plasmids for Inducible Effector Protein Expression
4.3. Generation of Constitutive Expression Vectors
4.4. Gene Expression Analysis via Quantitative RT-PCR
4.5. Morphological Characterization
4.6. Confocal Laser Scanning Microscopy
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hogenhout, S.A.; Oshima, K.; Ammar, D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Beanland, L. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef]
- Power, A.G. Host plant dispersion, leafhopper movement and disease transmission. Ecol. Entomol. 1992, 17, 63–68. [Google Scholar] [CrossRef]
- Tscharntke, T.; Brandl, R. Plant insect interactions in fragmented landscapes. Annu. Rev. Entomol. 2004, 49, 405–430. [Google Scholar] [CrossRef]
- Bai, X.; Correa, V.R.; Toruño, T.Y.; Ammar, E.D.; Kamoun, S.; Hogenhout, S.A. AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant Microbe Interact. 2009, 22, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maejima, K.; Oshima, K.; Namba, S. Exploring the phytoplasmas, plant pathogenic bacteria. J. Gen. Plant Pathol. 2014, 80, 210–221. [Google Scholar] [CrossRef] [Green Version]
- MacLean, A.M.; Sugio, A.; Makarova, O.V.; Findlay, K.C.; Grieve, V.M.; Toth, R.; Nicolaisen, M.; Hogenhout, S.A. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 2011, 157, 831–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLean, A.M.; Orlovskis, Z.; Kowitwanich, K.; Zdziarska, A.M.; Angenent, G.C.; Immink, R.G.; Hogenhout, S.A. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014, 12, e1001835. [Google Scholar] [CrossRef]
- Maejima, K.; Iwai, R.; Himeno, M.; Komatsu, K.; Kitazawa, Y.; Fujita, N.; Ishikawa, K.; Fukuoka, M.; Minato, N.; Yamaji, Y.; et al. Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J. 2014, 78, 541–554. [Google Scholar] [CrossRef]
- Maejima, K.; Kitazawa, Y.; Tomomitsu, T.; Yusa, A.; Neriya, Y.; Himeno, M.; Yamaji, Y.; Oshima, K.; Namba, S. Degradation of class E MADS-domain transcription factors in Arabidopsis by a phytoplasmal effector, phyllogen. Plant Signal. Behav. 2015, 10, e1042635. [Google Scholar] [CrossRef] [Green Version]
- Sugio, A.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugio, A.; Maclean, A.M.; Hogenhout, S.A. The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. N. Phytol. 2014, 202, 838–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Yang, H.; Yin, Z.; Liu, W.; Sun, L.; Wu, Y. Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol. 2018, 19, 2623–2634. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2009, 15, 31–39. [Google Scholar] [CrossRef]
- Dhaka, N.; Bhardwaj, V.; Sharma, M.K.; Sharma, R. Evolving tale of TCPs: New paradigms and old lacunae. Front. Plant Sci. 2017, 8, 479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshi, A.; Oshima, K.; Kakizawa, S.; Ishii, Y.; Ozeki, J.; Hashimoto, M.; Komatsu, K.; Kagiwada, S.; Yamaji, Y.; Namba, S. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. USA 2009, 106, 6416–6421. [Google Scholar] [CrossRef] [Green Version]
- Minato, N.; Himeno, M.; Hoshi, A.; Maejima, K.; Komatsu, K.; Takebayashi, Y.; Kasahara, H.; Yusa, A.; Yamaji, Y.; Oshima, K.; et al. The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep. 2014, 4, 7399. [Google Scholar] [CrossRef] [Green Version]
- Sugawara, K.; Honma, Y.; Komatsu, K.; Himeno, M.; Oshima, K.; Namba, S. The alteration of plant morphology by small peptides released from the proteolytic processing of the bacterial peptide TENGU. Plant Physiol. 2013, 162, 2005–2014. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Li, Y.; Chen, W.; Yang, H.; Zhang, P.; Wu, Y. Identification of wheat blue dwarf phytoplasma effectors targeting plant proliferation and defence responses. Plant Pathol. 2018, 67, 603–609. [Google Scholar] [CrossRef]
- Pecher, P.; Moro, G.; Canale, M.C.; Capdevielle, S.; Singh, A.; MacLean, A.; Sugio, A.; Kuo, C.; Lopes, J.R.S.; Hogenhout, S.A. Phytoplasma SAP11 effector destabilization of TCP transcription factors differentially impact development and defence of Arabidopsis versus maize. PLoS Pathog. 2019, 15, e1008035. [Google Scholar] [CrossRef] [Green Version]
- Iwabuchi, N.; Kitazawa, Y.; Maejima, K.; Koinuma, H.; Miyazaki, A.; Matsumoto, O.; Suzuki, T.; Nijo, T.; Oshima, K.; Namba, S.; et al. Functional variation in phyllogen, a phyllody-inducing phytoplasma effector family, attributable to a single amino acid polymorphism. Mol. Plant Pathol. 2020, 21, 1322–1336. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Davis, R.E.; Nuss, D.L.; Zhao, Y. Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc. Natl. Acad. Sci. USA 2013, 110, 19149–19154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Niu, Q.W.; Chua, N.H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000, 24, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Siligato, R.; Wang, X.; Yadav, S.R.; Lehesranta, S.; Ma, G.; Ursache, R.; Sevilem, I.; Zhang, J.; Gorte, M.; Prasad, K.; et al. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 2016, 170, 627–641. [Google Scholar] [CrossRef]
- Orlovskis, Z.; Hogenhout, S.A. A bacterial parasite effector mediates insect vector attraction in host plants independently of developmental changes. Front. Plant Sci. 2016, 7, 885. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Aurin, M.B.; Haupt, M.; Görlach, M.; Rümpler, F.; Theißen, G. Structural requirements of the phytoplasma effector protein SAP54 for causing homeotic transformation of floral organs. Mol. Plant Microbe Interact. 2020, 33, 1129–1141. [Google Scholar] [CrossRef]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; De Meyer, B.; Hilson, P. Modular cloning in plant cells. Trends Plant Sci. 2005, 10, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 2008, 1, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lilly, S.T.; Drummond, R.S.M.; Pearson, M.N.; MacDiarmid, R.M. Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol. Plant Microbe Interact. 2011, 24, 294–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekkers, B.J.; Willems, L.; Bassel, G.W.; van Bolderen-Veldkamp, R.P.; Ligterink, W.; Hilhorst, H.W.; Bentsink, L. Identification of reference genes for RT–qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 2012, 53, 28–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omenge, K.M.; Rümpler, F.; Kathalingam, S.S.; Furch, A.C.U.; Theißen, G. Studying the Function of Phytoplasma Effector Proteins Using a Chemical-Inducible Expression System in Transgenic Plants. Int. J. Mol. Sci. 2021, 22, 13582. https://doi.org/10.3390/ijms222413582
Omenge KM, Rümpler F, Kathalingam SS, Furch ACU, Theißen G. Studying the Function of Phytoplasma Effector Proteins Using a Chemical-Inducible Expression System in Transgenic Plants. International Journal of Molecular Sciences. 2021; 22(24):13582. https://doi.org/10.3390/ijms222413582
Chicago/Turabian StyleOmenge, Keziah M., Florian Rümpler, Subha Suvetha Kathalingam, Alexandra C. U. Furch, and Günter Theißen. 2021. "Studying the Function of Phytoplasma Effector Proteins Using a Chemical-Inducible Expression System in Transgenic Plants" International Journal of Molecular Sciences 22, no. 24: 13582. https://doi.org/10.3390/ijms222413582
APA StyleOmenge, K. M., Rümpler, F., Kathalingam, S. S., Furch, A. C. U., & Theißen, G. (2021). Studying the Function of Phytoplasma Effector Proteins Using a Chemical-Inducible Expression System in Transgenic Plants. International Journal of Molecular Sciences, 22(24), 13582. https://doi.org/10.3390/ijms222413582






