Screening of Self-Assembling of Collagen IV Fragments into Stable Structures Potentially Useful in Regenerative Medicine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of Collagen IV Fragments Reconstructing the Inner Sphere of the Native Protein
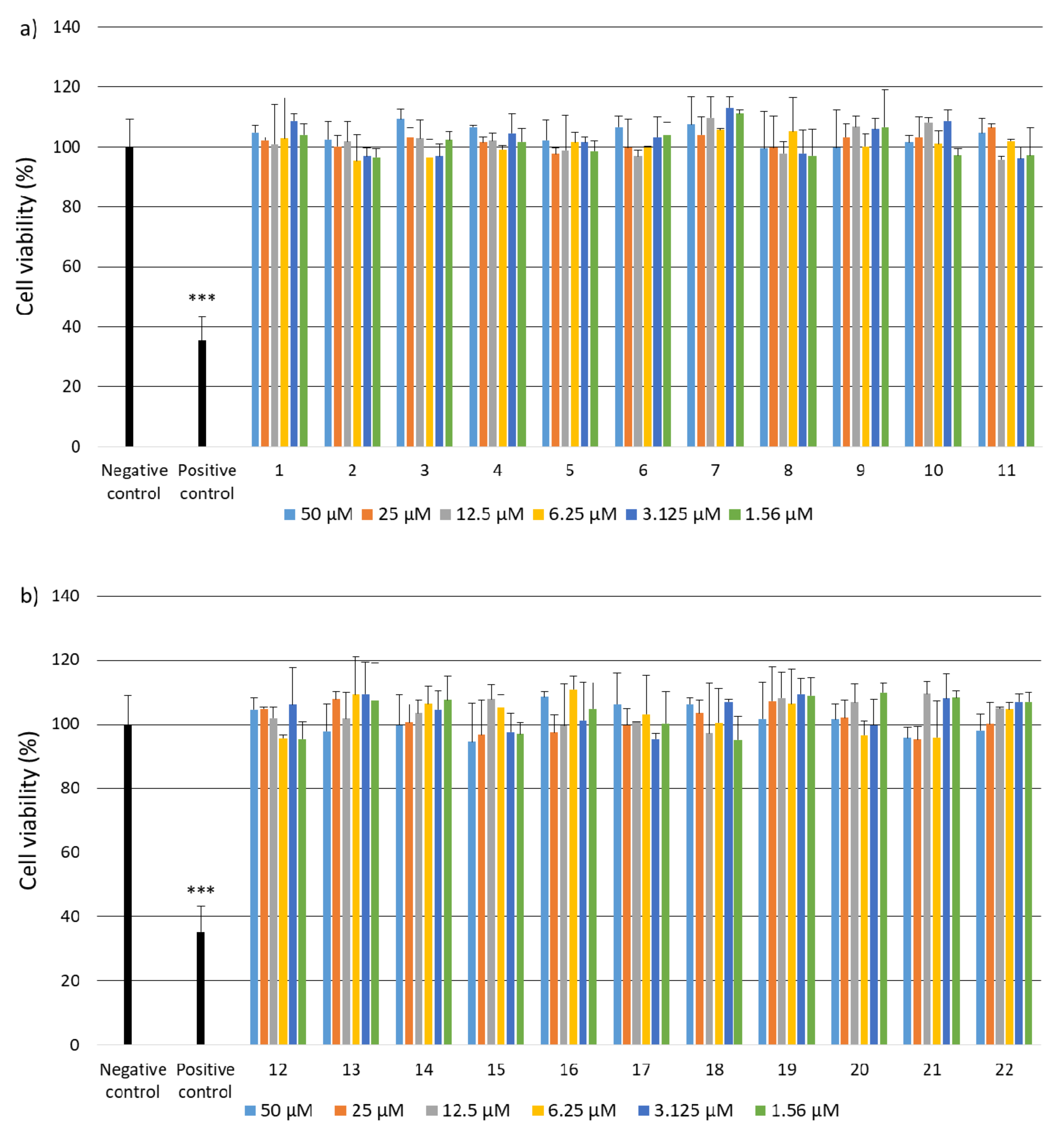
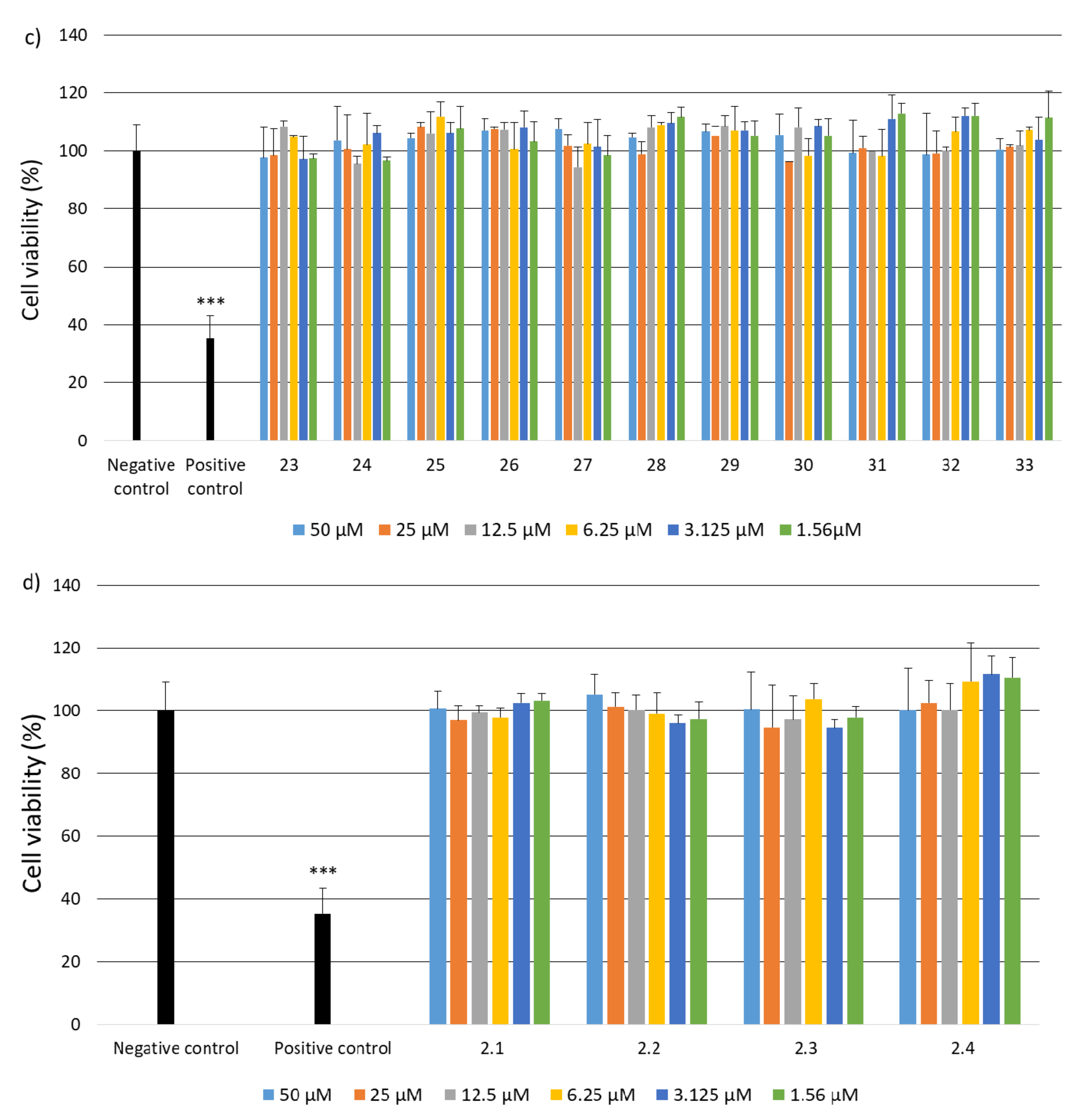
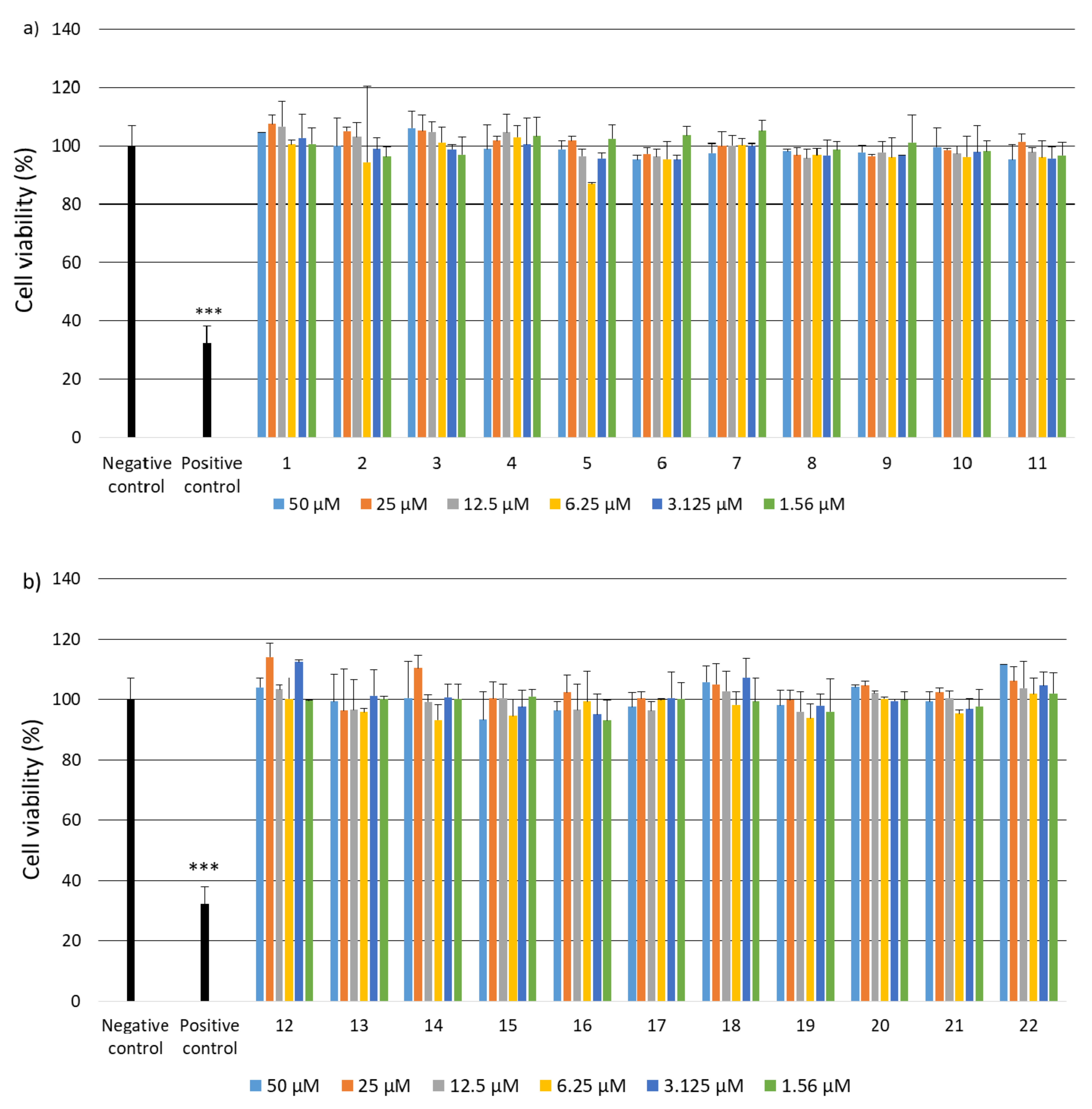
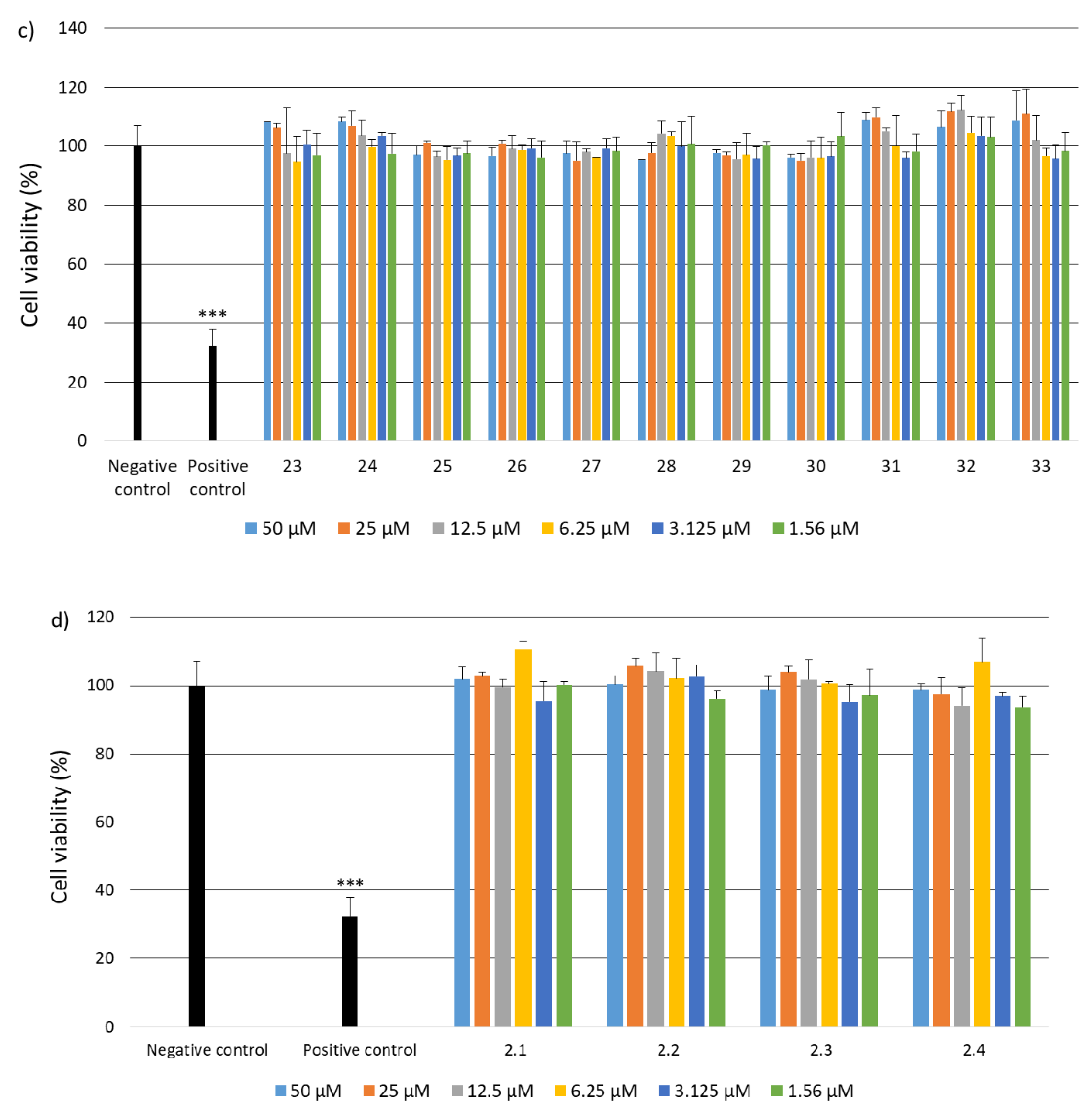
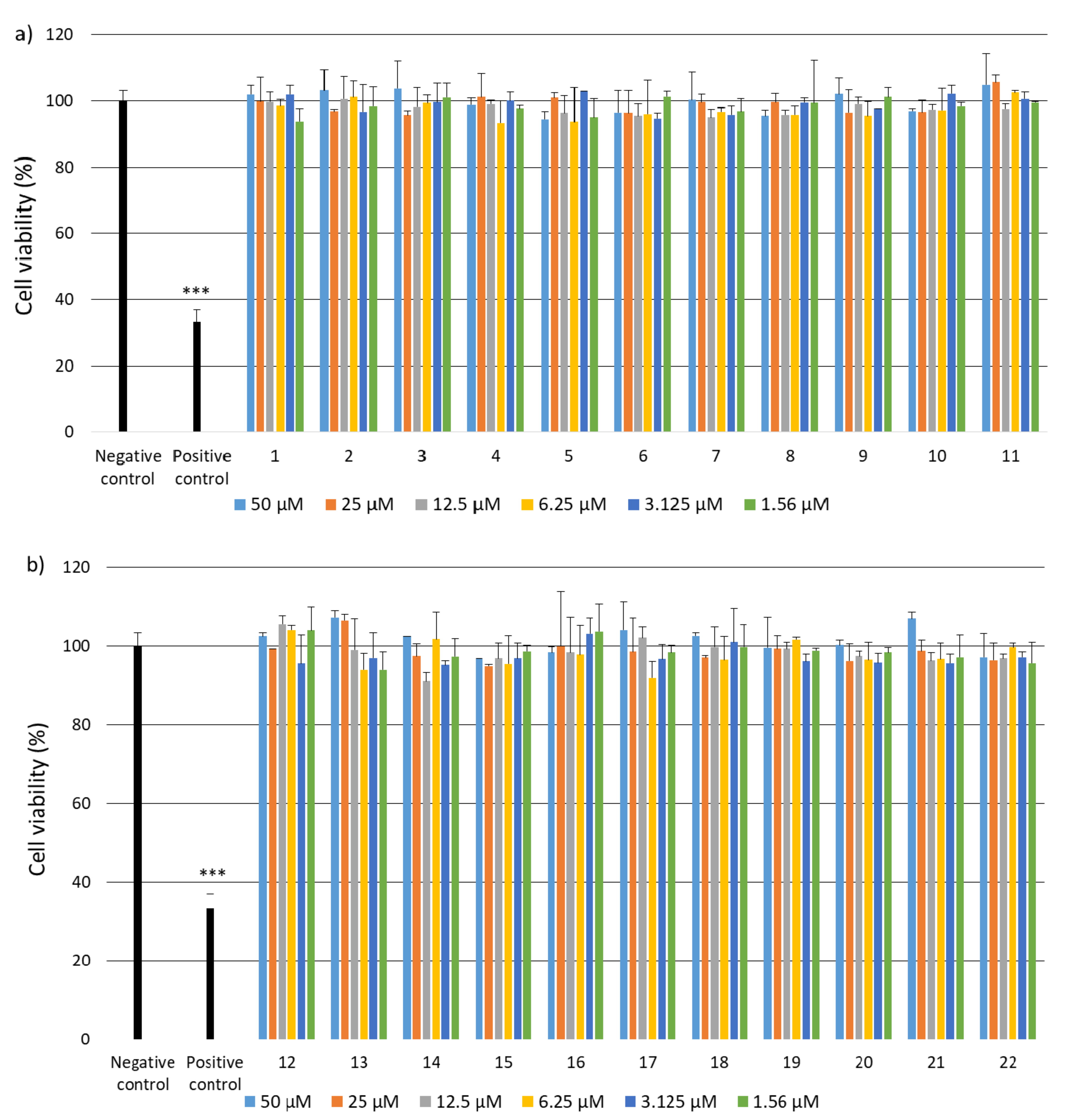
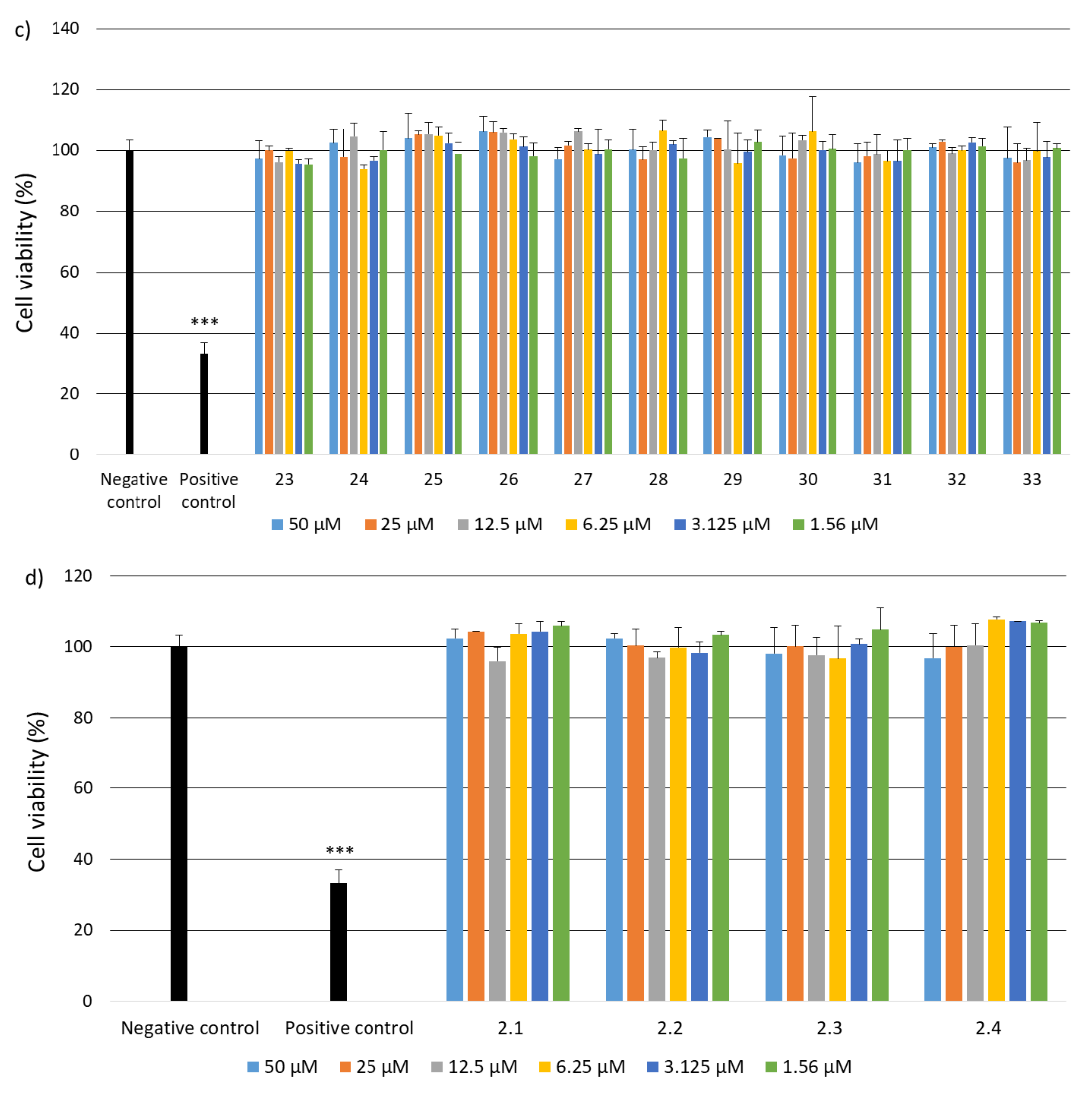
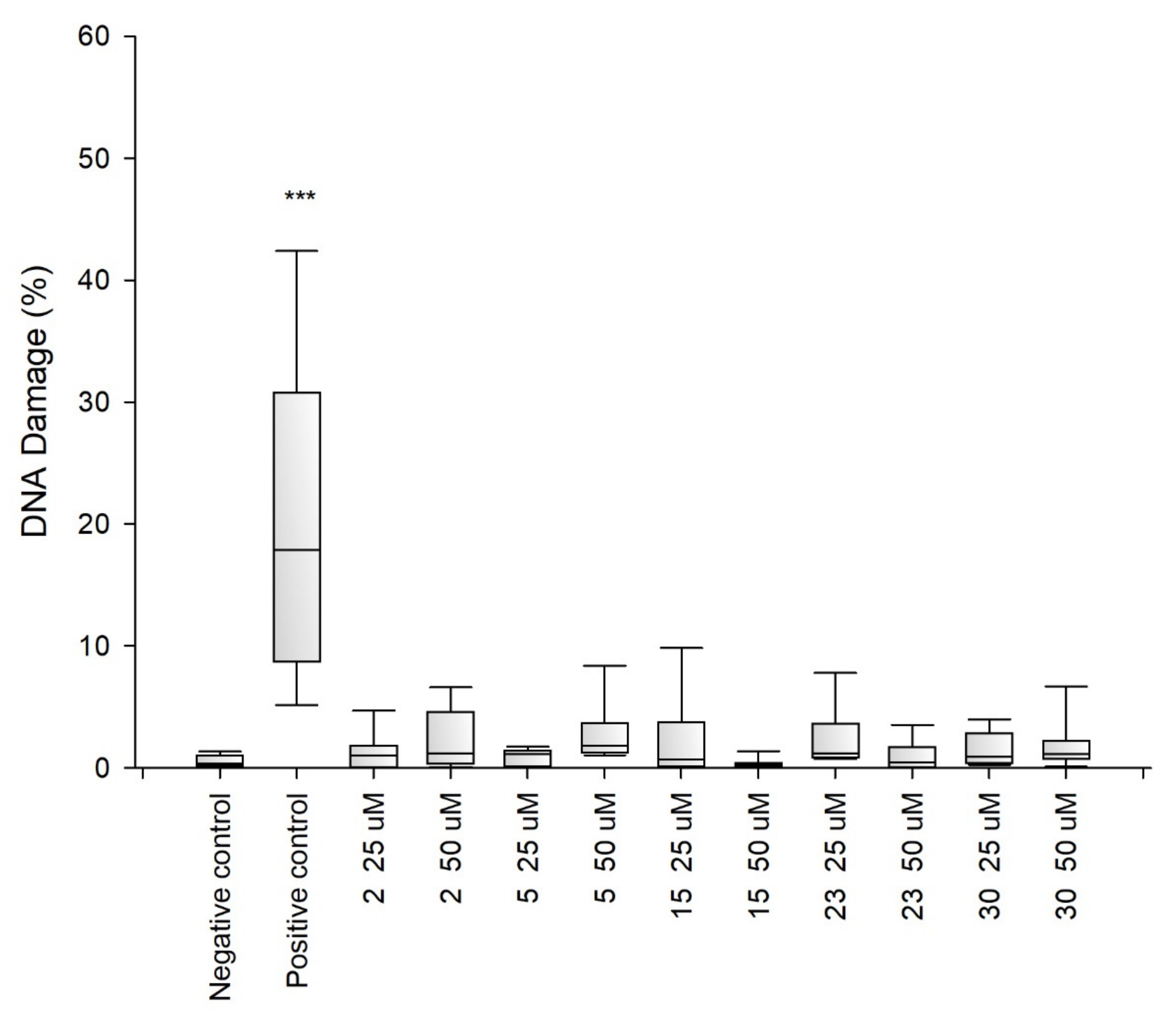
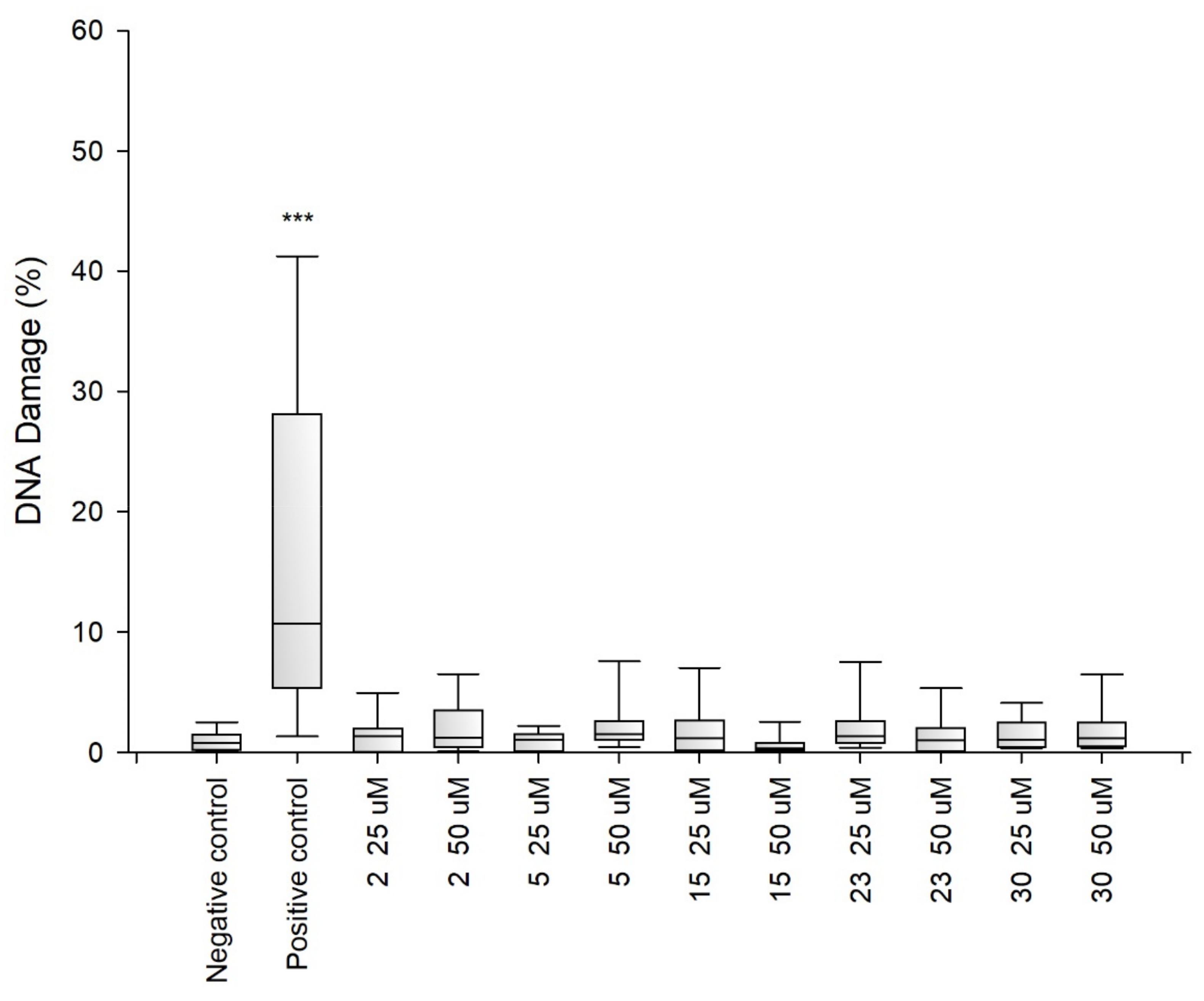
2.2. Research on Biological Activity of Collagen IV Fragments
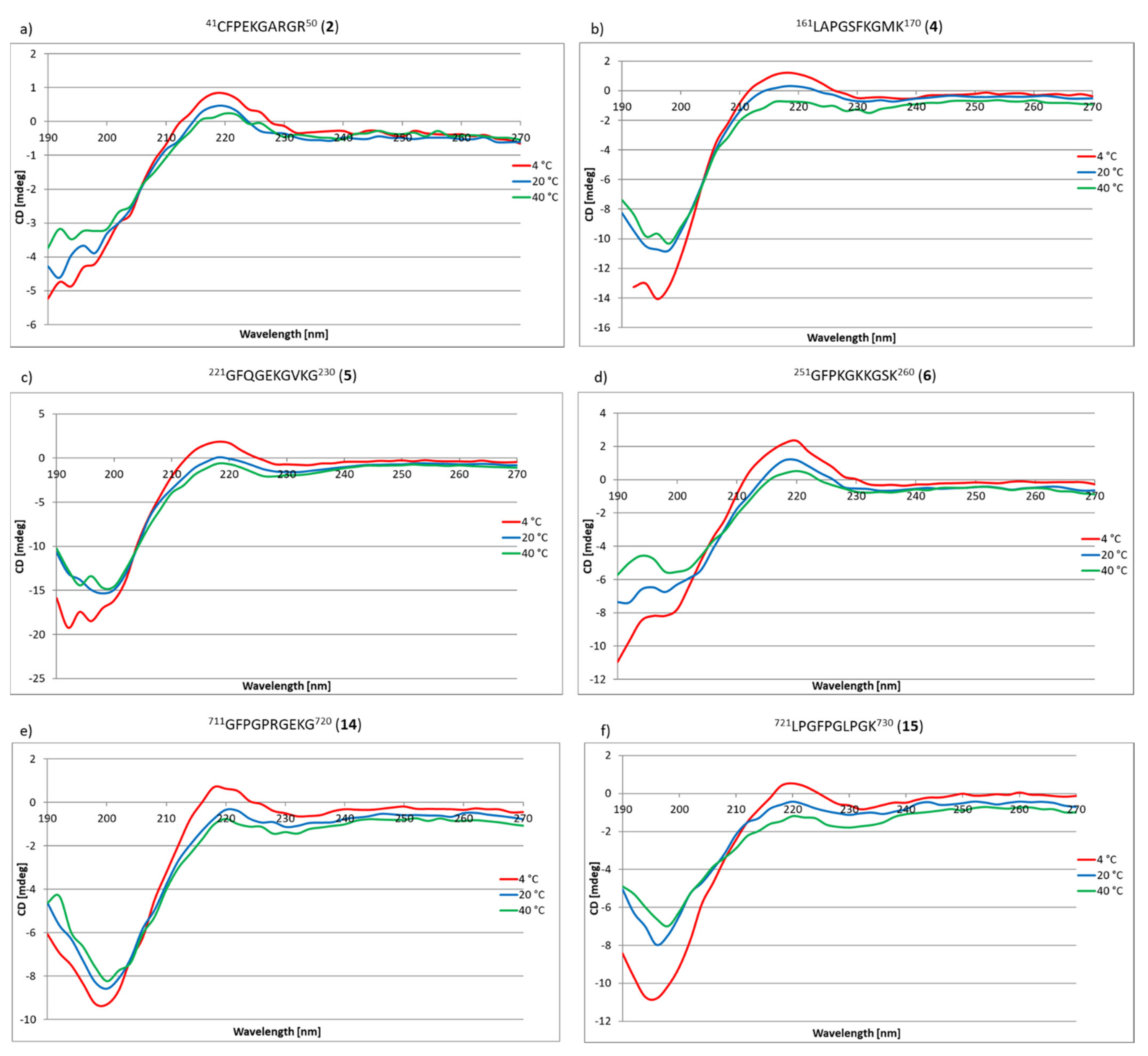
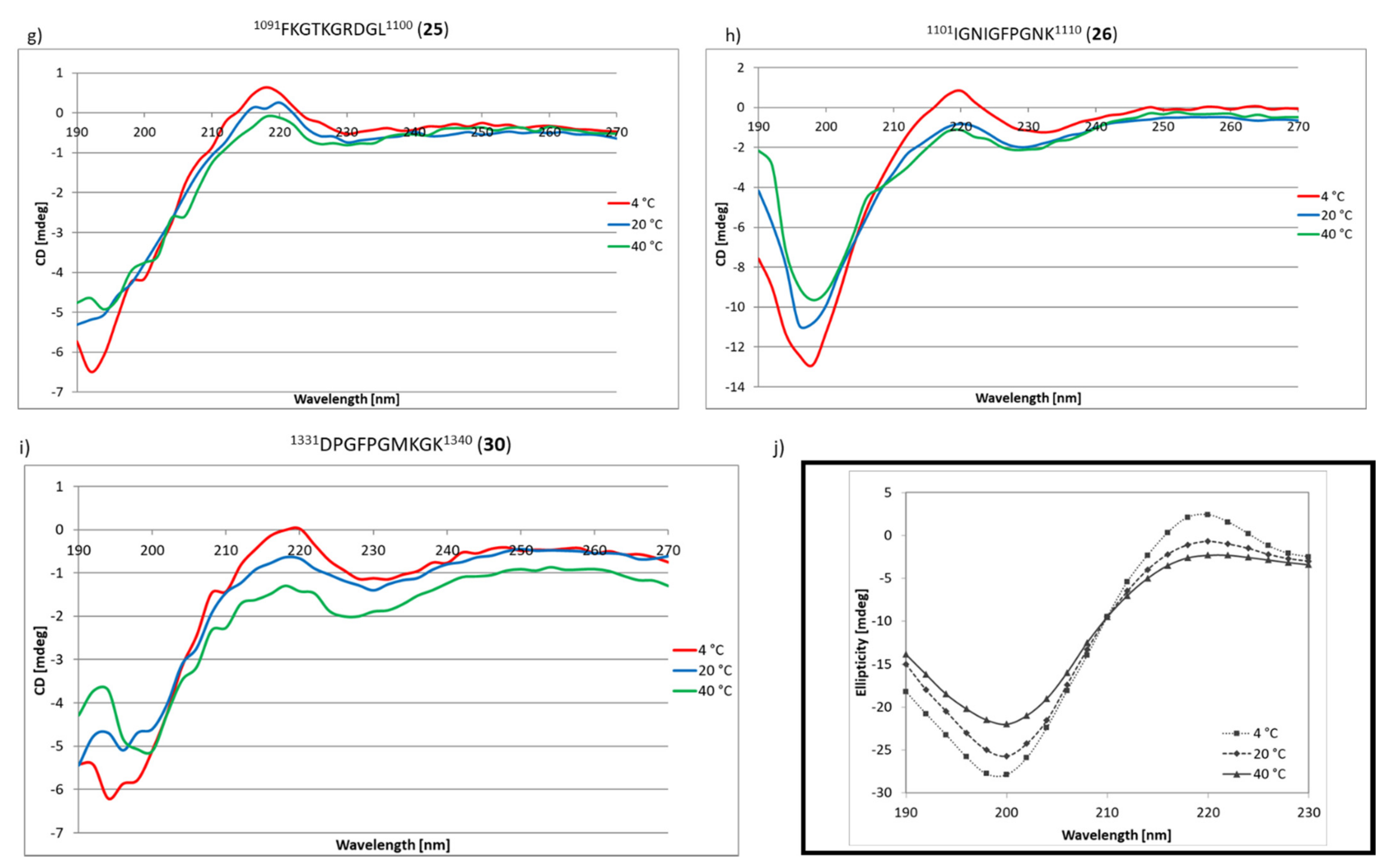
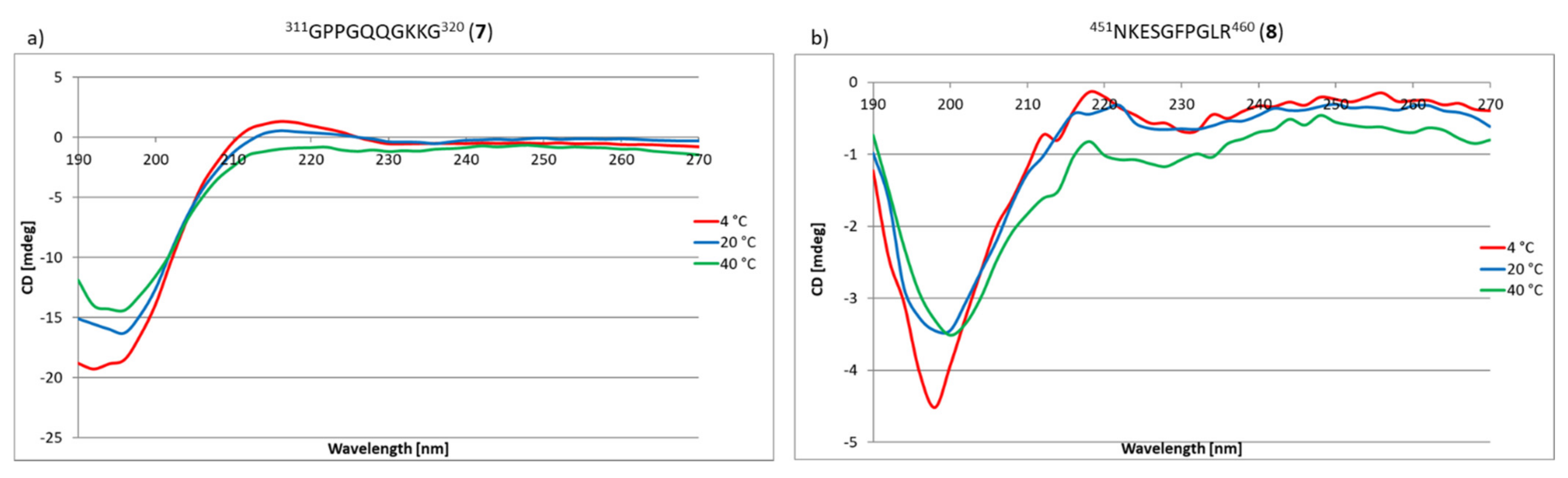
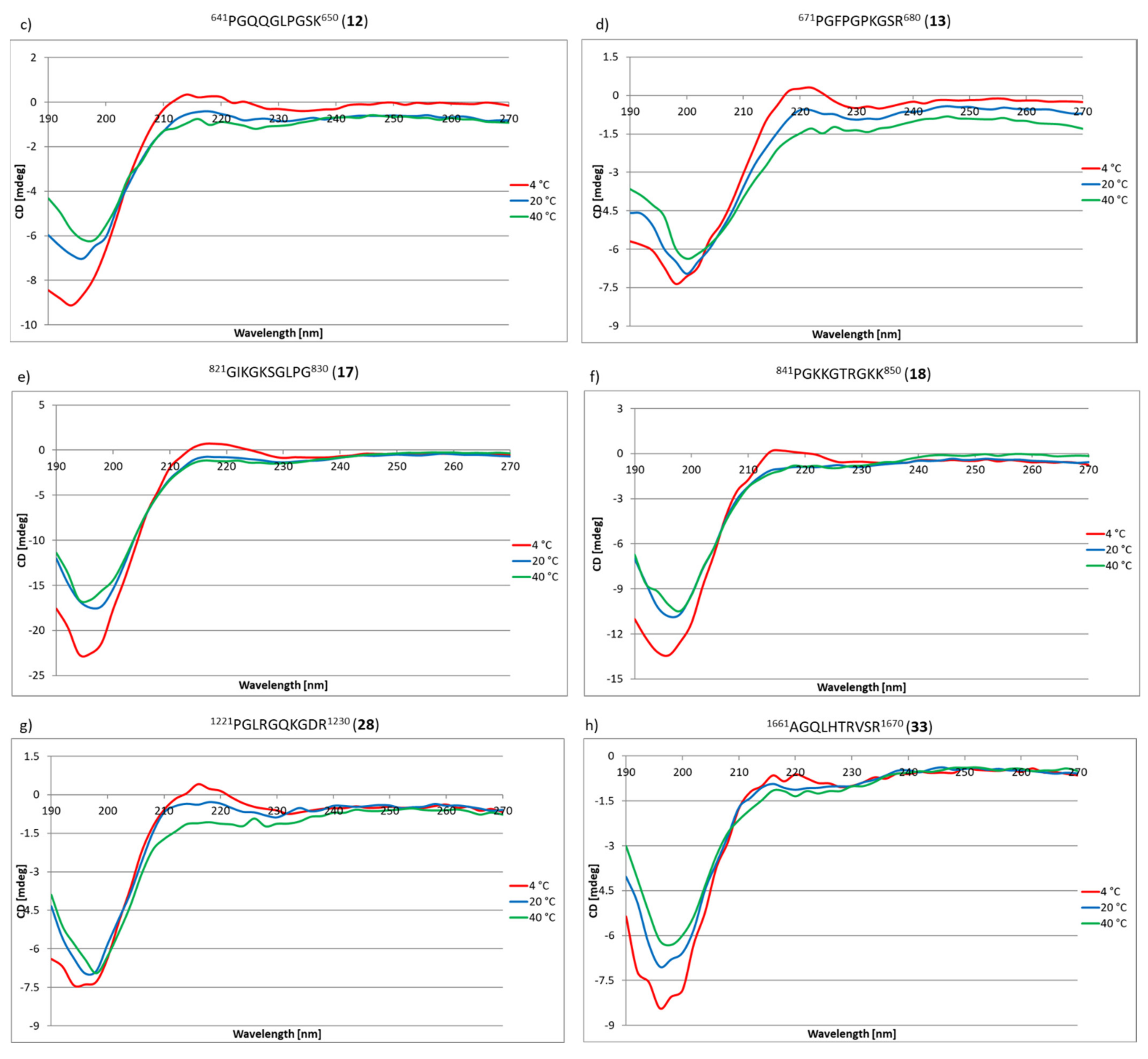
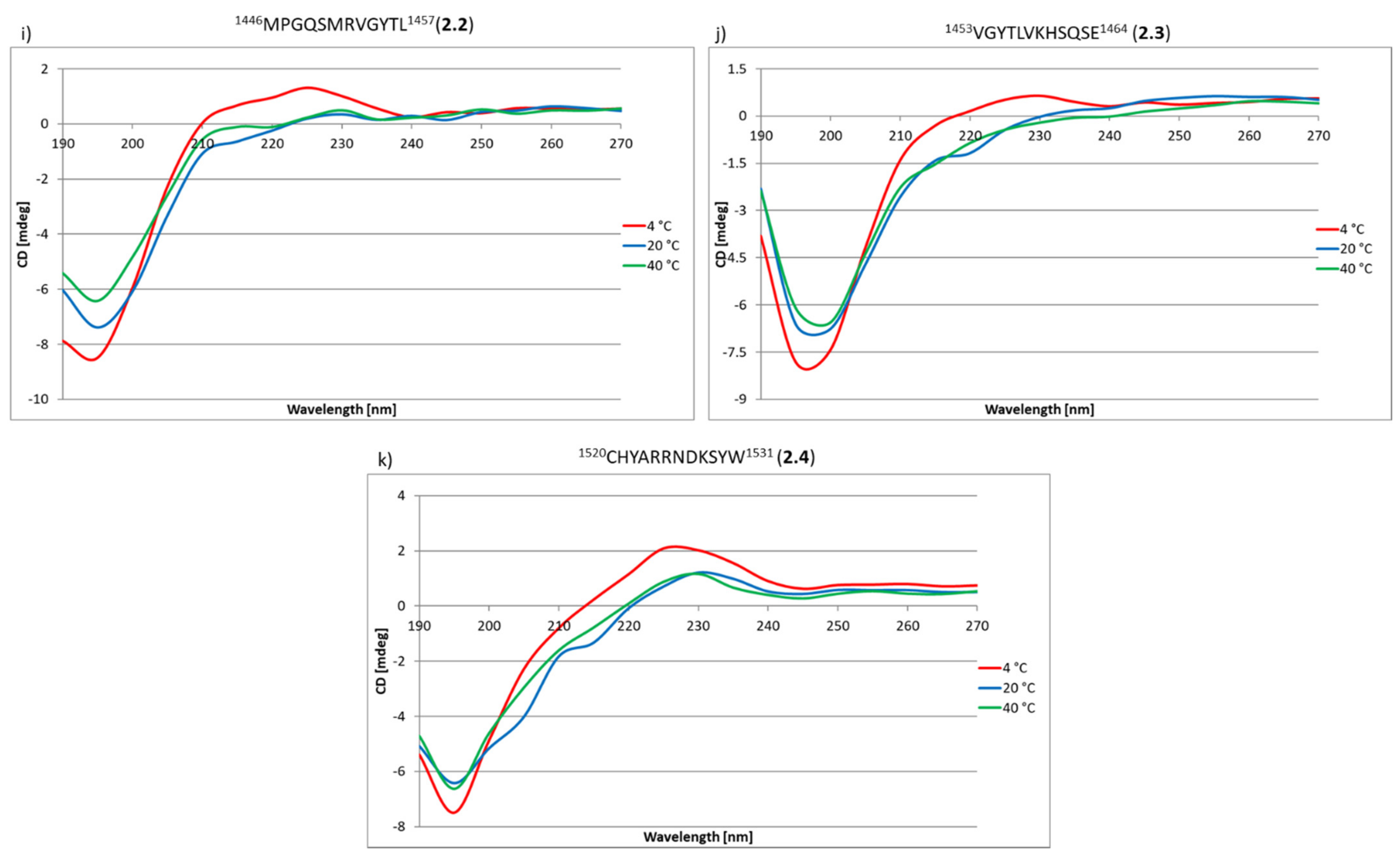
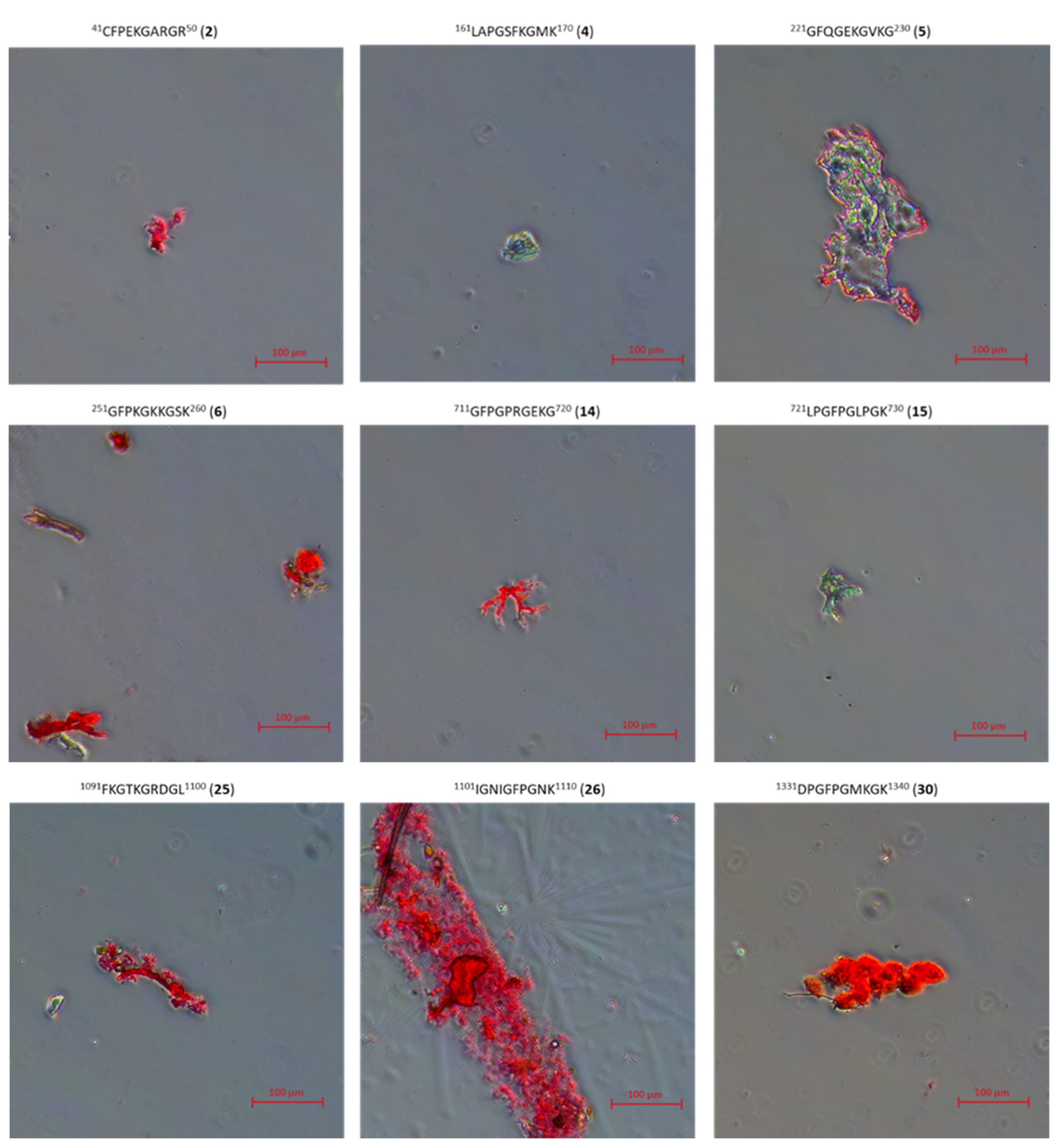
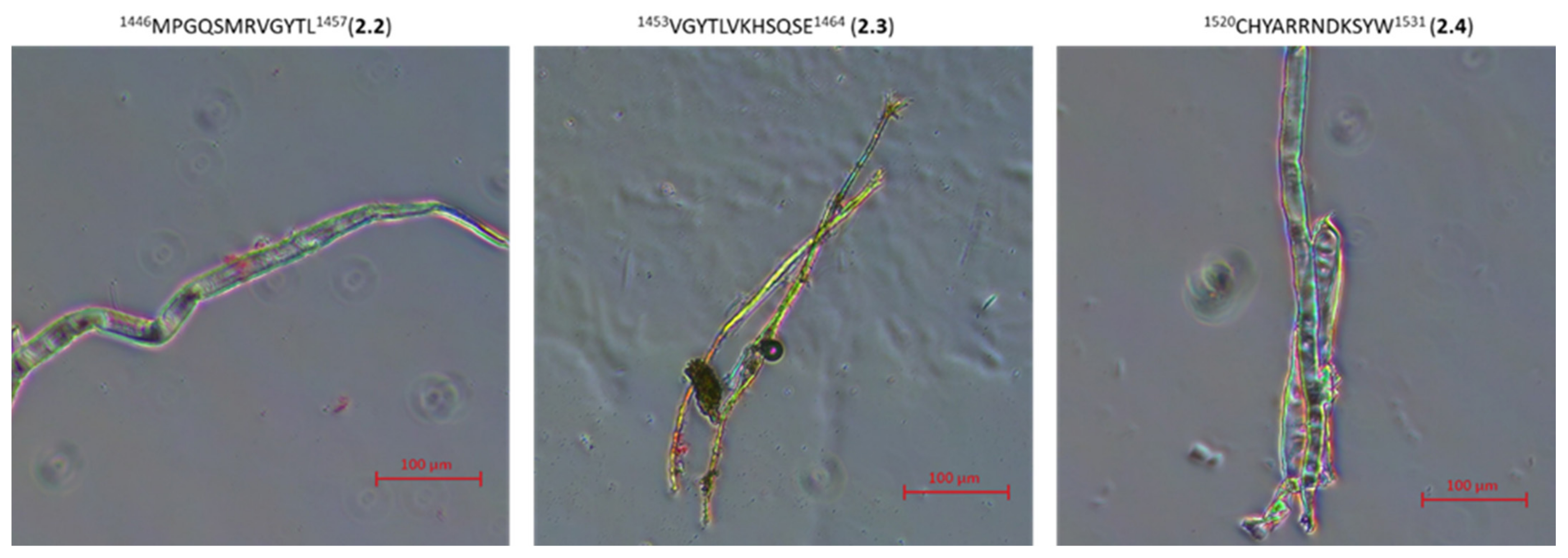
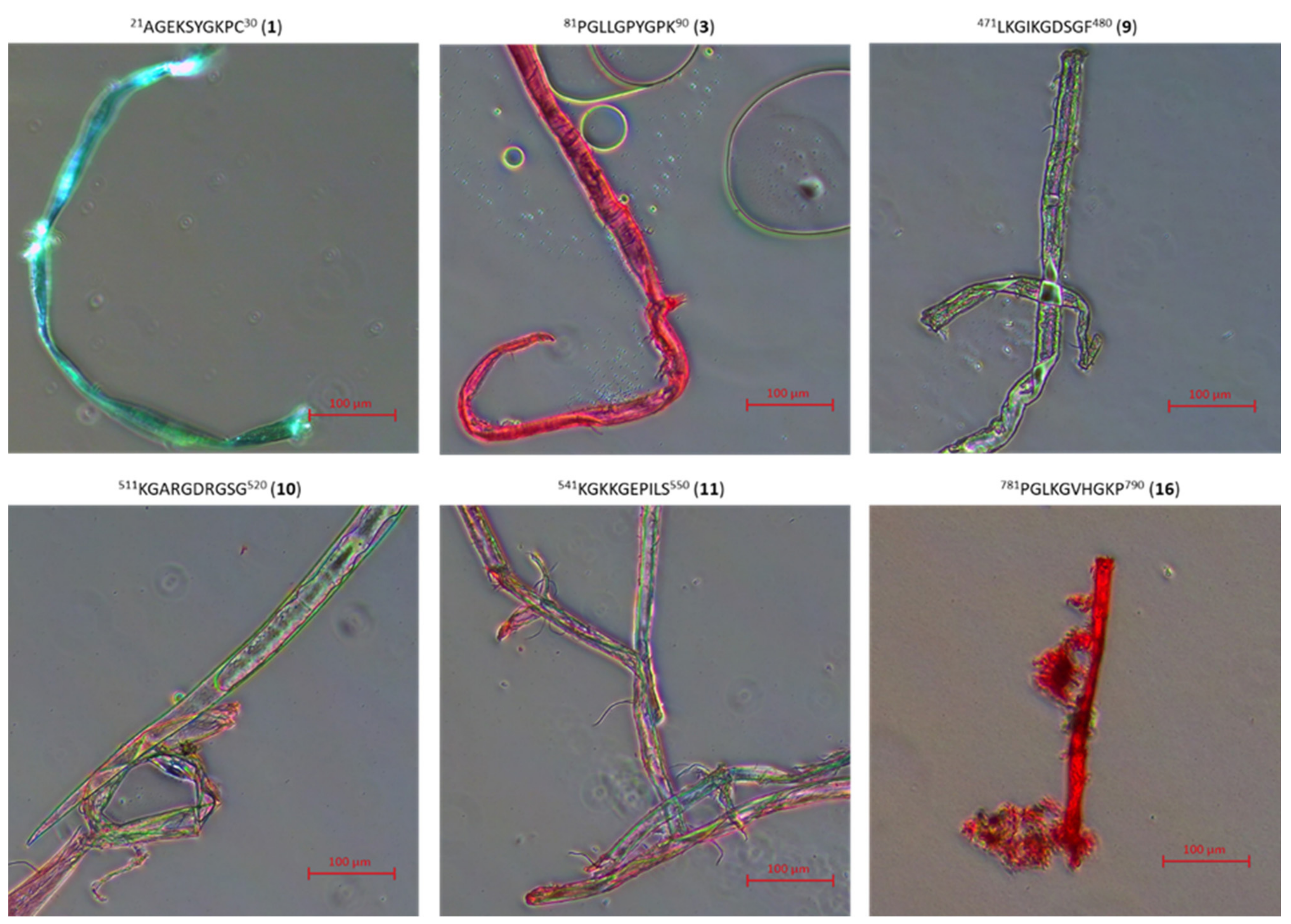
2.3. Research on the Ability to Self-Organization of Collagen IV Fragments
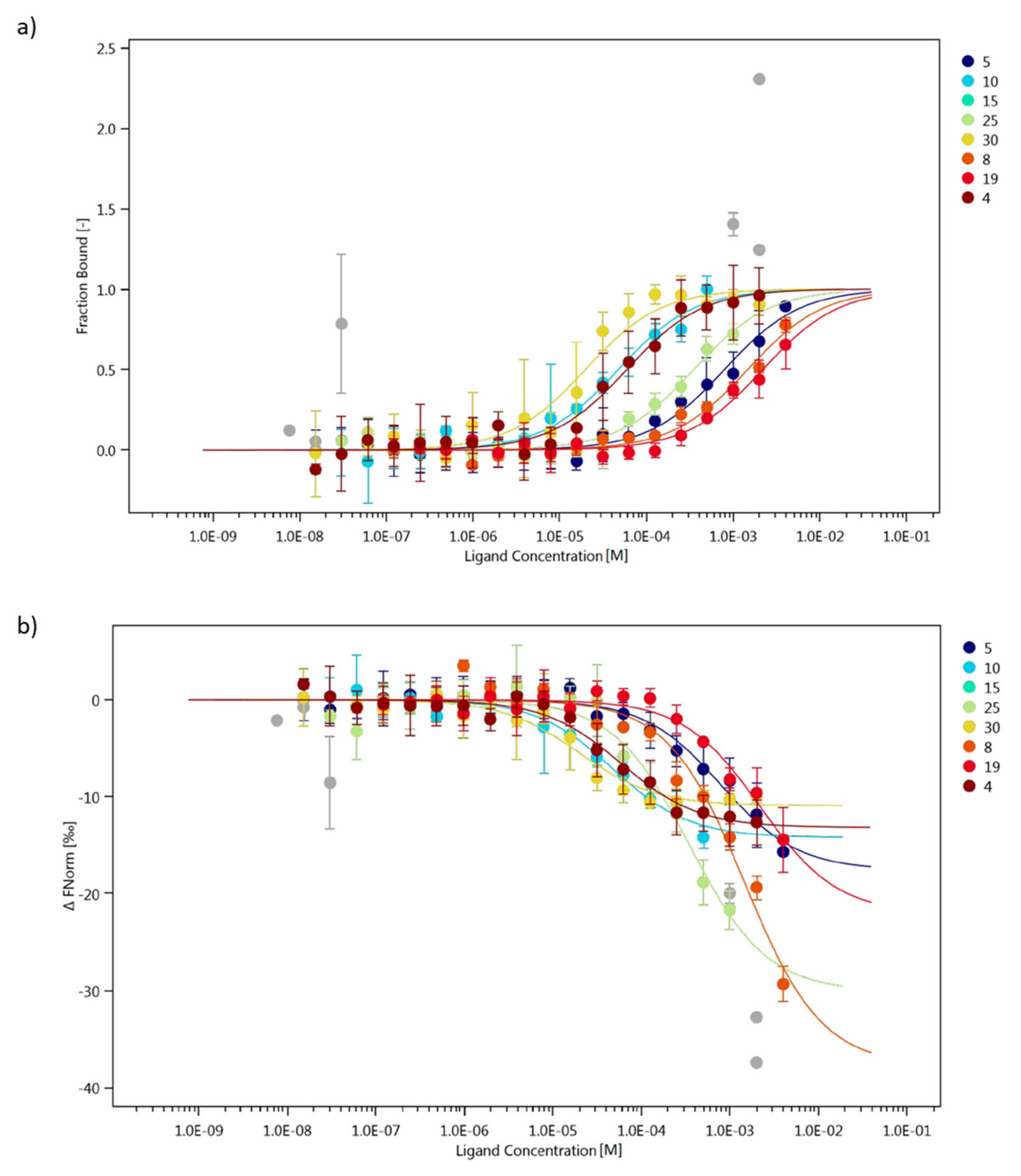
2.4. Interaction Study of Collagen IV Fragments with α1β1 Integrin (ITGα1β1)
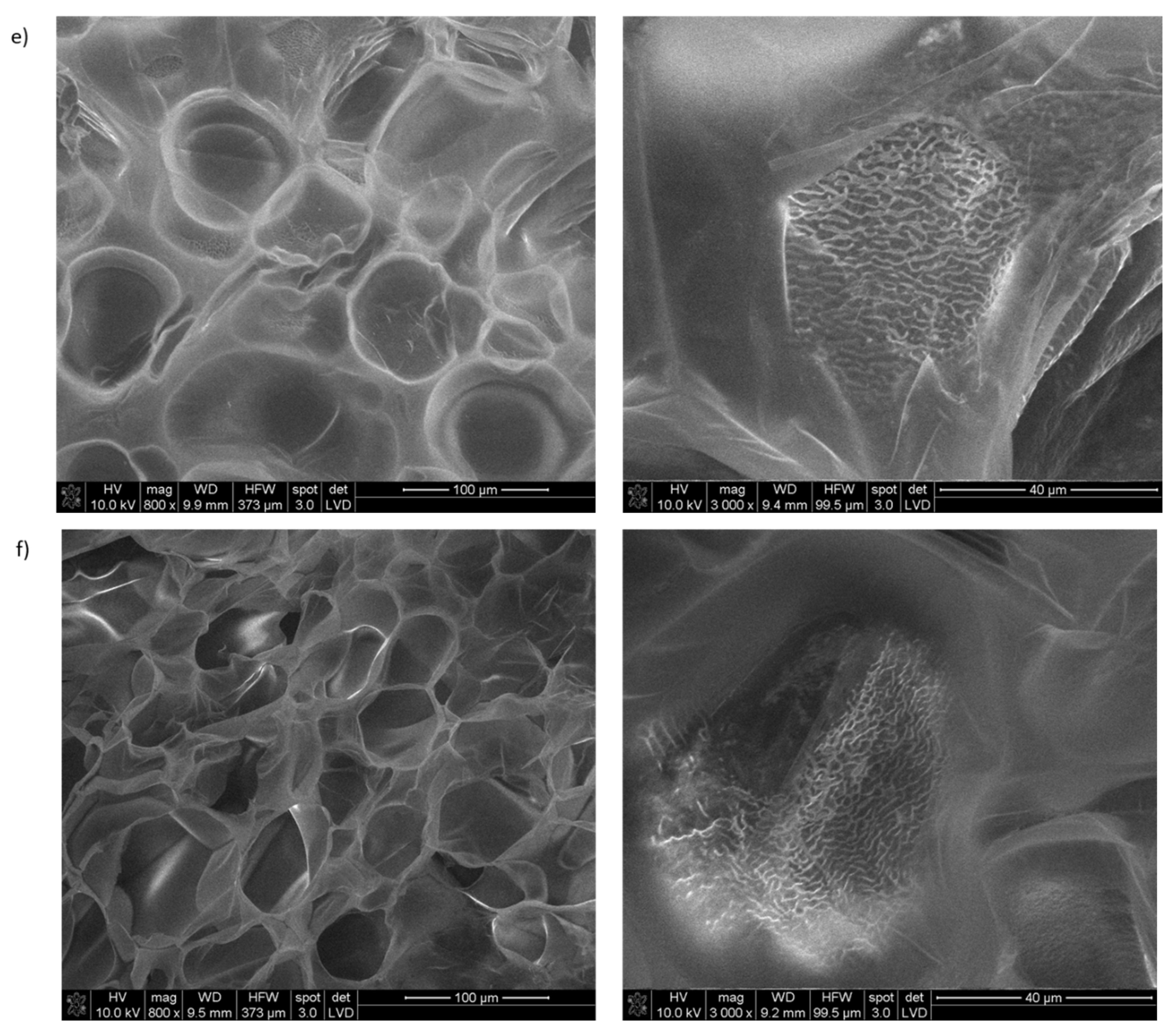
2.5. Study of the Usefulness of Selected Collagen IV Fragments in Obtaining Porous Materials
3. Materials and Methods
3.1. General Information
- Devices used in determining the purity and confirming the structure of the tested collagen fragments IV
- General Procedures for Synthesis of Peptides 1–33 and 2.1–2.4.
3.2. Peptide Synthesis
- Synthesis of 21AGEKSYGKPC30 (1)
- Synthesis of 41CFPEKGARGR50 (2)
- Synthesis of 81PGLLGPYGPK90 (3)
- Synthesis of 161LAPGSFKGMK170 (4)
- Synthesis of 221GFQGEKGVKG230 (5)
- Synthesis of 251GFPKGKKGSK260 (6)
- Synthesis of 311GPPGQQGKKG320 (7)
- Synthesis of 451NKESGFPGLR460 (8)
- Synthesis of 471LKGIKGDSGF480 (9)
- Synthesis of 511KGARGDRGSG520 (10)
- Synthesis of 541KGKKGEPILS550 (11)
- Synthesis of 641PGQQGLPGSK650 (12)
- Synthesis of 671PGFPGPKGSR680 (13)
- Synthesis of 711GFPGPRGEKG720 (14)
- Synthesis of 721LPGFPGLPGK730 (15)
- Synthesis of 781PGLKGVHGKP790 (16)
- Synthesis of 821GIKGKSGLPG830 (17)
- Synthesis of 841PGKKGTRGKK850 (18)
- Synthesis of 851GPPGSIVKKG860 (19)
- Synthesis of 891LSGPKGEKGS900 (20)
- Synthesis of 921LKGIPGSTGK930 (21)
- Synthesis of 951PVGIPSPRRP960 (22)
- Synthesis of 961MSNLWLKGDK970 (23)
- Synthesis of 1001GAPGLPGIIK1010 (24)
- Synthesis of 1091FKGTKGRDGL1100 (25)
- Synthesis of 1101IGNIGFPGNK1110 (26)
- Synthesis of 1211PGIGIGAPGK1220 (27)
- Synthesis of 1221PGLRGQKGDR1230 (28)
- Synthesis of 1311KGMRGEPGFM1320 (29)
- Synthesis of 1331DPGFPGMKGK1340 (30)
- Synthesis of 1451MRVGYTLVKH1460 (31)
- Synthesis of 1521HYARRNDKSY1530 (32)
- Synthesis of 1661AGQLHTRVSR1670 (33)
- Synthesis of 23EKSYGKPCGGQDC35 (2.1)
- Synthesis of 1446MPGQSMRVGYTL1457 (2.2)
- Synthesis of 1453VGYTLVKHSQSE1464 (2.3)
- Synthesis of 1520CHYARRNDKSYW1531 (2.4)
3.3. CD Studies of Peptides 1–33 and 2.1–2.4
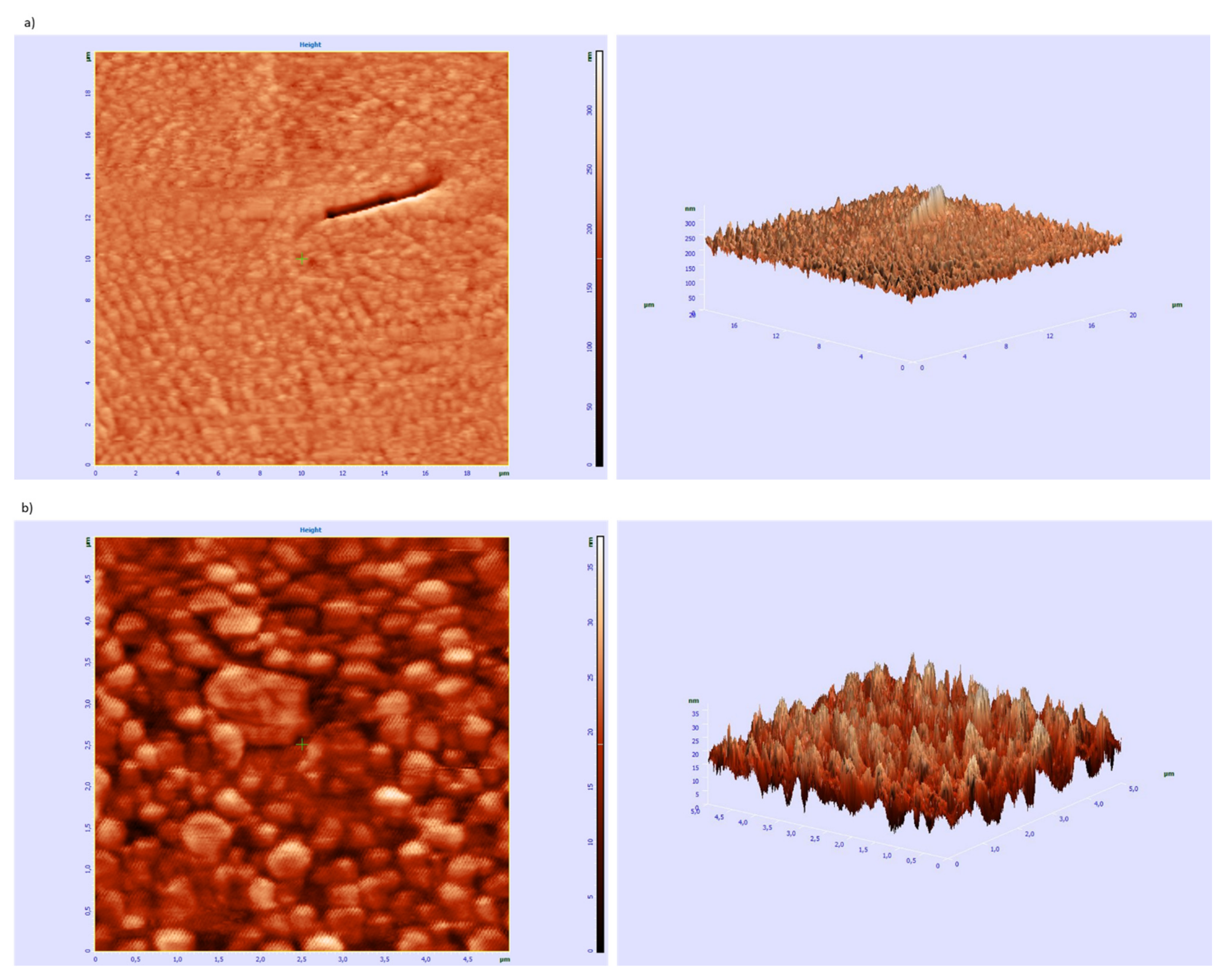
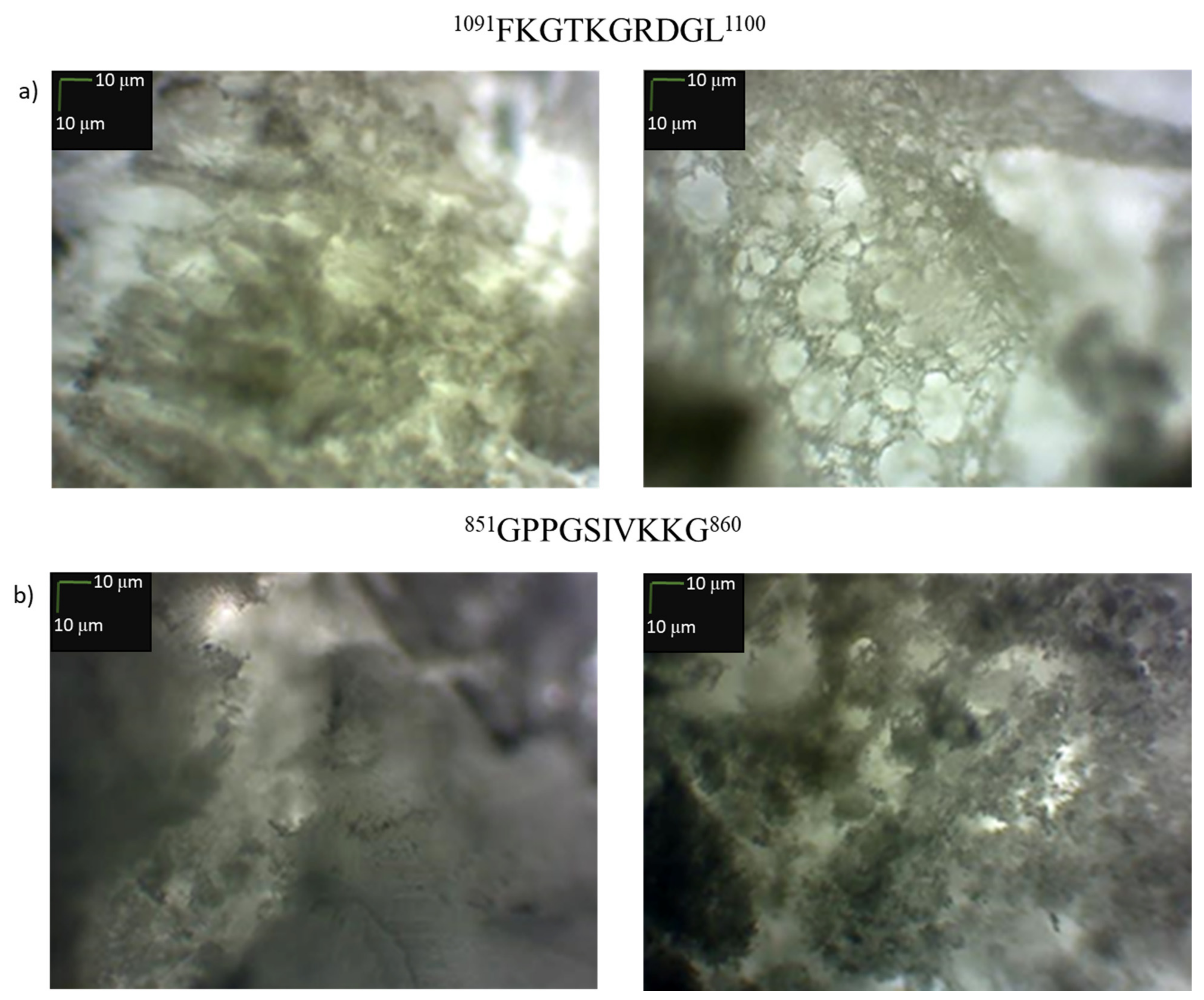
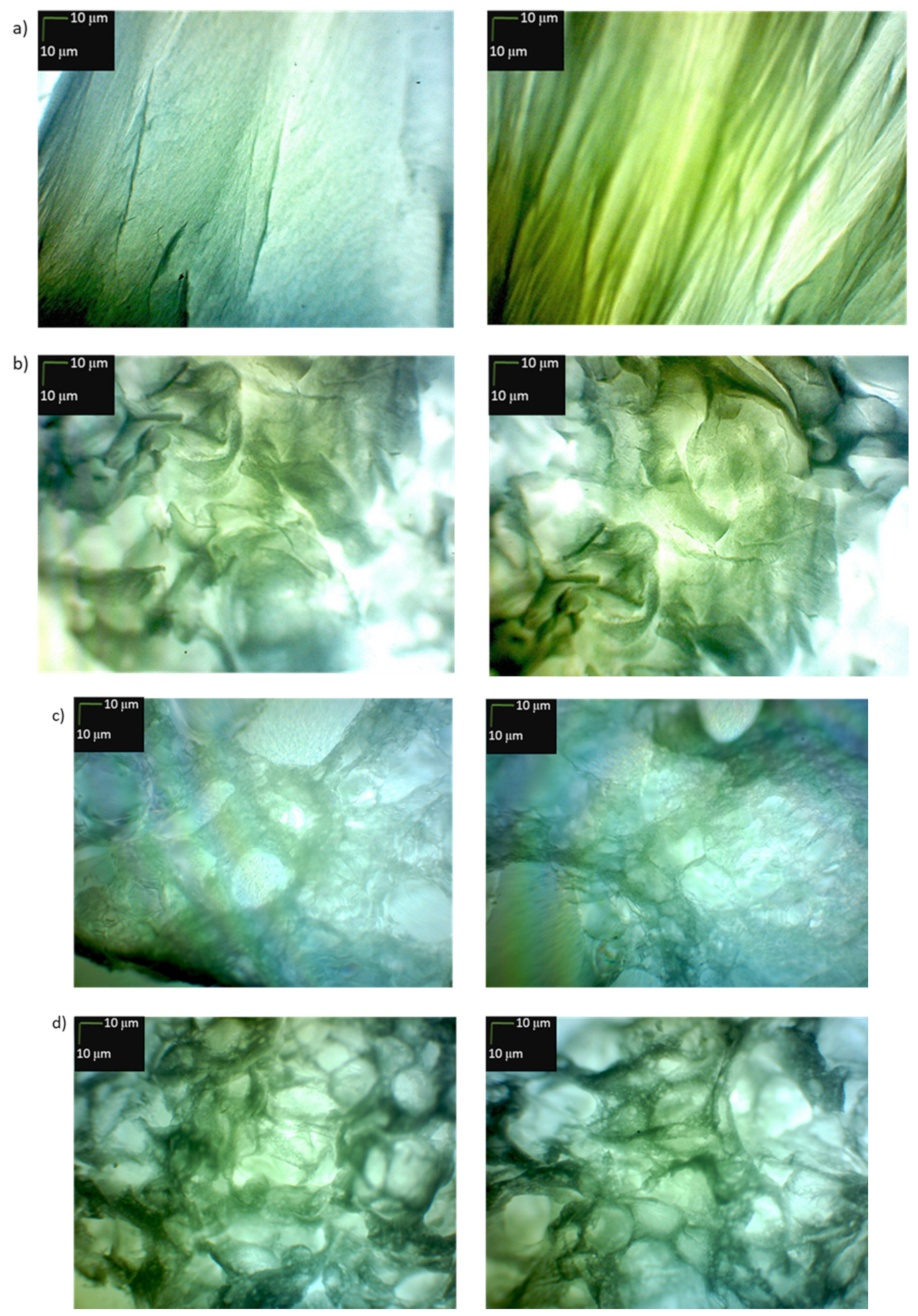
3.4. Microscopic Examination of Aggregates Formed by Collagen IV Fragments
- Microscopic Examination of Collagen IV Fragments Stained with Congo Red
- AFM Studies of the Ability of Peptide 8 to Aggregation
- SEM Studies of the Porous Materials
3.5. Studies on the Ability of Collagen IV Fragments to Interact with ITGα1β1 by Using MST Method
- Preparation of Materials for MST Analysis
- (1)
- ITGα1β1—a solution with a concentration of 9.3 µM was prepared. For this, 50 µL of sterile PBS (filtered through a 0.22 µm filter) was added to the protein. The solution was divided into 4 µL aliquots. The protein was His-tag labeled at C-end.
- (2)
- Marker/Dye—100 µg (HIS Lite™ OG488-Tris NTA-Ni Complex (AAT Bioquest, Inc., Sunnyvale, CA, USA) https://www.aatbio.com/products/his-lite-og488-tris-nta-ni-complex?unit=12615 (accessed on 22 April 2021)) was dissolved in 54.4 µL of H2O to obtain a 1 mM solution. The solution was divided into 2 µL (stock solution) aliquots. To obtain a 5 µM stock solution, 398 µL PBST was added to 2 µL of dye. The fluorescent dye OG488 (ex = 498 nm, ex = 526 nm) provide an efficient method for site-specific and stable noncovalent fluorescence labeling of polyhistidine-tagged proteins.
- (3)
- A 0.05% Tween20 solution in PBS (PBST) was prepared.
- Testing the Affinity of the Dye to the Protein
- Protein Labeling before Studying the Ability to Interact with Peptides
- Studies of the Affinity of Peptides to ITGα1β1
3.6. Preparation of Porous Materials from Collagen IV Derivatives
- Preparation of porous materials from 1091FKGTKGRDGL1100 (25) and 851GPPGSIVKKG860 (19)
- Preparation of porous materials with an equimolar mixture of peptides 2, 4, 5, 6, 14, 15, 25, 26 and 30
3.7. Biological Activity Studies
- Cell Culture
- Cell Viability
- Genotoxicity Analysis
- Statistical Analysis
- Cell Culture
- Human Inflammation Panel Assay
- Secondary Structure Degradation and Enzymatic Hydrolysis of Collagen IV
- Examination of cytotoxicity of cross-linked porous materials based on the equimolar mixture of peptides 2, 4, 5, 6, 14, 15, 25, 26 and 30
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mullen, L.M.; Best, S.M.; Brooks, R.A.; Ghose, S.; Gwynne, J.H.; Wardale, J.; Rushton, N.; Cameron, R.E. Binding and release characteristics of insulin-like growth factor-1 from a collagen-glycosaminoglycan scaffold. Tissue Eng. Part C 2010, 16, 1439–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawelec, K.M.; Best, S.M.; Cameron, R.E. Collagen: A network for regenerative medicine. J. Mater. Chem. B 2016, 4, 6484–6496. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.-B.; Su, D.-H.; Liu, P.; Ma, Y.-Q.; Shao, Z.-Z.; Dong, J. Shape-memory collagen scaffold for enhanced cartilage regeneration: Native collagen versus denatured collagen. Osteoarthr. Cartil. 2018, 26, 1389–1399. [Google Scholar] [CrossRef] [Green Version]
- Mackey, A.L.; Kjaer, M. Connective tissue regeneration in skeletal muscle after eccentric contraction-induced injury. J. Appl. Physiol. 2017, 122, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Louis, F.; Piantino, M.; Liu, H.; Kang, D.-H.; Sowa, Y.; Kitano, S.; Matsusaki, M. Bioprinted Vascularized Mature Adipose Tissue with Collagen Microfibers for Soft Tissue Regeneration. AAAS Cyborg. Bionic Syst. 2021, 2021, 1412542. [Google Scholar] [CrossRef]
- Mahoney, C.M.; Imbarlina, C.; Yates, C.C.; Marra, K.G. Current Therapeutic Strategies for Adipose Tissue Defects/Repair Using Engineered Biomaterials and Biomolecule Formulations. Front. Pharmacol. 2018, 9, 507. [Google Scholar] [CrossRef] [Green Version]
- Puls, T.J.; Fisher, C.S.; Cox, A.; Plantenga, J.M.; McBride, E.L.; Anderson, J.L.; Goergen, C.J.; Bible, M.; Moller, T.; Voytik-Harbin, S.L. Regenerative tissue filler for breast conserving surgery and other soft tissue restoration and reconstruction needs. Sci. Rep. 2021, 11, 2711. [Google Scholar] [CrossRef] [PubMed]
- Copes, F.; Pien, N.; van Vlierberghe, S.; Boccafoschi, F.; Mantovani, D. Collagen-Based Tissue Engineering Strategies for Vascular Medicine. Front. Bioeng. Biotechnol. 2019, 7, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, S.; Yamamoto, K.; Ikeda, T.; Yanagiguchi, K.; Hayashi, Y. Potency of Fish Collagen as a Scaffold for Regenerative Medicine. BioMed Res. Int. 2014, 2014, 302932. [Google Scholar] [CrossRef]
- Jency, G.; Jegan, S.R.; Mahija, S.P. Collagen and Its Therapeutical Applications in Regenerative Medicine. Int. J. Sci. Res. Dev. 2018, 6, 268–277. [Google Scholar]
- Dong, C.; Lv, Y. Application of Collagen Scaffold in Tissue Engineering: Recent Advances and New Perspectives. Polymers 2016, 8, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubert, L.; Dubus, M.; Rammal, H.; Bour, C.; Mongaret, C.; Boulagnon-Rombi, C.; Garnotel, R.; Schneider, C.; Rahouadj, R.; Laurent, C.; et al. Collagen-Based Medical Device as a Stem Cell Carrier for Regenerative Medicine. Int. J. Mol. Sci. 2017, 18, 2210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zata, H.F.; Chiquita, P.; Shafira, K. Collagen from marine source for regenerative therapy: A literature review. AIP Conf. Proc. 2020, 2314, 050017. [Google Scholar]
- Goren, S.; Koren, Y.; Xu, X.; Lesman, A. Elastic Anisotropy Governs the Range of Cell-Induced Displacements. Biophys. J. 2020, 118, 1152–1164. [Google Scholar] [CrossRef]
- Park, D.; Wershof, E.; Boeing, S.; Labernadie, A.; Jenkins, R.P.; George, S.; Trepat, X.; Bates, P.A.; Sahai, E. Extracellular matrix anisotropy is determined by TFAP2Cdependent regulation of cell collisions. Nat. Mater. 2020, 19, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Rasoulianboroujeni, M.; Kiaie, N.; Tabatabaei, F.S.; Yadegari, A.; Fahimipour, F.; Khoshroo, K.; Tayebi, L. Dual Porosity Protein-based Scaffolds with Enhanced Cell Infiltration and Proliferation. Sci. Rep. 2018, 8, 14889. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, J.C.; Mehr, M.; Buxton, P.G.; Best, S.M.; Cameron, R. Parameterizing the transport pathways for cell invasion in complex scaffold architectures. Tissue Eng. Part C 2016, 22, 409–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.M.; Blanco, M.D.; Davidenko, N.; Cameron, R.E. Tailoring chitosan/collagen scaffolds for tissue engineering: Effect of composition and different crosslinking agents on scaffold properties. Carbohydr. Polym. 2015, 132, 606–619. [Google Scholar] [CrossRef] [Green Version]
- Grover, C.N.; Cameron, R.E.; Best, S.M. Early plastic deformation behaviour and energy absorption in porous β-type biomedical titanium produced by selective laser melting. J. Mech. Behav. Biomed. Mater. 2012, 10, 62–74. [Google Scholar] [CrossRef]
- Nicolas, J.; Magli, S.; Rabbachin, L.; Sampaolesi, S.; Nicotra, F.; Russo, L. 3D Extracellular Matrix Mimics: Fundamental Concepts and Role of Materials Chemistry to Influence Stem Cell Fate. Biomacromolecules 2020, 21, 1968–1994. [Google Scholar] [CrossRef]
- Ntege, E.H.; Sunami, H.; Shimizu, Y. Advances in regenerative therapy: A review of the literature and future directions. Regen. Ther. 2020, 14, 136–153. [Google Scholar] [CrossRef] [PubMed]
- Shpichka, A.; Butnaru, D.; Bezrukov, E.A.; Sukhanov, R.B.; Atala, A.; Burdukovskii, V.; Zhang, Y.; Timashev, P. Skin tissue regeneration for burn injury. Stem Cell Res. Ther. 2019, 10, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimek, K.; Ginalska, G. Proteins and Peptides as Important Modifiers of the Polymer Scaffolds for Tissue Engineering Applications—A Review. Polymers 2020, 12, 844. [Google Scholar] [CrossRef] [Green Version]
- Cruz Walma, D.A.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147, 175596. [Google Scholar] [CrossRef]
- Kim, S.-H.; Turnbull, J.; Guimond, S. Extracellular matrix and cell signalling: The dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011, 209, 139–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valiente-Alandi, I.; Schafer, A.E.; Blaxall, B.C. Extracellular matrix-mediated cellular communication in the heart. J. Mol. Cell Cardiol. 2016, 91, 228–237. [Google Scholar] [CrossRef] [Green Version]
- Jansen, K.A.; Atherton, P.; Ballestrem, C. Mechanotransduction at the cell-matrix interface. Semin. Cell Dev. Biol. 2017, 71, 75–83. [Google Scholar] [CrossRef]
- Dhavalikar, P.; Robinson, A.; Lan, Z.; Jenkins, D.; Chwatko, M.; Salhadar, K.; Jose, A.; Kar, R.; Shoga, E.; Kannapiran, A.; et al. Review of Integrin-Targeting Biomaterials in Tissue Engineering. Adv. Healthc. Mater. 2020, 9, 2000795. [Google Scholar] [CrossRef]
- Bachmann, M.; Kukkurainen, S.; Hytönen, V.P.; Wehrle-Haller, B. Cell Adhesion by Integrins. Physiol. Rev. 2019, 99, 1655–1699. [Google Scholar] [CrossRef]
- Witjas, F.M.R.; van den Berg, B.M.; van den Berg, C.W.; Engelse, M.A.; Rabelink, T.J. Concise Review: The Endothelial Cell Extracellular Matrix Regulates Tissue Homeostasis and Repair. Stem Cells Transl. Med. 2019, 8, 375–382. [Google Scholar] [CrossRef] [Green Version]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, J.; Chiquet, M. An overview of extracellular matrix structure and function. In The Extracellular Matrix: An Overview; Mecham, R.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–39. [Google Scholar]
- Birk, D.E.; Bruckner, P. Collagens, suprastructures, and collagen fibril assembly. In The Extracellular Matrix: An Overview; Mecham, R.P., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 77–115. [Google Scholar]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoshnoodi, J.; Pedchenko, V.; Hudson, B. Mammalian Collagen IV. Microsc. Res. Tech. 2008, 71, 357–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abreu-Velez, A.M.; Howard, M.S. Collagen IV in Normal Skin and in Pathological Processes. N. Am. J. Med. Sci. 2012, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. Basement membranes: Structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer 2003, 3, 422–433. [Google Scholar] [CrossRef]
- Yurchenco, P.D. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 2011, 3, a004911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pöschl, E.; Schlötzer-Schrehardt, U.; Brachvogel, B.; Saito, K.; Ninomiya, Y.; Mayer, U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 2004, 131, 1619–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruegel, J.; Miosge, N. Basement membrane components are key players in specialized extracellular matrices. Cell Mol. Life Sci. 2010, 67, 2879–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, K.M.; Mei, R. Basement Membrane Type IV Collagen and Laminin: An Overview of Their Biology and Value as Fibrosis Biomarkers of Liver Disease. Anat. Rec. 2017, 300, 1371–1390. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, N.J.I.; Lee, D.D.H.; Gowers, K.H.C.; Butler, C.R.; Maughan, E.F.; Jevans, B.; Orr, J.C.; McCann, C.J.; Burns, A.J.; MacNeil, S.; et al. Bioengineered airway epithelial grafts with mucociliary function based on collagen IV- and laminin-containing extracellular matrix scaffolds. Eur. Respir. J. 2020, 55, 1901200. [Google Scholar] [CrossRef]
- Steffensen, L.B.; Rasmussen, L.M. Extracellular Matrix in Cardiovascular Pathophysiology. A role for collagen type IV in cardiovascular disease? Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H610–H625. [Google Scholar] [CrossRef] [PubMed]
- Fidler, A.L.; Darris, C.E.; Chetyrkin, S.V.; Pedchenko, V.K.; Boudko, S.P.; Brown, K.L.; Jerome, W.G.; Hudson, J.K.; Rokas, A.; Hudson, B.G. Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. eLife 2017, 6, e24176. [Google Scholar] [CrossRef]
- Bella, J.; Hulmes, D.J. Fibrillar collagens. Subcell. Biochem. 2017, 82, 457–490. [Google Scholar] [PubMed]
- Sillat, T.; Saat, R.; Pöllänen, R.; Hukkanen, M.; Takagi, M.; Konttinen, Y.T. Basement membrane collagen type IV expression by human mesenchymal stem cells during adipogenic differentiation. J. Cell. Mol. Med. 2012, 16, 1485–1495. [Google Scholar] [CrossRef]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Kisling, A.; Lust, R.M.; Katwa, L.C. What is the role of peptide fragments of collagen I and IV in health and disease? Life Sci. 2019, 228, 30–34. [Google Scholar] [CrossRef]
- Helling, A.L.; Tsekoura, E.K.; Biggs, M.; Bayon, Y.; Pandit, A.; Zeugolis, D.I. In Vitro Enzymatic Degradation of Tissue Grafts and Collagen Biomaterials by Matrix Metalloproteinases: Improving the Collagenase Assay. ACS Biomater. Sci. Eng. 2017, 3, 1922–1932. [Google Scholar] [CrossRef]
- Xu, Y.; Kirchner, M. Collagen Mimetic Peptides. Bioengineering 2021, 8, 5. [Google Scholar] [CrossRef]
- Becht, A.; Menaszek, E.; Slowiak, K.; Kolesinska, B. Searching for the fragments of collagen IV forming the external sphere of the native protein. Chem. Biodivers. 2021. submitted. [Google Scholar]
- Marcotte, E.M.; Pellegrini, M.; Yeates, T.O.; Eisenberg, D. A census of protein repeats. J. Mol. Biol. 1999, 293, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Headd, J.J.; Ban, Y.E.A.; Brown, P.; Edelsbrunner, H.; Vaidya, M.; Rudolph, J. Protein—Protein Interfaces: Properties, Preferences, and Projections research articles. J. Proteome Res. 2007, 6, 2576–2586. [Google Scholar] [CrossRef] [PubMed]
- Ngounou Wetie, A.G.; Sokolowska, I.; Woods, A.G.; Roy, U.; Loo, J.A.; Darie, C.C. Investigation of stable and transient protein-protein interactions: Past, present, and future. Proteomics 2013, 13, 538–557. [Google Scholar] [CrossRef] [Green Version]
- Kolesinska, B.; Rozniakowski, K.K.; Fraczyk, J.; Relich, I.; Papini, A.M.; Kaminski, Z.J. The Effect of Counterion and Tertiary Amine on the Efficiency of N-Triazinylammonium Sulfonates in Solution and Solid-Phase Peptide Synthesis. Eur. J. Org. Chem. 2015, 2015, 401–408. [Google Scholar] [CrossRef]
- Gadhave, K.; Bhardwaj, T.; Uversky, V.N.; Vendruscolo, M.; Giri, R. The signal peptide of the amyloid precursor protein forms amyloid-like aggregatesand enhances Ab42 aggregation. Cell Rep. Phys. Sci. 2021, 2, 100599. [Google Scholar] [CrossRef]
- Edwards, I.A.; Elliott, A.G.; Kavanagh, A.M.; Zuegg, J.; Blaskovich, M.A.T.; Cooper, M.A. Contribution of Amphipathicity and Hydrophobicity to the Antimicrobial Activity and Cytotoxicity of β-Hairpin Peptides. ACS Infect. Dis. 2016, 2, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Thybaud, V.; Kasper, P.; Sobol, Z.; Elhajouji, A.; Fellows, M.; Guerard, M.; Lynch, A.M.; Sutter, A.; Tanir, J.Y. Genotoxicity assessment of peptide/protein-related biotherapeutics: Points to consider before testing. Mutagenesis 2016, 31, 375–384. [Google Scholar] [CrossRef]
- Saladin, P.M.; Zhang, B.D.; Reichert, J.M. Current trends in the clinical development of peptide therapeutics. IDrugs 2009, 12, 779–784. [Google Scholar]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef]
- ISO (Ed.) Biological Evaluation of Medical Devices—Part 1: Evaluation and Testing within a Risk Management Process; ISO: Geneva, Switzerland, 2009. [Google Scholar]
- Hulsart-Billstrom, G.; Dawson, J.I.; Hofmann, S.; Müller, R.; Stoddart, M.J.; Alini, M.; Redl, H.; El Haj, A.; Brown, R.; Salih, V.; et al. A surprisingly poor correlation between in vitro and in vivo testing of biomaterials for bone regeneration: Results of a multicentre analysis. Eur. Cells Mater. 2016, 31, 312–322. [Google Scholar] [CrossRef]
- Lock, A.; Cornish, J.; Musson, D.S. The Role of In Vitro Immune Response Assessment for Biomaterials. J. Funct. Biomater. 2019, 10, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.M.; McNally, A.K. Biocompatibility of implants: Lymphocyte/macrophage interactions. Semin. Immunopathol. 2011, 33, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Radović, S.; Selak, I.; Babić, M.; Knezević, Z.; Vukobrat-Bijedić, Z. Type IV collagen immunoreactivity of basement membrane in inflammatory-regenerative and dysplastic lesions of the flat colonic mucosa. Bosn. J. Basic Med. Sci. 2003, 3, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, J.; Dunér, P.; To, F.; Engelbertsen, D.; Gonçalves, I.; Nilsson, J.; Bengtsson, E. Activation of immune responses against the basement membrane component collagen type IV does not affect the development of atherosclerosis in ApoE-deficient mice. Sci. Rep. 2019, 9, 5964. [Google Scholar] [CrossRef]
- Veidal, S.S.; Karsdal, M.A.; Nawrocki, A.; Larsen, M.R.; Dai, Y.; Zheng, Q.; Hägglund, P.; Vainer, B.; Skjøt-Arkil, H.; Leeming, D.J. Assessment of proteolytic degradation of the basement membrane: A fragment of type IV collagen as a biochemical marker for liver fibrosis. Fibrogenesis Tissue Repair 2011, 4, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murawaki, Y.; Ikuta, Y.; Koda, M.; Yamada, S.; Kawasaki, H. Comparison of serum 7s fragment of type IV collagen and serum central triple-helix of type IV collagen for assessment of liver fibrosis in patients with chronic viral liver disease. J. Hepatol. 1996, 24, 148–154. [Google Scholar] [CrossRef]
- Katsumata, K.; Ishihara, J.; Mansurov, A.; Ishihara, A.; Raczy, M.M.; Yuba, E.; Hubbell, J.A. Targeting inflammatory sites through collagen affinity enhances the therapeutic efficacy of anti-inflammatory antibodies. Sci. Adv. 2019, 5, eaay1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marneros, A.G.; Olsen, B.R. The role of collagen-derived proteolytic fragments in angiogenesis. Matrix Biol. 2001, 20, 337–345. [Google Scholar] [CrossRef]
- Edgar, S.; Hopley, B.; Genovese, L.; Sibilla, S.; Laight, D.; Shute, J. Effects of collagen-derived bioactive peptides and natural antioxidant compounds on proliferation and matrix protein synthesis by cultured normal human dermal fibroblasts. Sci. Rep. 2018, 8, 10474. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, T.; Kawakami, K.; Uraji, M. Inhibitory effect of collagen-derived tripeptides on dipeptidylpeptidase-IV activity. J. Enzyme Inhib. Med. Chem. 2014, 29, 823–828. [Google Scholar] [CrossRef] [Green Version]
- Rosca, E.V.; Koskimaki, J.E.; Pandey, N.B.; Wolff, A.C.; Popel, A.S. Development of a biomimetic peptide derived from collagen IV with anti-angiogenic activity in breast cancer. Cancer Biol. Ther. 2011, 12, 808–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Ding, C.; Tang, L.; Deng, F.; Yang, Q.; Wu, H.; Chen, L.; Ni, Y.; Huang, L.; Zhang, M. Novel Modification of Collagen: Realizing Desired Water Solubility and Thermostability in a Conflict-Free Way. ACS Omega 2020, 5, 5772–5780. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 3095–3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]
- Pelc, D.; Marion, S.; Prozek, M.; Basletić, M. Role of microscopic phase separation in gelation of aqueous gelatin solutions. Soft Matter 2014, 10, 348–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, T.M.; Heima, G.P.; Kubiak, C.P. Effects of electron transfer on the stability of hydrogen bonds. Chem. Sci. 2017, 8, 7324–7329. [Google Scholar]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Schell, D.; Thurlkill, R.L.; Imura, S.; Scholtz, J.M.; Gajiwala, K.; et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014, 23, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Klunk, W.E.; Pettegrew, J.W.; Abraham, D.J. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. 1989, 37, 1273–1281. [Google Scholar] [CrossRef]
- Abedini, A.; Meng, F.; Raleigh, D.P. A Single-Point Mutation Converts the Highly Amyloidogenic Human Islet Amyloid Polypeptide into a Potent Fibrillization Inhibitor. J. Am. Chem. Soc. 2007, 129, 11300–11301. [Google Scholar] [CrossRef]
- Carter, D.B.; Chou, K.C. A model for structure-dependent binding of Congo red to Alzheimer beta-amyloid fibrils. Neurobiol. Aging 1998, 19, 37–40. [Google Scholar] [CrossRef]
- Anazco, C.; Lopez-Jimenez, A.J.; Rafi, M.; Vega-Montoto, L.; Zhang, M.Z.; Hudson, B.G.; Vanacore, R.M. Lysyl oxidase-like-2 cross-links collagen IV of glomerular basement membrane. J. Biol. Chem. 2016, 291, 25999–26012. [Google Scholar] [CrossRef] [Green Version]
- Mirando, A.C.; Shen, J.; Lima e Silva, R.; Chu, Z.; Sass, N.C.; Lorenc, V.E.; Green, J.J.; Campochiaro, P.A.; Popel, A.S.; Pandey, N.B. A collagen IV-derived peptide disrupts α5β1 integrin and potentiates Ang2/Tie2 signaling. JCI Insight 2019, 4, e122043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidel, S.A.; Dijkman, P.M.; Lea, W.A.; van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Neungseon, S.; Russell, B.H.; Rivera, J.J.; Liang, X.; Xu, X.; Afshar-Kharghan, V.; Höök, M. An Engineered α1 Integrin-binding Collagenous Sequence. J. Biol. Chem. 2010, 285, 31046–31054. [Google Scholar]
- Pieper, J.S.; Oosterhof, A.; Dijkstra, P.J.; Veerkamp, J.H.; van Kuppevelt, T.H. Preparation and characterization of porous crosslinked collagenous matrices containing bioavailable chondroitin sulphate. Biomaterials 1999, 20, 847–858. [Google Scholar] [CrossRef]
- Pietrucha, K. Changes in denaturation and rheological properties of collagen-hyaluronic acid scaffolds as a result of temperature dependencies. Int. J. Biol. Macromol. 2005, 36, 299–304. [Google Scholar] [CrossRef]
- Fraczyk, J.; Wasko, J.; Walczak, M.; Kaminski, Z.J.; Puchowicz, D.; Kaminska, I.; Bogun, M.; Kolasa, M.; Stodolak-Zych, E.; Scislowska-Czarnecka, A.; et al. Conjugates of Copper Alginate with Arginine-Glycine-Aspartic Acid (RGD) for Potential Use in Regenerative Medicine. Materials 2020, 13, 337. [Google Scholar] [CrossRef] [Green Version]
- Wasko, J.; Fraczyk, J.; Becht, A.; Kaminski, Z.J.; Flinčec Grgac, S.; Tarbuk, A.; Kaminska, M.; Dudek, M.; Gliscinska, E.; Draczynski, Z.; et al. Conjugates of Chitosan and Calcium Alginate with Oligoproline and Oligohydroxyproline Derivatives for Potential Use in Regenerative Medicine. Materials 2020, 13, 307. [Google Scholar] [CrossRef] [PubMed]
- Brochocka, A.; Nowak, A.; Majchrzycka, K.; Puchalski, M.; Sztajnowski, S. Multifunctional Polymer Composites Produced by Melt-Blown Technique to Use in Filtering Respiratory Protective Devices. Materials 2020, 13, 712. [Google Scholar] [CrossRef] [Green Version]
- Pieklarz, K.; Galita, G.; Tylman, M.; Maniukiewicz, W.; Kucharska, E.; Majsterek, I.; Modrzejewska, Z. Physico-Chemical Properties and Biocompatibility of Thermosensitive Chitosan Lactate and Chitosan Chloride Hydrogels Developed for Tissue Engineering Application. J. Funct. Biomater. 2021, 12, 37. [Google Scholar] [CrossRef]
- Rozpędek-Kamińska, W.; Galita, G.; Siwecka, N.; Carroll, S.L.; Diehl, J.A.; Kucharska, E.; Pytel, D.; Majsterek, I. The Potential Role of Small-Molecule PERK Inhibitor LDN-0060609 in Primary Open-Angle Glaucoma Treatment. Int. J. Mol. Sci. 2021, 22, 4494. [Google Scholar] [CrossRef] [PubMed]
- Khiari, Z.; Ndagijimana, M.; Betti, M. Low molecular weight bioactive peptides derived from the enzymatic hydrolysis of collagen after isoelectric solubilization/precipitation process of turkey by-products. Poult. Sci. 2014, 93, 2347–2362. [Google Scholar] [CrossRef] [PubMed]
- León-López, A.; Morales-Peñaloza, A.; Martínez-Juárez, V.M.; Vargas-Torres, A.; Zeugolis, D.I.; Aguirre-Álvarez, G. Hydrolyzed Collagen—Sources and Applications. Molecules 2019, 24, 4031. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Peptide | Fragment | Purity Based on HPLC | M Calculated [g/mol] | m/z Found |
|---|---|---|---|---|
| 1 | 21AGEKSYGKPC30 | 88% | 1038.47 | 529.2788 [M + 2H]2+ 1039.5492 [M + H]+ |
| 2 | 41CFPEKGARGR50 | 80% | 1119.55 | 374.2167 [M + 3H]3+ 1120.6136 [M + H]+ |
| 3 | 81PGLLGPYGPK90 | 86% | 997.55 | 499.8145 [M + 2H]2+ 998.6127 [M + H]+ |
| 4 | 161LAPGSFKGMK170 | 87% | 1034.55 | 1035.6251 [M + H]+ |
| 5 | 221GFQGEKGVKG230 | 98% | 1005.51 | 503.7991 [M + 2H]2+ 1006.5898 [M + H]+ |
| 6 | 251GFPKGKKGSK260 | 93% | 1032.59 | 345.2301 [M + 3H]3+ 1033.6762 [M + H]+ |
| 7 | 311GPPGQQGKKG320 | 90% | 952.49 | 953.5674 [M + H]+ |
| 8 | 451NKESGFPGLR460 | 95% | 1103.56 | 552.8254 [M + 2H]2+ |
| 9 | 471LKGIKGDSGF480 | 85% | 1020.55 | 511.2875 [M + 2H]2+ 1021.5612 [M + H]+ |
| 10 | 511KGARGDRGSG520 | 99% | 959.47 | 320.8502 [M + 3H]3+ 480.7737 [M + 2H]2+ 960.5373 [M + H]+ |
| 11 | 541KGKKGEPILS550 | 85% | 1055.62 | 528.8563 [M + 2H]2+ 1056.7046 [M + H]+ |
| 12 | 641PGQQGLPGSK650 | 99% | 967.49 | 484.7949 [M + 2H]2+ 968.5737 [M + H]+ |
| 13 | 671PGFPGPKGSR680 | 87% | 998.51 | 333.8715 [M + 3H]3+ 999.5955 [M + H]+ |
| 14 | 711GFPGPRGEKG720 | 84% | 1000.49 | 501.2775 [M + 2H]2+ 1001.5502 [M + H]+ |
| 15 | 721LPGFPGLPGK730 | 91% | 981.55 | 491.8182 [M + 2H]2+ 982.6217 [M + H]+ |
| 16 | 781PGLKGVHGKP790 | 97% | 988.57 | 330.5547 [M + 3H]3+ 989.6496 [M + H]+ |
| 17 | 821GIKGKSGLPG830 | 99% | 912.52 | 457.3051 [M + 2H]2+ 913.6027 [M + H]+ |
| 18 | 841PGKKGTRGKK850 | 81% | 1055.64 | 352.9130 [M + 3H]3+ 1056.7249 [M + H]+ |
| 19 | 851GPPGSIVKKG860 | 95% | 938.54 | 470.3106 [M + 2H]2+ 939.6105 [M + H]+ |
| 20 | 891LSGPKGEKGS900 | 82% | 958,49 | 480,2892 [M + 2H]2+ 959,5691 [M + H]+ |
| 21 | 921LKGIPGSTGK930 | 91% | 956.55 | 479.3194 [M + 2H]2+ 957.6297 [M + H]+ |
| 22 | 951PVGIPSPRRP960 | 92% | 1074.62 | 359.2365 [M + 3H]3+ 538.3456 [M + 2H]2+ 1075.6860 [M + H]+ |
| 23 | 961MSNLWLKGDK970 | 91% | 1190.60 | 397.9016 [M + 3H]3+ 596.3429 [M + 2H]2+ 1191.6721 [M + H]+ |
| 24 | 1001GAPGLPGIIK1010 | 80% | 921.55 | 461.8178 [M + 2H]2+ 922.6209 [M + H]+ |
| 25 | 1091FKGTKGRDGL1100 | 92% | 1077.58 | 360.2275 [M + 3H]3+ 1078.6536 [M + H]+ |
| 26 | 1101IGNIGFPGNK1110 | 80% | 1015.53 | 508.8136 [M + 2H]2+ 1016.6166 [M + H]+ |
| 27 | 1211PGIGIGAPGK1220 | 95% | 865.49 | 433.7870 [M + 2H]2+ 866.5585 [M + H]+ |
| 28 | 1221PGLRGQKGDR1230 | 80% | 1082.58 | 361.8937 [M + 3H]3+ 1083.6613 [M + H]+ |
| 29 | 1311KGMRGEPGFM1320 | 92% | 1108.51 | 555.3018 [M + 2H]2+ 1109.5893 [M + H]+ |
| 30 | 1331DPGFPGMKGK1340 | 92% | 1032.49 | 517.2877 [M + 2H]2+ 1033.5643 [M + H]+ |
| 31 | 1451MRVGYTLVKH1460 | 80% | 1202.65 | 401.8919 [M + 3H]3+ 1203.6398 [M + H]+ |
| 32 | 1521HYARRNDKSY1530 | 82% | 1308.62 | 437.2168 [M + 3H]3+ 655.3173 [M + 2H]2+ 1309.6100 [M + H]+ |
| 33 | 1661AGQLHTRVSR1670 | 92% | 1123.61 | 375.5688 [M + 3H]3+ 562.8491 [M + 2H]2+ 1124.6773 [M + H]+ |
| Peptide | Fragment | Ability to Mimic the Structure of the Collagen Helix | Kd [μM] | Maximum Concentration |
|---|---|---|---|---|
| 4 | 161LAPGSFKGMK170 | +++ | 59.2 ± 12.5 | 2 mM |
| 5 | 221GFQGEKGVKG230 | +++ | 790.3 ± 174.2 | 4 mM |
| 8 | 451NKESGFPGLR460 | + | 1465.3 ± 424.2 | 4 mM |
| 10 | 511KGARGDRGSG520 | - | 47.3 ± 10.3 | 0.5 mM |
| 15 | 721LPGFPGLPGK730 | +++ | no binding | 2 mM |
| 19 | 851GPPGSIVKKG860 | - | 2191.9 ± 741.9 | 4 mM |
| 25 | 1091FKGTKGRDGL1100 | +++ | 350.9 ± 109.9 | 1 mM |
| 30 | 1331DPGFPGMKGK1340 | +++ | 19.7 ± 5.6 | 2 mM |
| Peptide | Fragment | Purity Based on HPLC | M Calculated [g/mol] | m/z Found |
|---|---|---|---|---|
| 2.1 | 23EKSYGKPCGGQDC35 | 99% | 1371.52 | 1372.5602 [M + H]+ |
| 2.2 | 1446MPGQSMRVGYTL1457 | 99% | 1339.60 | 1340.6140 [M + H]+ |
| 2.3 | 1453VGYTLVKHSQSE1464 | 99% | 1347.49 | 1349.6571 [M + H]+ |
| 2.4 | 1520CHYARRNDKSYW1531 | 99% | 1598.77 | 1600.6767 [M + H]+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolasa, M.; Galita, G.; Majsterek, I.; Kucharska, E.; Czerczak, K.; Wasko, J.; Becht, A.; Fraczyk, J.; Gajda, A.; Pietrzak, L.; et al. Screening of Self-Assembling of Collagen IV Fragments into Stable Structures Potentially Useful in Regenerative Medicine. Int. J. Mol. Sci. 2021, 22, 13584. https://doi.org/10.3390/ijms222413584
Kolasa M, Galita G, Majsterek I, Kucharska E, Czerczak K, Wasko J, Becht A, Fraczyk J, Gajda A, Pietrzak L, et al. Screening of Self-Assembling of Collagen IV Fragments into Stable Structures Potentially Useful in Regenerative Medicine. International Journal of Molecular Sciences. 2021; 22(24):13584. https://doi.org/10.3390/ijms222413584
Chicago/Turabian StyleKolasa, Marcin, Grzegorz Galita, Ireneusz Majsterek, Ewa Kucharska, Katarzyna Czerczak, Joanna Wasko, Angelika Becht, Justyna Fraczyk, Anna Gajda, Lukasz Pietrzak, and et al. 2021. "Screening of Self-Assembling of Collagen IV Fragments into Stable Structures Potentially Useful in Regenerative Medicine" International Journal of Molecular Sciences 22, no. 24: 13584. https://doi.org/10.3390/ijms222413584
APA StyleKolasa, M., Galita, G., Majsterek, I., Kucharska, E., Czerczak, K., Wasko, J., Becht, A., Fraczyk, J., Gajda, A., Pietrzak, L., Szymanski, L., Krakowiak, A., Draczynski, Z., & Kolesinska, B. (2021). Screening of Self-Assembling of Collagen IV Fragments into Stable Structures Potentially Useful in Regenerative Medicine. International Journal of Molecular Sciences, 22(24), 13584. https://doi.org/10.3390/ijms222413584







