Tal2c Activates the Expression of OsF3H04g to Promote Infection as a Redundant TALE of Tal2b in Xanthomonas oryzae pv. oryzicola
Abstract
:1. Introduction
2. Results
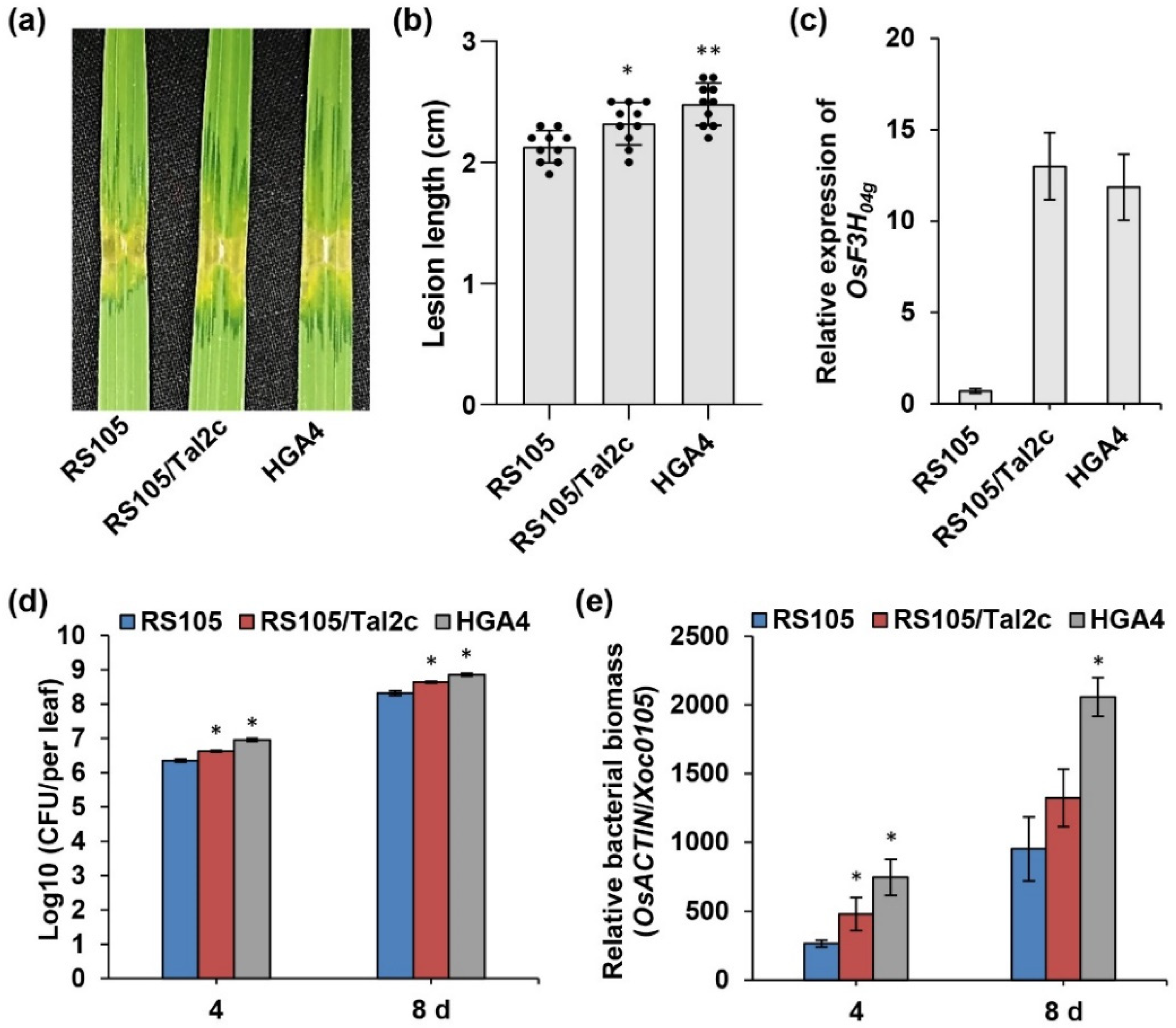
2.1. Tal2c Acts as a Virulence Factor
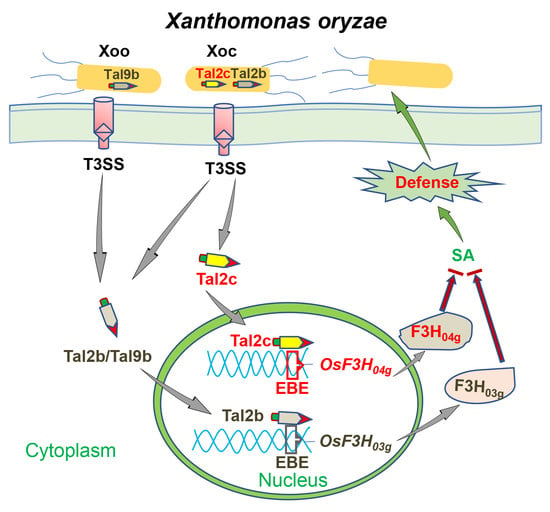
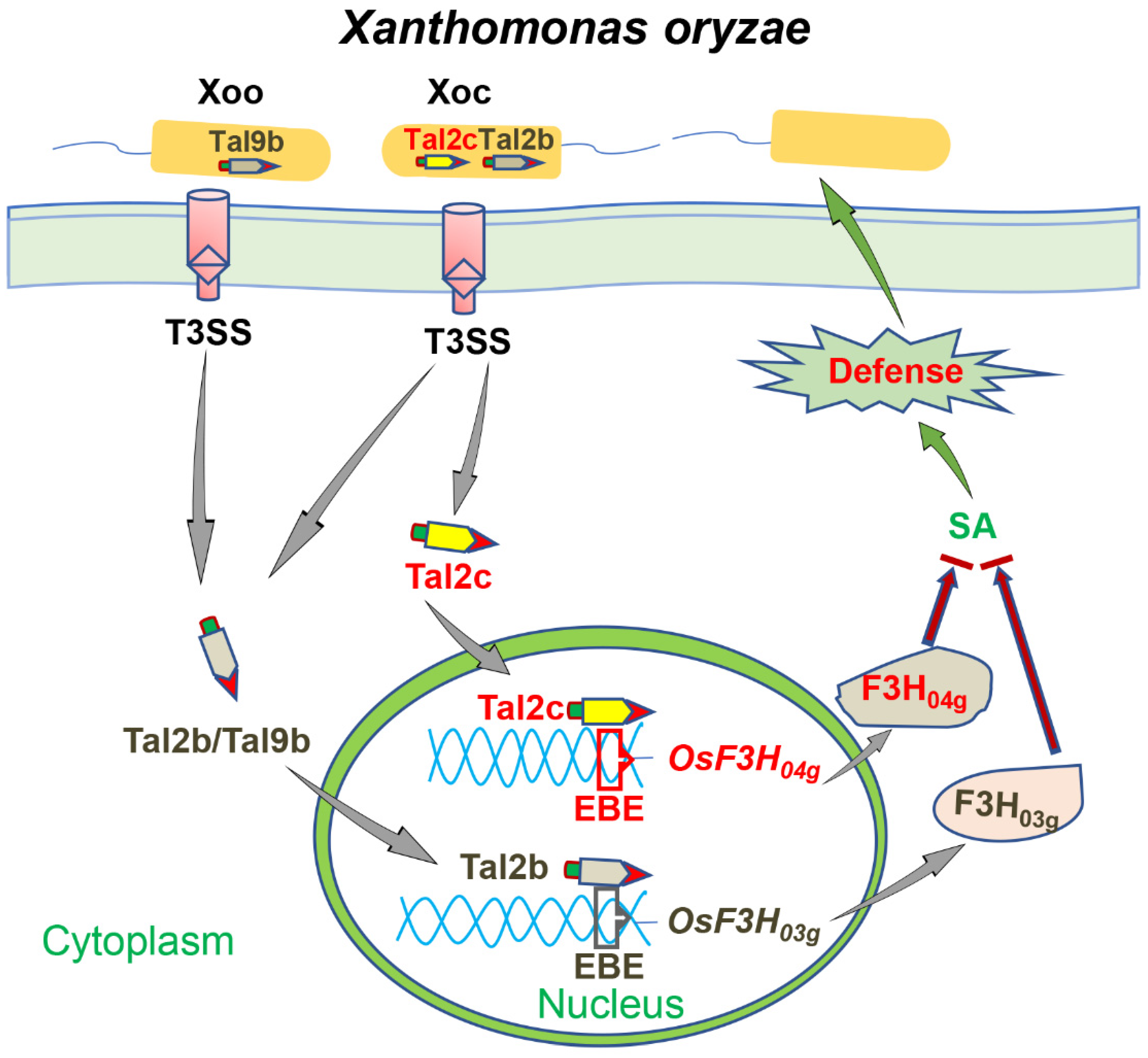
2.2. Tal2c Targets OsF3H04g and Tandem Pairs of Tal2b in Xoc
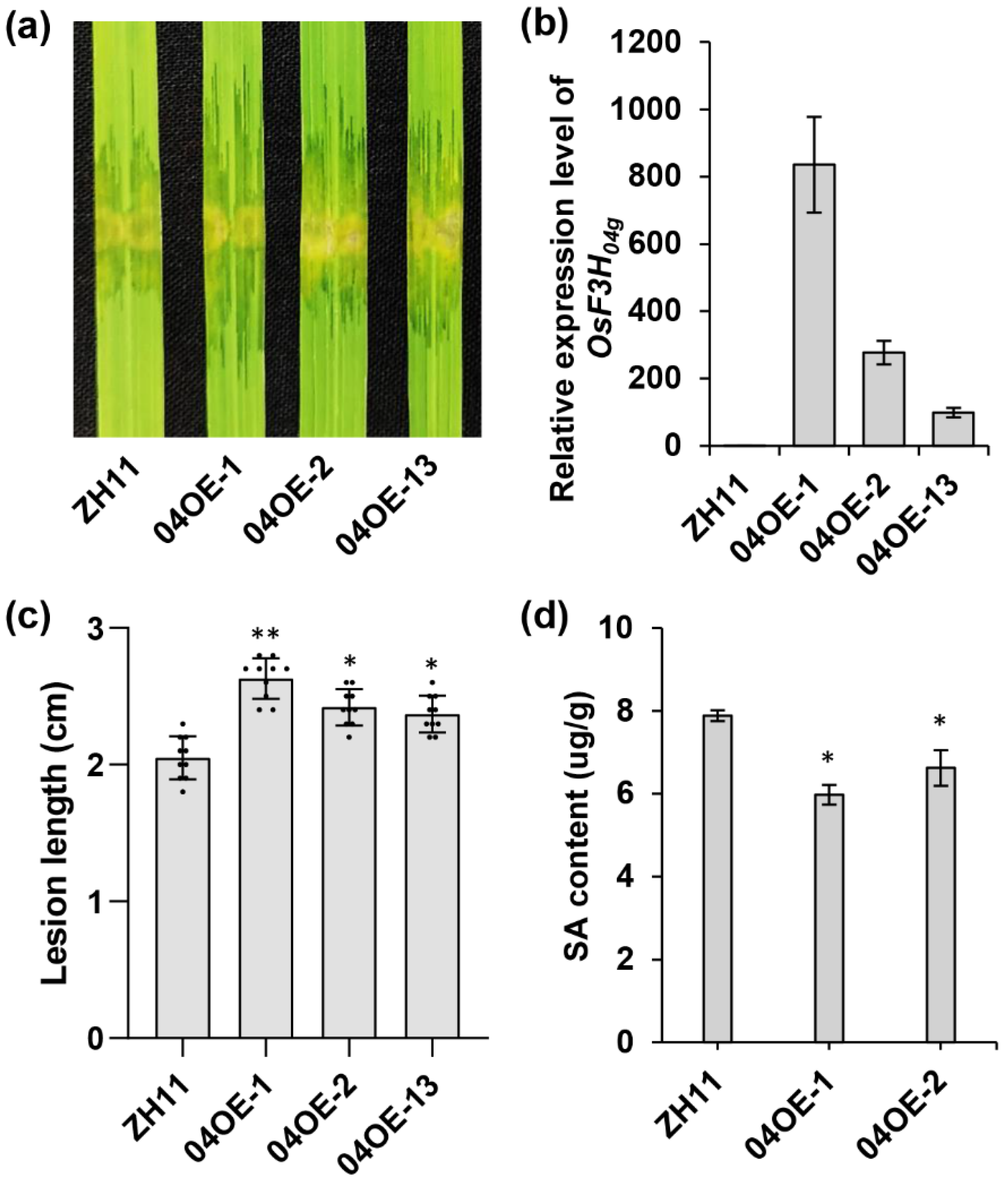
2.3. Overexpression of OsF3H04g Increases Susceptibility to Xoc in Rice
2.4. Editing EBE of the OsF3H04g Promoter Compromised Tal2c-Mediated Susceptibility in Rice
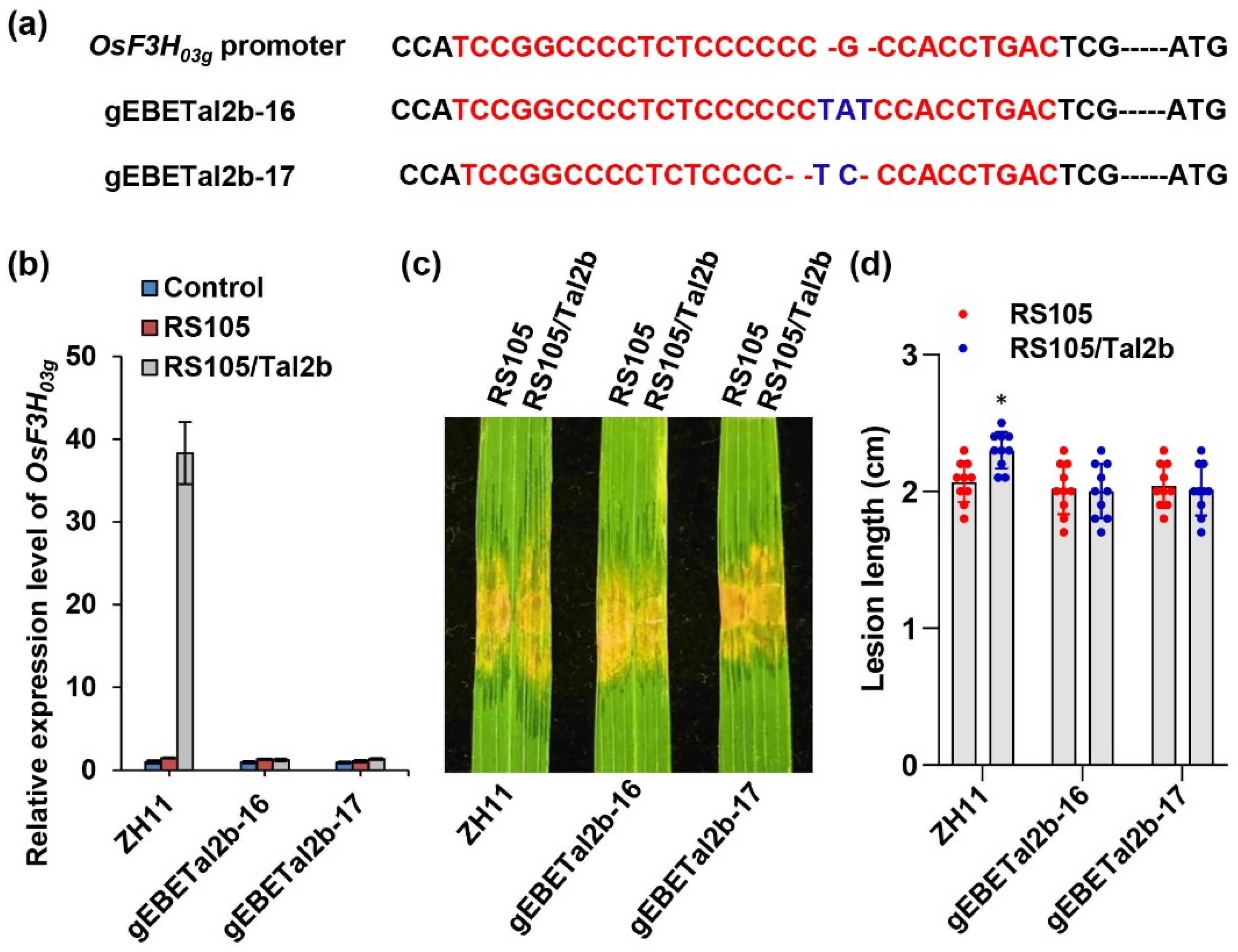
2.5. Gene Editing of EBE in the OsF3H03g Promoter Specifically Attenuates Rice Susceptibility to RS105/Tal2b
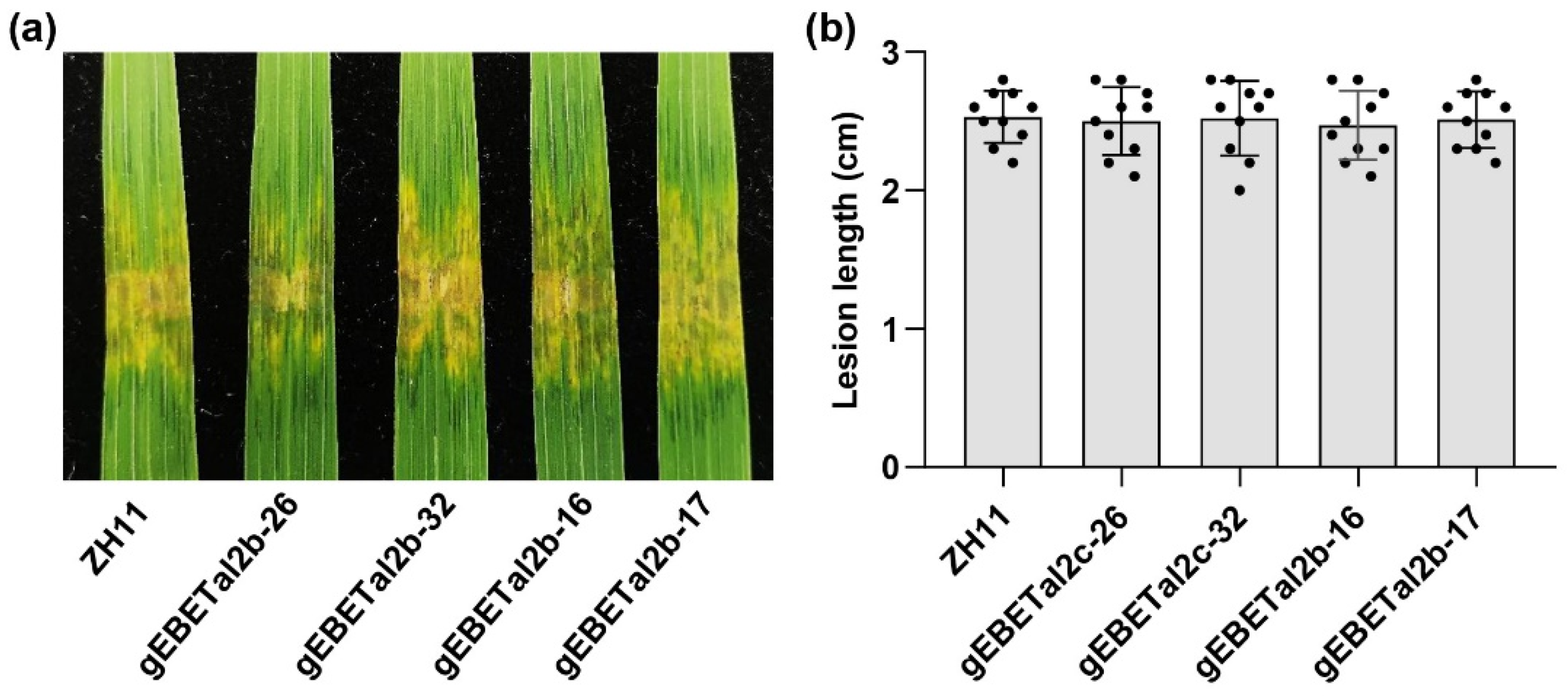
2.6. Single Gene Editing of EBE in OsF3H04g or OsF3H03g Does Not Affect Resistance to Xoc HGA4
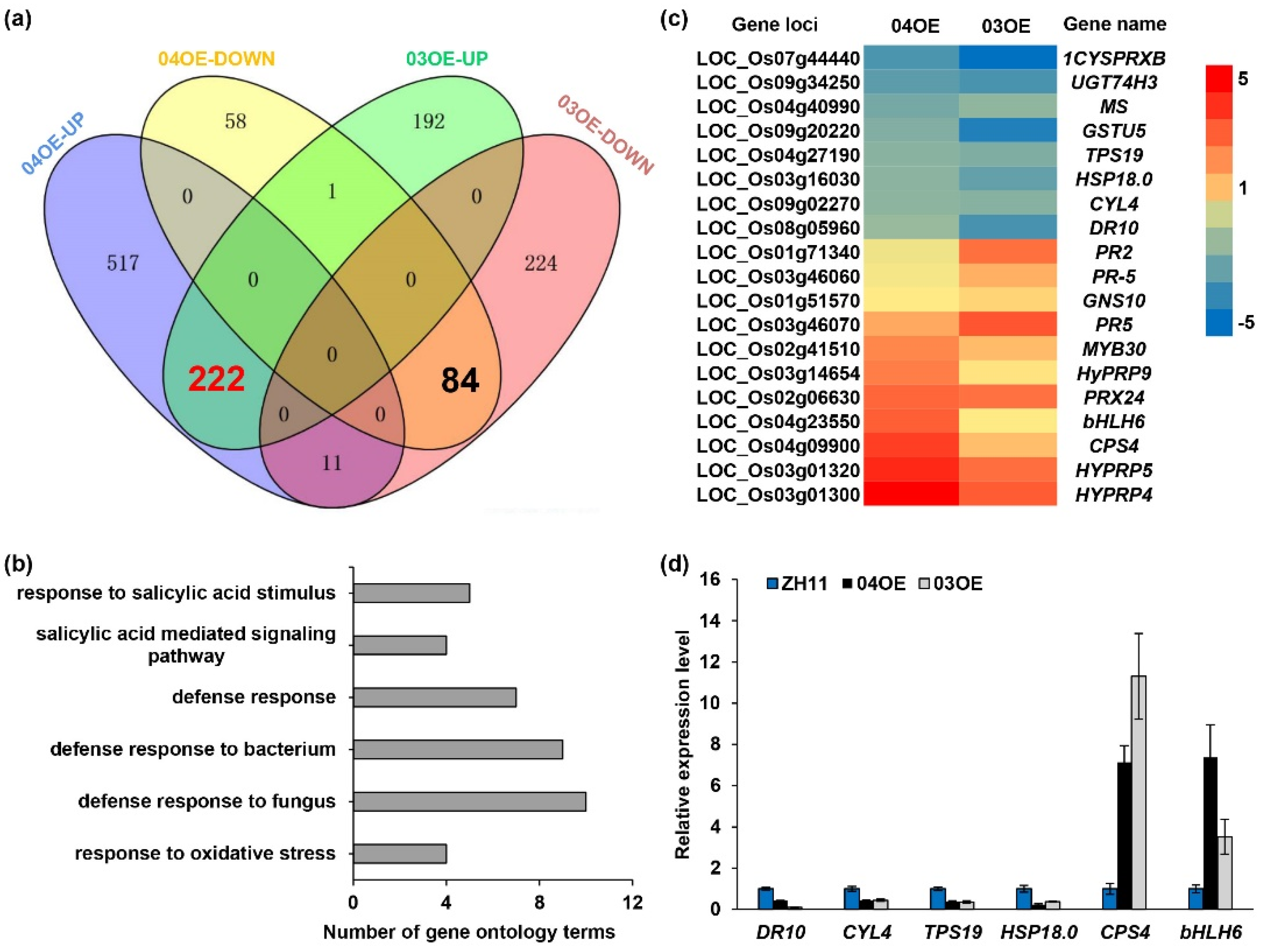
2.7. Comparative Analysis of the Transcriptome Profiles of the OsF3H04g and OsF3H03g Overexpression Lines
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Transient Expression
4.3. Construction of the RS105/Tal2c Strain and Bacterial Inoculation
4.4. RNA Manipulation and RNA-seq
4.5. Vector Construction and Rice Transformation for OsF3H04g Overexpression and EBE Gene Editing Plasmids
4.6. Hormone Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Deng, H.; Wang, S. Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 2017, 90, 738–748. [Google Scholar] [CrossRef]
- Jiang, N.; Yan, J.; Liang, Y.; Shi, Y.; He, Z.; Wu, Y.; Zeng, Q.; Liu, X.; Peng, J. Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)—An updated review. Rice 2020, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Zhang, H.; Yuan, B.; Liu, H.; Kong, L.; Chu, Z.; Ding, X. Tal2b targets and activates the expression of OsF3H03g to hijack OsUGT74H4 and synergistically interfere with rice immunity. New Phytol. 2021, nph17877. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S. Disease resistance. In Genetics and Genomics of Rice; Zhang, Q., Wing, R.A., Eds.; Springer: New York, NY, USA, 2013; pp. 161–175. [Google Scholar] [CrossRef]
- White, F.F.; Potnis, N.; Jones, J.B.; Koebnik, R. The type III effectors of Xanthomonas. Mol. Plant Pathol. 2009, 10, 749–766. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Mak, A.N.; Bradley, P.; Cernadas, R.A.; Bogdanove, A.J.; Stoddard, B.L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 2012, 335, 716–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Chang, Q.; Feng, W.; Zhang, B.; Wu, T.; Li, N.; Yao, F.; Ding, X.; Chu, Z. Domain dissection of AvrRxo1 for suppressor, avirulence and cytotoxicity functions. PLoS ONE 2014, 9, e113875. [Google Scholar] [CrossRef] [PubMed]
- Schuebel, F.; Rocker, A.; Edelmann, D.; Schessner, J.; Brieke, C.; Meinhart, A. 3′-NADP and 3′-NAADP, two metabolites formed by the bacterial type III effector AvrRxo1. J. Biol. Chem. 2016, 291, 22868–22880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Li, S.; Wang, J.; Li, Q.; Xin, X.F.; Zhou, S.; Wang, Y.; Li, D.; Xu, J.; Luo, Z.Q.; et al. A bacterial kinase phosphorylates OSK1 to suppress stomatal immunity in rice. Nat. Commun. 2021, 12, 5479. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Quintero, A.L.; Rodriguez-R, L.M.; Dereeper, A.; López, C.; Koebnik, R.; Szurek, B.; Cunnac, S. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS ONE 2013, 8, e68464. [Google Scholar] [CrossRef]
- Cernadas, R.A.; Doyle, E.L.; Niño-Liu, D.O.; Wilkins, K.E.; Bancroft, T.; Wang, L.; Schmidt, C.L.; Caldo, R.; Yang, B.; White, F.F.; et al. Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 2014, 10, e1003972. [Google Scholar] [CrossRef]
- Read, A.C.; Rinaldi, F.C.; Hutin, M.; He, Y.; Triplett, L.R.; Bogdanove, A.J. Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front. Plant Sci. 2016, 7, 1516. [Google Scholar] [CrossRef] [Green Version]
- Read, A.C.; Hutin, M.; Moscou, M.J.; Rinaldi, F.C.; Bogdanove, A.J. Cloning of the rice Xo1 resistance gene and interaction of the Xo1 protein with the defense-suppressing Xanthomonas effector Tal2h. Mol. Plant Microbe Interact. 2020, 33, 1189–1195. [Google Scholar] [CrossRef]
- Cai, L.; Cao, Y.; Xu, Z.; Ma, W.; Zakria, M.; Zou, L.; Cheng, Z.; Chen, G. A transcription activator-like effector Tal7 of Xanthomonas oryzae pv. oryzicola activates rice gene Os09g29100 to suppress rice immunity. Sci. Rep. 2017, 7, 5089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hummel, A.W.; Wilkins, K.E.; Wang, L.; Cernadas, R.A.; Bogdanove, A.J. A transcription activator-like effector from Xanthomonas oryzae pv. oryzicola elicits dose-dependent resistance in rice. Mol. Plant Pathol. 2017, 18, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Mücke, S.; Reschke, M.; Erkes, A.; Schwietzer, C.A.; Becker, S.; Streubel, J.; Morgan, R.D.; Wilson, G.G.; Grau, J.; Boch, J. Transcriptional reprogramming of rice cells by Xanthomonas oryzae TALEs. Front. Plant Sci. 2019, 10, 162. [Google Scholar] [CrossRef]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Nadi, R.; Mateo-Bonmatí, E.; Juan-Vicente, L.; Micol, J.L. The 2OGD superfamily: Emerging functions in plant epigenetics and hormone metabolism. Mol. Plant 2018, 11, 1222–1224. [Google Scholar] [CrossRef] [Green Version]
- Farrow, S.C.; Facchini, P.J. Functional diversity of 2-oxoglutarate/Fe (II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 2014, 5, 524. [Google Scholar] [CrossRef] [Green Version]
- Qi, G.; Chen, J.; Chang, M.; Chen, H.; Hall, K.; Korin, J.; Liu, F.; Wang, D.; Fu, Z.Q. Pandemonium breaks out: Disruption of salicylic acid-mediated defense by plant pathogens. Mol. Plant 2018, 11, 1427–1439. [Google Scholar] [CrossRef] [Green Version]
- van Damme, M.; Huibers, R.P.; Elberse, J.; Van den Ackerveken, G. Arabidopsis DMR6 encodes a putative 2OG-Fe (II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008, 54, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Van den Ackerveken, G. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015, 81, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.J.; Zhang, K. S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.; Gan, S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieu, N.P.; Lenman, M.; Wang, E.S.; Petersen, B.L.; Andreasson, E. Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 2021, 11, 4487. [Google Scholar] [CrossRef]
- Hasley, J.A.R.; Navet, N.; Tian, M. CRISPR/Cas9-mediated mutagenesis of sweet basil candidate susceptibility gene ObDMR6 enhances downy mildew resistance. PLoS ONE 2021, 16, e0253245. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Shah, T.; Tripathi, L. CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol. J. 2021, 19, 1291–1293. [Google Scholar] [CrossRef]
- Thomazella, D.P.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118. [Google Scholar] [CrossRef]
- Dai, Z.; Tan, J.; Zhou, C.; Yang, X.; Yang, F.; Zhang, S.; Sun, S.; Miao, X.; Shi, Z. The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 2019, 17, 1657–1669. [Google Scholar] [CrossRef] [Green Version]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas system: Recent advances and future prospects for genome editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef] [Green Version]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, W.; Zhou, Z.; Chen, H.; Xie, C.; Lin, Y. A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 2020, 18, 313–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Xu, Z.; Li, Z.; Zakria, M.; Zou, L.; Chen, G. Increasing resistance to bacterial leaf streak in rice by editing the promoter of susceptibility gene OsSULRT3;6. Plant Biotechnol. J. 2021, 19, 1101–1103. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Cao, Y.; Jin, X.; Fu, Z.; Li, J.; Mo, X.; He, Y.; Tang, J.; Huang, S. Engineering resistance to bacterial blight and bacterial leaf streak in rice. Rice 2021, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, K.E.; Booher, N.J.; Wang, L.; Bogdanove, A.J. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 2015, 6, 536. [Google Scholar] [CrossRef]
- Xiao, W.; Liu, H.; Li, Y.; Li, X.; Xu, C.; Long, M.; Wang, S. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS ONE 2009, 4, e4603. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Shen, X.; Wang, N.; Ding, X. Characterization of a novel cyclase-like gene family involved in controlling stress tolerance in rice. J. Plant Physiol. 2015, 181, 30–41. [Google Scholar] [CrossRef]
- Meng, F.; Yang, C.; Cao, J.; Chen, H.; Pang, J.; Zhao, Q.; Wang, Z.; Qing, F.Z.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef]
- Ju, Y.; Tian, H.; Zhang, R.; Zuo, L.; Jin, G.; Xu, Q.; Ding, X.; Li, X.; Chu, Z. Overexpression of OsHSP18.0-CI enhances resistance to bacterial leaf streak in rice. Rice 2017, 10, 12. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Ju, Y.; Zuo, L.; Shang, L.; Li, X.; Li, X.; Feng, S.; Ding, X.; Chu, Z. OsHsfB4d binds the promoter and regulates the expression of OsHsp18.0-CI to resistant against Xanthomonas oryzae. Rice 2020, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, H.; Yuan, J.S.; Köllner, T.G.; Chen, Y.; Guo, Y.; Zhuang, X.; Chen, X.; Zhang, Y.J.; Fu, J.; et al. The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 2018, 16, 1778–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Zhang, J.; Brown, B.; Li, R.; Rodríguez-Romero, J.; Berasategui, A.; Liu, B.; Xu, M.; Luo, D.; Pan, Z.; et al. Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell 2018, 30, 1119–1131. [Google Scholar] [CrossRef] [Green Version]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef]
- Yang, B.; Sugio, A.; White, F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 10503–10508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streubel, J.; Pesce, C.; Hutin, M.; Koebnik, R.; Boch, J.; Szurek, B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013, 200, 808–819. [Google Scholar] [CrossRef]
- Antony, G.; Zhou, J.; Huang, S.; Li, T.; Liu, B.; White, F.F.; Yang, B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 2010, 22, 3864–3876. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Streubel, J.; Balzergue, S.; Champion, A.; Boch, J.; Koebnik, R.; Feng, J.; Verdier, V.; Szurek, B. Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant Microbe Interact. 2011, 24, 1102–1113. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.S.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B.; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef]
- Sugio, A.; Yang, B.; Zhu, T.; White, F.F. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. USA 2007, 104, 10720–10725. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; Wang, Z.; Chen, X.; Liu, Y.; Zhang, M.; Song, C.; Dong, H. Identification of a TAL effector in Xanthomonas oryzae pv. oryzicola enhancing pathogen growth and virulence in plants. Physiol. Mol. Plant Pathol. 2021, 114, 101620. [Google Scholar] [CrossRef]
- Doucouré, H.; Pérez-Quintero, A.L.; Reshetnyak, G.; Tekete, C.; Auguy, F.; Thomas, E.; Koebnik, R.; Szurek, B.; Koita, O.; Verdier, V.; et al. Functional and genome sequence-driven characterization of tal effector gene repertoires reveals novel variants with altered specificities in closely related Malian Xanthomonas oryzae pv. oryzae strains. Front. Microbiol. 2018, 9, 1657. [Google Scholar] [CrossRef] [Green Version]
- Ding, P.; Ding, Y. Stories of salicylic acid: A plant defense hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Jelenska, J.; van Hal, J.A.; Greenberg, J.T. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. USA 2010, 107, 13177–13182. [Google Scholar] [CrossRef] [Green Version]
- Jelenska, J.; Yao, N.; Vinatzer, B.A.; Wright, C.M.; Brodsky, J.L.; Greenberg, J.T. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 2007, 17, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Chen, J.; Li, M.; Chang, M.; Xu, K.; Shang, Z.; Zhao, Y.; Palmer, I.; Zhang, Y.; McGill, J.; et al. A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 2017, 22, 777–788. [Google Scholar] [CrossRef] [Green Version]
- Lowe-Power, T.M.; Jacobs, J.M.; Ailloud, F.; Fochs, B.; Prior, P.; Allen, C. Degradation of the plant defense signal salicylic acid protects Ralstonia solanacearum from toxicity and enhances virulence on tobacco. mBio 2016, 7, e00656-16. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Pang, Z.; Trivedi, P.; Zhou, X.; Ying, X.; Jia, H.; Wang, N. ‘Candidatus Liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant Microbe Interact. 2017, 30, 620–630. [Google Scholar] [CrossRef] [Green Version]
- Shih, C.; Chu, H.; Tang, L.; Sakamoto, W.; Maekawa, M.; Chu, I.; Wang, M.; Lo, C. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 2008, 228, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Songkumarn, P.; Liu, J.; Wang, G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009, 150, 1111–1121. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.; Geng, T.; Liu, H.; Yang, W.; Zhong, W.; Zhang, Z.; Zhu, C.; Chu, Z. Foliar application of silicon enhances resistance against Phytophthora infestans through the ET/JA- and NPR1- dependent signaling pathways in potato. Front. Plant Sci. 2021, 12, 609870. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Peng, C.; Li, B.; Wu, W.; Kong, L.; Li, F.; Chu, Z.; Liu, F.; Ding, X. OsPGIP1-mediated resistance to bacterial leaf streak in rice is beyond responsive to the polygalacturonase of Xanthomonas oryzae pv. oryzicola. Rice 2019, 12, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y. CRISPR/Cas9 platforms for genome editing in plants: Developments and applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Han, H.; Chen, M.; Yang, W.; Liu, L.; Li, N.; Ding, X.; Chu, Z. Overexpression of OsDT11, which encodes a novel cysteine-rich peptide, enhances drought tolerance and increases ABA concentration in rice. Plant Mol. Biol. 2017, 93, 21–34. [Google Scholar] [CrossRef]

| Xoc Strains | Tal2b Ortholog | Tal2c Ortholog | Area of Isolation |
|---|---|---|---|
| HGA4 | + | + | China |
| RS105 | − | − | China |
| L8 | + | + | China |
| B8-12 | + | + | China |
| GX01 | + | + | China |
| 0–9 | + | + | China |
| BLS256 | + | + | Philippines |
| BLS279 | + | + | Philippines |
| CFBP2286 | + | + | Malaysia |
| CFBP7331/MAI10 | + | + | Mali |
| CFBP7341/BAI5 | + | + | Burkina |
| CFBP2286/BAI11 | + | + | Burkina |
| BXOR1 | + | + | India |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Zhang, H.; Bi, Y.; Yu, Y.; Liu, H.; Yang, H.; Yuan, B.; Ding, X.; Chu, Z. Tal2c Activates the Expression of OsF3H04g to Promote Infection as a Redundant TALE of Tal2b in Xanthomonas oryzae pv. oryzicola. Int. J. Mol. Sci. 2021, 22, 13628. https://doi.org/10.3390/ijms222413628
Wu T, Zhang H, Bi Y, Yu Y, Liu H, Yang H, Yuan B, Ding X, Chu Z. Tal2c Activates the Expression of OsF3H04g to Promote Infection as a Redundant TALE of Tal2b in Xanthomonas oryzae pv. oryzicola. International Journal of Molecular Sciences. 2021; 22(24):13628. https://doi.org/10.3390/ijms222413628
Chicago/Turabian StyleWu, Tao, Haimiao Zhang, Yunya Bi, Yue Yu, Haifeng Liu, Hong Yang, Bin Yuan, Xinhua Ding, and Zhaohui Chu. 2021. "Tal2c Activates the Expression of OsF3H04g to Promote Infection as a Redundant TALE of Tal2b in Xanthomonas oryzae pv. oryzicola" International Journal of Molecular Sciences 22, no. 24: 13628. https://doi.org/10.3390/ijms222413628
APA StyleWu, T., Zhang, H., Bi, Y., Yu, Y., Liu, H., Yang, H., Yuan, B., Ding, X., & Chu, Z. (2021). Tal2c Activates the Expression of OsF3H04g to Promote Infection as a Redundant TALE of Tal2b in Xanthomonas oryzae pv. oryzicola. International Journal of Molecular Sciences, 22(24), 13628. https://doi.org/10.3390/ijms222413628