Comparative 2D and 3D Ultrastructural Analyses of Dendritic Spines from CA1 Pyramidal Neurons in the Mouse Hippocampus
Abstract
1. Introduction
2. Results
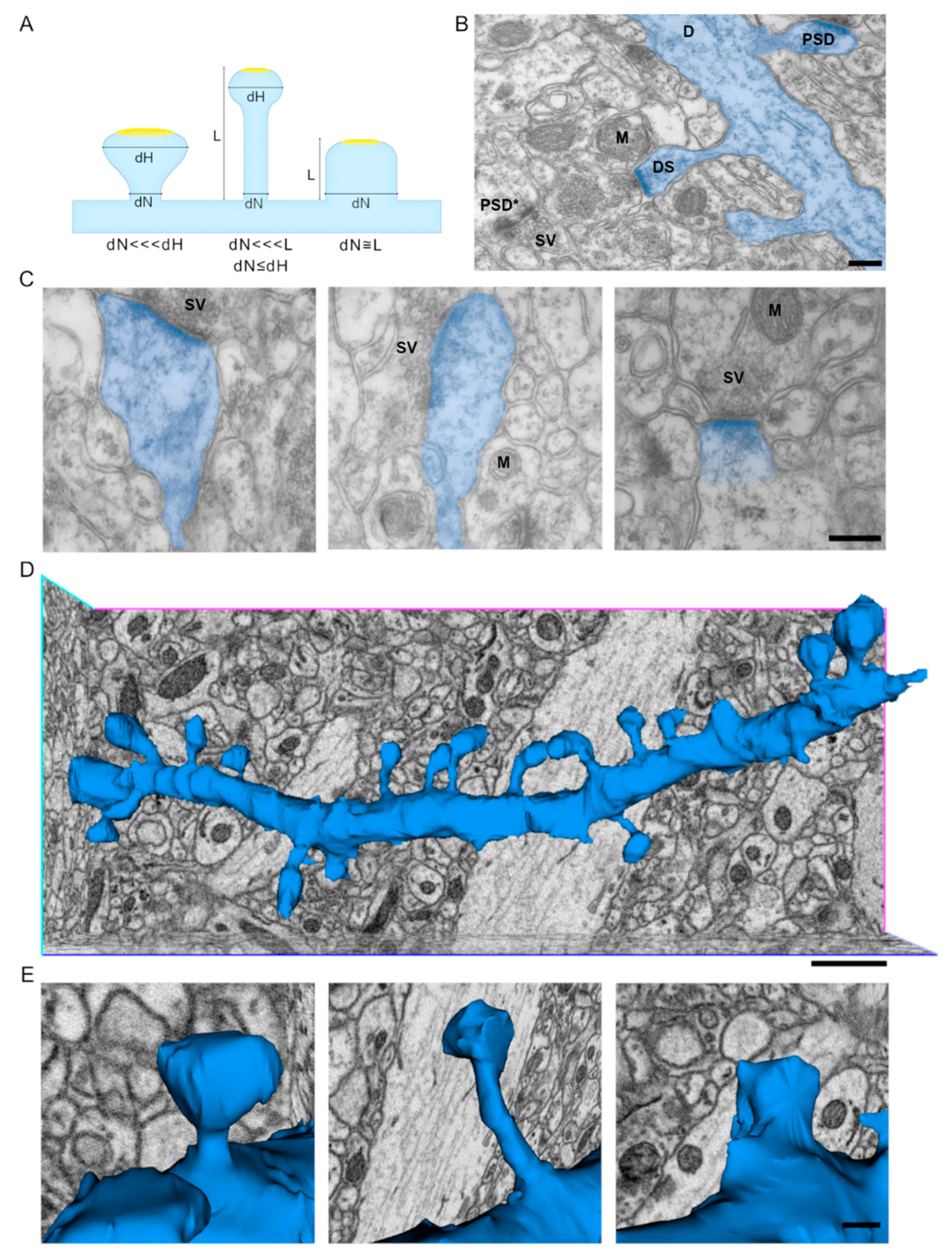
2.1. 3D Reconstruction of Dendrites Provides for a Better Picture of Spine Shape with Respect to Transmission Electron Microscopy 2D Projections
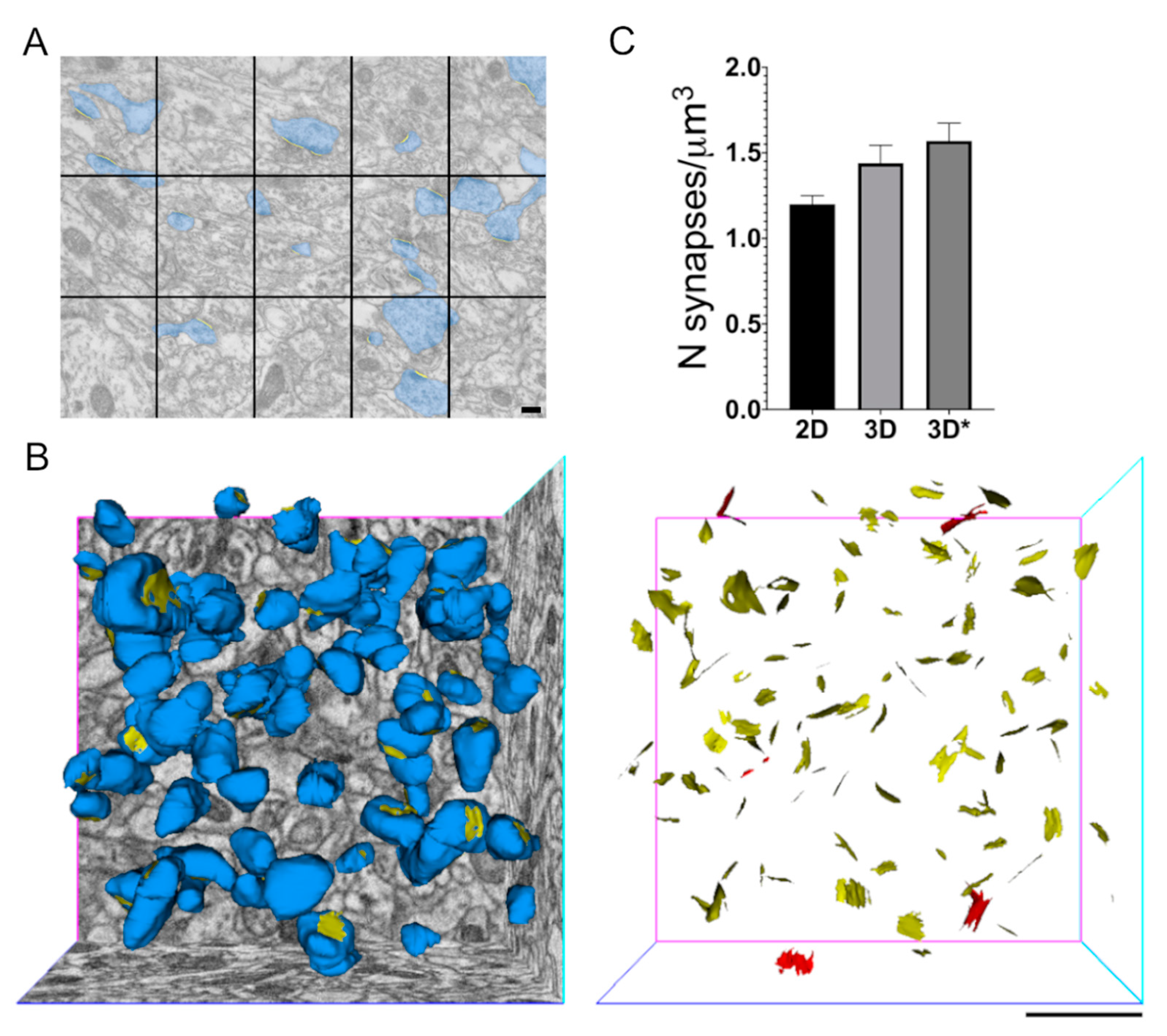
2.2. Stereology as a Favourable Approach for Spine and Synapse Density
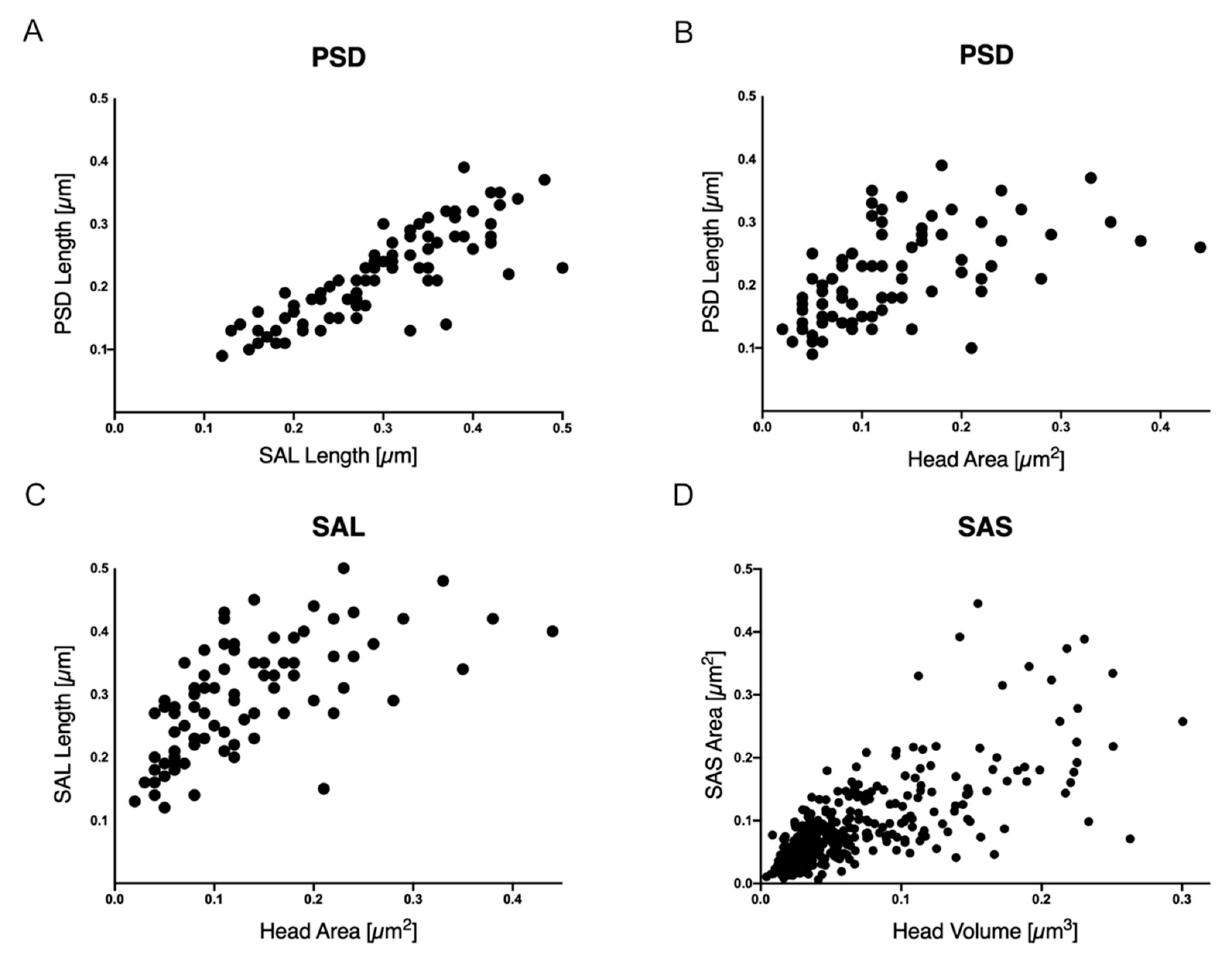
2.3. Spine Features Related to Their Function Can Be Evaluated with Both 2D and 3D EM Datasets
3. Discussion
4. Materials and Methods
4.1. Quantitative Morphometric Analysis of Dendritic Spines from 2D-TEM Micrographs
4.2. Reconstruction of Dendrites and Spine Heads from 3D Data-sets
4.3. Strategies to Compare the Data from 2D and 3D Objects
4.4. Data Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gray, E.G. Electron microscopy of synaptic contacts on dendrite spines of the cerebral cortex. Nature 1959, 183, 1592–1593. [Google Scholar] [CrossRef]
- Hering, H.; Sheng, M. Dendritic spines: Structure, dynamics and regulation. Nat. Rev. Neurosci. 2001, 2, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; Hoogenraad, C.C. The postsynaptic architecture of excitatory synapses: A more quantitative view. Annu. Rev. Biochem. 2007, 76, 823–847. [Google Scholar] [CrossRef] [PubMed]
- Rochefort, N.L.; Konnerth, A. Dendritic spines: From structure to in vivo function. EMBO Rep. 2012, 13, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Gipson, C.D.; Olive, M.F. Structural and functional plasticity of dendritic spines—root or result of behavior? Genes Brain Behav. 2017, 16, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.; Luebke, J.I.; Medalla, M. Comparative ultrastructural features of excitatory synapses in the visual and frontal cortices of the adult mouse and monkey. J. Comp. Neurol. 2017, 525, 2175–2191. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, D.H.; Zhang, S.; Gan, W.B. Dendritic spine dynamics. Annu. Rev. Physiol. 2009, 71, 261–282. [Google Scholar] [CrossRef]
- Harris, K.M.; Jensen, F.E.; Tsao, B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 1992, 12, 2685–2705. [Google Scholar] [CrossRef]
- Yuste, R.; Bonhoeffer, T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu. Rev. Neurosci. 2001, 24, 1071–1089. [Google Scholar] [CrossRef]
- Kasai, H.; Matsuzaki, M.; Noguchi, J.; Yasumatsu, N.; Nakahara, H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003, 26, 360–368. [Google Scholar] [CrossRef]
- Hayashi, Y.; Majewska, A.K. Dendritic spine geometry: Functional implication and regulation. Neuron 2005, 46, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.P.; Nedivi, E. Spine Dynamics: Are They All the Same? Neuron 2017, 96, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Runge, K.; Cardoso, C.; de Chevigny, A. Dendritic Spine Plasticity: Function and Mechanisms. Front. Synaptic Neurosci. 2020, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Bourne, J.N.; Harris, K.M. Nanoscale analysis of structural synaptic plasticity. Curr. Opin. Neurobiol. 2012, 22, 372–382. [Google Scholar] [CrossRef]
- Glebov, O.O.; Cox, S.; Humphreys, L.; Burrone, J. Neuronal activity controls transsynaptic geometry. Sci. Rep. 2016, 6, 22703. [Google Scholar] [CrossRef]
- Tao-Cheng, J.H. Stimulation induces gradual increases in the thickness and curvature of postsynaptic density of hippocampal CA1 neurons in slice cultures. Mol. Brain 2019, 12, 44. [Google Scholar] [CrossRef]
- Spacek, J.; Harris, K.M. Trans-endocytosis via spinules in adult rat hippocampus. J. Neurosci. 2004, 24, 4233–4241. [Google Scholar] [CrossRef]
- Herms, J.; Dorostkar, M.M. Dendritic Spine Pathology in Neurodegenerative Diseases. Annu. Rev. Pathol. 2016, 11, 221–250. [Google Scholar] [CrossRef]
- Chidambaram, S.B.; Rathipriya, A.G.; Bolla, S.R.; Bhat, A.; Ray, B.; Mahalakshmi, A.M.; Manivasagam, T.; Thenmozhi, A.J.; Essa, M.M.; Guillemin, G.J.; et al. Dendritic spines: Revisiting the physiological role. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 92, 161–193. [Google Scholar] [CrossRef]
- Bourne, J.N.; Harris, K.M. Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 2008, 31, 47–67. [Google Scholar] [CrossRef]
- Burette, A.; Collman, F.; Micheva, K.D.; Smith, S.J.; Weinberg, R.J. Knowing a synapse when you see one. Front. Neuroanat 2015, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Knott, G.; Genoud, C. Is EM dead? J. Cell Sci. 2013, 126 Pt 20, 4545–4552. [Google Scholar] [CrossRef] [PubMed]
- Kuwajima, M.; Spacek, J.; Harris, K.M. Beyond counts and shapes: Studying pathology of dendritic spines in the context of the surrounding neuropil through serial section electron microscopy. Neuroscience 2013, 251, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Kasthuri, N.; Hayworth, K.J.; Berger, D.R.; Schalek, R.L.; Conchello, J.A.; Knowles-Barley, S.; Lee, D.; Vázquez-Reina, A.; Kaynig, V.; Jones, T.R.; et al. Saturated Reconstruction of a Volume of Neocortex. Cell 2015, 162, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Swanson, L.W.; Lichtman, J.W. From Cajal to Connectome and Beyond. Annu. Rev. Neurosci. 2016, 39, 197–216. [Google Scholar] [CrossRef]
- Schmidt, H.; Gour, A.; Straehle, J.; Boergens, K.M.; Brecht, M.; Helmstaedter, M. Axonal synapse sorting in medial entorhinal cortex. Nature 2017, 549, 469–475. [Google Scholar] [CrossRef]
- Ohno, N.; Katoh, M.; Saitoh, Y.; Saitoh, S.; Ohno, S. Three-dimensional volume imaging with electron microscopy toward connectome. Microscopy (Oxf.) 2015, 64, 17–26. [Google Scholar] [CrossRef]
- Motta, A.; Schurr, M.; Staffler, B.; Helmstaedter, M. Big data in nanoscale connectomics, and the greed for training labels. Curr. Opin. Neurobiol. 2019, 55, 180–187. [Google Scholar] [CrossRef]
- Spano, G.M.; Banningh, S.W.; Marshall, W.; de Vivo, L.; Bellesi, M.; Loschky, S.S.; Tononi, G.; Cirelli, C. Sleep Deprivation by Exposure to Novel Objects Increases Synapse Density and Axon-Spine Interface in the Hippocampal CA1 Region of Adolescent Mice. J. Neurosci. 2019, 39, 6613–6625. [Google Scholar] [CrossRef]
- Borczyk, M.; Śliwińska, M.A.; Caly, A.; Bernas, T.; Radwanska, K. Neuronal plasticity affects correlation between the size of dendritic spine and its postsynaptic density. Sci. Rep. 2019, 9, 1693. [Google Scholar] [CrossRef]
- Südhof, T.C. The presynaptic active zone. Neuron 2012, 75, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Rodríguez, A.; Rodríguez, J.R.; Defelipe, J.; Merchán-Pérez, A. Characterization and extraction of the synaptic apposition surface for synaptic geometry analysis. Front. Neuroanat 2013, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.M.; Weinberg, R.J. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.M.; Stevens, J.K. Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: Serial electron microscopy with reference to their biophysical characteristics. J. Neurosci. 1989, 9, 2982–2997. [Google Scholar] [CrossRef]
- Schikorski, T.; Stevens, C.F. Quantitative ultrastructural analysis of hippocampal excitatory synapses. J. Neurosci. 1997, 17, 5858–5867. [Google Scholar] [CrossRef]
- Denk, W.; Horstmann, H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004, 2, e329. [Google Scholar] [CrossRef]
- Titze, B.; Genoud, C. Volume scanning electron microscopy for imaging biological ultrastructure. Biol. Cell 2016, 108, 307–323. [Google Scholar] [CrossRef]
- Xu, C.S.; Hayworth, K.J.; Lu, Z.; Grob, P.; Hassan, A.M.; García-Cerdán, J.G.; Niyogi, K.K.; Nogales, E.; Weinberg, R.J.; Hess, H.F. Enhanced FIB-SEM systems for large-volume 3D imaging. Elife 2017, 6. [Google Scholar] [CrossRef]
- Kubota, Y.; Sohn, J.; Hatada, S.; Schurr, M.; Straehle, J.; Gour, A.; Neujahr, R.; Miki, T.; Mikula, S.; Kawaguchi, Y. A carbon nanotube tape for serial-section electron microscopy of brain ultrastructure. Nat. Commun. 2018, 9, 437. [Google Scholar] [CrossRef]
- Helmstaedter, M. Cellular-resolution connectomics: Challenges of dense neural circuit reconstruction. Nat. Methods 2013, 10, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Pereira, U.; Rosa, M.G.; Kennedy, H. Brain connectomes come of age. Curr. Opin. Neurobiol. 2020, 65, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Baena, V.; Schalek, R.L.; Lichtman, J.W.; Terasaki, M. Serial-section electron microscopy using automated tape-collecting ultramicrotome (ATUM). Methods Cell Biol. 2019, 152, 41–67. [Google Scholar] [PubMed]
- Cohen, R.S.; Siekevitz, P. Form of the postsynaptic density. A serial section study. J. Cell Biol. 1978, 78, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Sampedro, M.; Hoff, S.F.; Cotman, C.W. Perforated postsynaptic densities: Probable intermediates in synapse turnover. Proc. Natl. Acad. Sci. USA 1982, 79, 5718–5722. [Google Scholar] [CrossRef] [PubMed]
- Santuy, A.; Rodriguez, J.R.; DeFelipe, J.; Merchan-Perez, A. Volume electron microscopy of the distribution of synapses in the neuropil of the juvenile rat somatosensory cortex. Brain Struct. Funct. 2018, 223, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Folci, A.; Murru, L.; Vezzoli, E.; Ponzoni, L.; Gerosa, L.; Moretto, E.; Longo, F.; Zapata, J.; Braida, D.; Pistillo, F.; et al. Myosin IXa Binds AMPAR and Regulates Synaptic Structure, LTP, and Cognitive Function. Front. Mol. Neurosci. 2016, 9, 1. [Google Scholar] [CrossRef]
- Murru, L.; Vezzoli, E.; Longatti, A.; Ponzoni, L.; Falqui, A.; Folci, A.; Moretto, E.; Bianchi, V.; Braida, D.; Sala, M.; et al. Pharmacological Modulation of AMPAR Rescues Intellectual Disability-Like Phenotype in Tm4sf2-/y Mice. Cereb. Cortex 2017, 27, 5369–5384. [Google Scholar] [CrossRef]
- Longaretti, A.; Forastieri, C.; Toffolo, E.; Caffino, L.; Locarno, A.; Misevičiūtė, I.; Marchesi, E.; Battistin, M.; Ponzoni, L.; Madaschi, L.; et al. LSD1 is an environmental stress-sensitive negative modulator of the glutamatergic synapse. Neurobiol. Stress 2020, 13, 100280. [Google Scholar] [CrossRef]
- Arellano, J.I.; Benavides-Piccione, R.; Defelipe, J.; Yuste, R. Ultrastructure of dendritic spines: Correlation between synaptic and spine morphologies. Front. Neurosci. 2007, 1, 131–143. [Google Scholar] [CrossRef]
- High, B.; Cole, A.A.; Chen, X.; Reese, T.S. Electron microscopic tomography reveals discrete transcleft elements at excitatory and inhibitory synapses. Front. Synaptic Neurosci. 2015, 7, 9. [Google Scholar] [CrossRef]
- Tao, C.L.; Liu, Y.T.; Sun, R.; Zhang, B.; Qi, L.; Shivakoti, S.; Tian, C.L.; Zhang, P.; Lau, P.M.; Zhou, Z.H.; et al. Differentiation and Characterization of Excitatory and Inhibitory Synapses by Cryo-electron Tomography and Correlative Microscopy. J. Neurosci. 2018, 38, 1493–1510. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.T.; Tao, C.L.; Zhang, X.; Xia, W.; Shi, D.Q.; Qi, L.; Xu, C.; Sun, R.; Li, X.W.; Lau, P.M.; et al. Mesophasic organization of GABA. Nat. Neurosci. 2020, 23, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Burnett, T.L.; Kelley, R.; Winiarski, B.; Contreras, L.; Daly, M.; Gholinia, A.; Burke, M.G.; Withers, P.J. Large volume serial section tomography by Xe Plasma FIB dual beam microscopy. Ultramicroscopy 2016, 161, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Soto, G.E.; Young, S.J.; Martone, M.E.; Deerinck, T.J.; Lamont, S.; Carragher, B.O.; Hama, K.; Ellisman, M.H. Serial section electron tomography: A method for three-dimensional reconstruction of large structures. Neuroimage 1994, 1, 230–243. [Google Scholar] [CrossRef]
- Raimondi, A.; Ferguson, S.M.; Lou, X.; Armbruster, M.; Paradise, S.; Giovedi, S.; Messa, M.; Kono, N.; Takasaki, J.; Cappello, V.; et al. Overlapping role of dynamin isoforms in synaptic vesicle endocytosis. Neuron 2011, 70, 1100–1114. [Google Scholar] [CrossRef]
- Menna, E.; Zambetti, S.; Morini, R.; Donzelli, A.; Disanza, A.; Calvigioni, D.; Braida, D.; Nicolini, C.; Orlando, M.; Fossati, G.; et al. Eps8 controls dendritic spine density and synaptic plasticity through its actin-capping activity. EMBO J. 2013, 32, 1730–1744. [Google Scholar] [CrossRef]
- DeFelipe, J.; Marco, P.; Busturia, I.; Merchán-Pérez, A. Estimation of the number of synapses in the cerebral cortex: Methodological considerations. Cereb. Cortex 1999, 9, 722–732. [Google Scholar] [CrossRef]
- Morales, J.; Alonso-Nanclares, L.; Rodríguez, J.R.; Defelipe, J.; Rodríguez, A.; Merchán-Pérez, A. Espina: A tool for the automated segmentation and counting of synapses in large stacks of electron microscopy images. Front. Neuroanat 2011, 5, 18. [Google Scholar] [CrossRef]
- Merchán-Pérez, A.; Rodriguez, J.R.; Alonso-Nanclares, L.; Schertel, A.; Defelipe, J. Counting Synapses Using FIB/SEM Microscopy: A True Revolution for Ultrastructural Volume Reconstruction. Front. Neuroanat 2009, 3, 18. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colombo, M.N.; Maiellano, G.; Putignano, S.; Scandella, L.; Francolini, M. Comparative 2D and 3D Ultrastructural Analyses of Dendritic Spines from CA1 Pyramidal Neurons in the Mouse Hippocampus. Int. J. Mol. Sci. 2021, 22, 1188. https://doi.org/10.3390/ijms22031188
Colombo MN, Maiellano G, Putignano S, Scandella L, Francolini M. Comparative 2D and 3D Ultrastructural Analyses of Dendritic Spines from CA1 Pyramidal Neurons in the Mouse Hippocampus. International Journal of Molecular Sciences. 2021; 22(3):1188. https://doi.org/10.3390/ijms22031188
Chicago/Turabian StyleColombo, Maria Nicol, Greta Maiellano, Sabrina Putignano, Lucrezia Scandella, and Maura Francolini. 2021. "Comparative 2D and 3D Ultrastructural Analyses of Dendritic Spines from CA1 Pyramidal Neurons in the Mouse Hippocampus" International Journal of Molecular Sciences 22, no. 3: 1188. https://doi.org/10.3390/ijms22031188
APA StyleColombo, M. N., Maiellano, G., Putignano, S., Scandella, L., & Francolini, M. (2021). Comparative 2D and 3D Ultrastructural Analyses of Dendritic Spines from CA1 Pyramidal Neurons in the Mouse Hippocampus. International Journal of Molecular Sciences, 22(3), 1188. https://doi.org/10.3390/ijms22031188




