Advanced Multi-Dimensional Cellular Models as Emerging Reality to Reproduce In Vitro the Human Body Complexity
Abstract
:1. Introduction
2. Multicellular Spheroids
2.1. Micro-Molded Non-Adherent Surfaces and Hydrogels
2.2. Magnetic Levitation
2.3. Microfluidic Devices
2.4. Multi-Approached Methods
3. Organoids and Organs-on-a-Chip
3.1. Organoids
3.2. Organs-on-a-Chip
4. Nanostructured Biomaterials
4.1. Decellularization Process
4.2. Electrospinning Method
4.3. Freeze-Casting Method
4.4. 3D Bioprinting Technology
4.5. Microfluidic Spinning Technology
4.6. Soft Robots
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ekert, J.E.; Deakyne, J.; Pribul-Allen, P.; Terry, R.; Schofield, C.; Jeong, C.G.; Storey, J.; Mohamet, L.; Francis, J.; Naidoo, A.; et al. Recommended Guidelines for Developing, Qualifying, and Implementing Complex In Vitro Models (CIVMs) for Drug Discovery. SLAS Discov. Adv. Life Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Eurostat. First Estimates of Research & Development Expenditure. R&D Expenditure in the EU Increased Slightly to 2.07% of GDP in 2017. Available online: https://ec.europa.eu/eurostat/documents/2995521/9483597/9-10012019-AP-EN.pdf/856ce1d3-b8a8-4fa6-bf00-a8ded6dd1cc1?t=1546960959000 (accessed on 7 January 2019).
- (PhRMA) Pharmaceutical Research and Manufacturers of America. 2016 Biopharmaceutical Research Industry Profile. Available online: http://phrma-docs.phrma.org/sites/default/files/pdf/biopharmaceutical-industry-profile.pdf (accessed on 8 January 2020).
- Unger, C.; Kramer, N.; Walzl, A.; Scherzer, M.; Hengstschläger, M.; Dolznig, H. Modeling human carcinomas: Physiologically relevant 3D models to improve anti-cancer drug development. Adv. Drug Deliv. Rev. 2014, 79–80, 50–67. [Google Scholar] [CrossRef] [PubMed]
- Ashammakhi, N.; Ahadian, S.; Darabi, M.A.; El Tahchi, M.; Lee, J.; Suthiwanich, K.; Sheikhi, A.; Dokmeci, M.R.; Oklu, R.; Khademhosseini, A. Minimally Invasive and Regenerative Therapeutics. Adv. Mater. 2019, 31, e1804041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Jin, G.-Z.; Han, C.-M.; Kim, H.-W. In vitro co-culture strategies to prevascularization for bone regeneration: A brief update. Tissue Eng. Regen. Med. 2015, 12, 69–79. [Google Scholar] [CrossRef]
- Skardal, A.; Mack, D.; Atala, A.; Soker, S. Substrate elasticity controls cell proliferation, surface marker expression and motile phenotype in amniotic fluid-derived stem cells. J. Mech. Behav. Biomed. Mater. 2013, 17, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Rietjens, I.M.; Alink, G.M. Future of toxicology--low-dose toxicology and risk--benefit analysis. Chem. Res. Toxicol. 2006, 19, 977–981. [Google Scholar] [CrossRef]
- Purchase, I.F. Fraud, errors and gamesmanship in experimental toxicology. Toxicology 2004, 202, 1–20. [Google Scholar] [CrossRef]
- Bolognin, S.; Fossépré, M.; Qing, X.; Jarazo, J.; Ščančar, J.; Moreno, E.L.; Nickels, S.L.; Wasner, K.; Ouzren, N.; Walter, J.; et al. 3D Cultures of Parkinson’s Disease-Specific Dopaminergic Neurons for High Content Phenotyping and Drug Testing. Adv. Sci. 2019, 6, 1800927. [Google Scholar] [CrossRef]
- Fennema, E.; Rivron, N.; Rouwkema, J.; van Blitterswijk, C.; de Boer, J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013, 31, 108–115. [Google Scholar] [CrossRef]
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in multicellular spheroids formation. J. R. Soc. Interface 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ao, Z.; Bin, C.; Muhsen, M.; Bondesson, M.; Lu, X.; Guo, F. Acoustic assembly of cell spheroids in disposable capillaries. Nanotechnology 2018, 29, 504006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimes, D.R.; Fletcher, A.G.; Partridge, M. Oxygen consumption dynamics in steady-state tumour models. R. Soc. Open Sci. 2014, 1, 140080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaganathan, H.; Gage, J.; Leonard, F.; Srinivasan, S.; Souza, G.R.; Dave, B.; Godin, B. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci. Rep. 2014, 4, 6468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laschke, M.W.; Menger, M.D. Life Is 3D: Boosting Spheroid Function for Tissue Engineering. Trends Biotechnol. 2017, 35, 133–144. [Google Scholar] [CrossRef]
- Powell, D.J.; Hertzberg, R.P.; Macarrόn, R. Design and Implementation of High-Throughput Screening Assays. Methods Mol. Biol. 2016, 1439, 1–32. [Google Scholar] [CrossRef]
- Dang, P.N.; Dwivedi, N.; Phillips, L.M.; Yu, X.; Herberg, S.; Bowerman, C.; Solorio, L.D.; Murphy, W.L.; Alsberg, E. Controlled Dual Growth Factor Delivery From Microparticles Incorporated Within Human Bone Marrow-Derived Mesenchymal Stem Cell Aggregates for Enhanced Bone Tissue Engineering via Endochondral Ossification. Stem Cells Transl. Med. 2016, 5, 206–217. [Google Scholar] [CrossRef]
- Mattix, B.; Olsen, T.R.; Gu, Y.; Casco, M.; Herbst, A.; Simionescu, D.T.; Visconti, R.P.; Kornev, K.G.; Alexis, F. Biological magnetic cellular spheroids as building blocks for tissue engineering. Acta Biomater. 2014, 10, 623–629. [Google Scholar] [CrossRef] [Green Version]
- Urbanczyk, M.; Zbinden, A.; Layland, S.L.; Duffy, G.; Schenke-Layland, K. Controlled Heterotypic Pseudo-Islet Assembly of Human β-Cells and Human Umbilical Vein Endothelial Cells Using Magnetic Levitation. Tissue Eng. Part A 2020, 26, 387–399. [Google Scholar] [CrossRef] [Green Version]
- Lewis, N.S.; Lewis, E.E.; Mullin, M.; Wheadon, H.; Dalby, M.J.; Berry, C.C. Magnetically levitated mesenchymal stem cell spheroids cultured with a collagen gel maintain phenotype and quiescence. J. Tissue Eng. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, T.; Shin, H.J.; Lee, J.; Shin, Y.M.; Perikamana, S.K.M.; Park, S.Y.; Jung, H.S.; Shin, H. Fabrication of in vitro 3D mineralized tissue by fusion of composite spheroids incorporating biomineral-coated nanofibers and human adipose-derived stem cells. Acta Biomater. 2018, 74, 464–477. [Google Scholar] [CrossRef]
- Shin, J.Y.; Park, J.; Jang, H.K.; Lee, T.J.; La, W.G.; Bhang, S.H.; Kwon, I.K.; Kwon, O.H.; Kim, B.S. Efficient formation of cell spheroids using polymer nanofibers. Biotechnol. Lett. 2012, 34, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Heo, D.N.; Hospodiuk, M.; Ozbolat, I.T. Synergistic interplay between human MSCs and HUVECs in 3D spheroids laden in collagen/fibrin hydrogels for bone tissue engineering. Acta Biomater. 2019, 95, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.S.; Hung, B.P.; Heyrani, N.; Lee, M.A.; Leach, J.K. Hypoxic Preconditioning of Mesenchymal Stem Cells with Subsequent Spheroid Formation Accelerates Repair of Segmental Bone Defects. Stem Cells 2018, 36, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Imaninezhad, M.; Hill, L.; Kolar, G.; Vogt, K.; Zustiak, S.P. Templated Macroporous Polyethylene Glycol Hydrogels for Spheroid and Aggregate Cell Culture. Bioconjug. Chem. 2019, 30, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Akagi, T.; Asaoka, T.; Eguchi, H.; Sasaki, K.; Iwagami, Y.; Yamada, D.; Noda, T.; Kawamoto, K.; Gotoh, K.; et al. Layer-by-layer cell coating technique using extracellular matrix facilitates rapid fabrication and function of pancreatic β-cell spheroids. Biomaterials 2018, 160, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Amaral, A.J.; Pasparakis, G. Rapid Formation of Cell Aggregates and Spheroids Induced by a “Smart” Boronic Acid Copolymer. Acs Appl. Mater. Interfaces 2016, 8, 22930–22941. [Google Scholar] [CrossRef]
- Metzger, W.; Sossong, D.; Bächle, A.; Pütz, N.; Wennemuth, G.; Pohlemann, T.; Oberringer, M. The liquid overlay technique is the key to formation of co-culture spheroids consisting of primary osteoblasts, fibroblasts and endothelial cells. Cytotherapy 2011, 13, 1000–1012. [Google Scholar] [CrossRef]
- Landry, J.; Bernier, D.; Ouellet, C.; Goyette, R.; Marceau, N. Spheroidal aggregate culture of rat liver cells: Histotypic reorganization, biomatrix deposition, and maintenance of functional activities. J. Cell Biol. 1985, 101, 914–923. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Yun, H.W.; Park, D.Y.; Choi, B.H.; Min, B.H. Three-Dimensional Spheroid Culture Increases Exosome Secretion from Mesenchymal Stem Cells. Tissue Eng. Regen. Med. 2018, 15, 427–436. [Google Scholar] [CrossRef]
- Ferreira, L.P.; Gaspar, V.M.; Monteiro, M.V.; Freitas, B.; Silva, N.J.O.; Mano, J.F. Screening of dual chemo-Photothermal cellular Nanotherapies in Organotypic breast Cancer 3D spheroids. J. Control. Release Off. J. Control. Release Soc. 2020. [Google Scholar] [CrossRef]
- Yakavets, I.; Francois, A.; Benoit, A.; Merlin, J.L.; Bezdetnaya, L.; Vogin, G. Advanced co-culture 3D breast cancer model for investigation of fibrosis induced by external stimuli: Optimization study. Sci. Rep. 2020, 10, 21273. [Google Scholar] [CrossRef]
- Costa, E.C.; de Melo-Diogo, D.; Moreira, A.F.; Carvalho, M.P.; Correia, I.J. Spheroids Formation on Non-Adhesive Surfaces by Liquid Overlay Technique: Considerations and Practical Approaches. Biotechnol. J. 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, A.P.; Chai, P.; Dean, D.M.; Morgan, J.R. Dynamics of the self-assembly of complex cellular aggregates on micromolded nonadhesive hydrogels. Tissue Eng. 2007, 13, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Jiang, J.; Yanase, T.; Nishi, Y.; Morgan, J.R. Connexon-mediated cell adhesion drives microtissue self-assembly. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2011, 25, 255–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaralı, Z.B.; Onak, G.; Karaman, O. Effect of Integrin Binding Peptide on Vascularization of Scaffold-Free Microtissue Spheroids. Tissue Eng. Regen. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Günter, J.; Wolint, P.; Bopp, A.; Steiger, J.; Cambria, E.; Hoerstrup, S.P.; Emmert, M.Y. Microtissues in Cardiovascular Medicine: Regenerative Potential Based on a 3D Microenvironment. Stem Cells Int. 2016, 2016, 9098523. [Google Scholar] [CrossRef] [Green Version]
- Safari, Z.; Soudi, S.; Jafarzadeh, N.; Hosseini, A.Z.; Vojoudi, E.; Sadeghizadeh, M. Promotion of angiogenesis by M13 phage and RGD peptide in vitro and in vivo. Sci. Rep. 2019, 9, 11182. [Google Scholar] [CrossRef] [Green Version]
- Nicosia, R.F.; Bonanno, E.; Smith, M. Fibronectin promotes the elongation of microvessels during angiogenesis in vitro. J. Cell. Physiol. 1993, 154, 654–661. [Google Scholar] [CrossRef]
- Schraa, A.J.; Kok, R.J.; Berendsen, A.D.; Moorlag, H.E.; Bos, E.J.; Meijer, D.K.; de Leij, L.F.; Molema, G. Endothelial cells internalize and degrade RGD-modified proteins developed for tumor vasculature targeting. J. Control. Release Off. J. Control. Release Soc. 2002, 83, 241–251. [Google Scholar] [CrossRef]
- Lam, C.R.; Wong, H.K.; Nai, S.; Chua, C.K.; Tan, N.S.; Tan, L.P. A 3D biomimetic model of tissue stiffness interface for cancer drug testing. Mol. Pharm. 2014, 11, 2016–2021. [Google Scholar] [CrossRef] [PubMed]
- Utama, R.H.; Atapattu, L.; O’Mahony, A.P.; Fife, C.M.; Baek, J.; Allard, T.; O’Mahony, K.J.; Ribeiro, J.C.C.; Gaus, K.; Kavallaris, M.; et al. A 3D Bioprinter Specifically Designed for the High-Throughput Production of Matrix-Embedded Multicellular Spheroids. iScience 2020, 23, 101621. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Buckle, A.M.; Markx, G.H. Tissue engineering with electric fields: Immobilization of mammalian cells in multilayer aggregates using dielectrophoresis. Biotechnol. Bioeng. 2007, 98, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Ino, K.; Okochi, M.; Honda, H. Application of magnetic force-based cell patterning for controlling cell-cell interactions in angiogenesis. Biotechnol. Bioeng. 2009, 102, 882–890. [Google Scholar] [CrossRef]
- Liu, J.; Kuznetsova, L.A.; Edwards, G.O.; Xu, J.; Ma, M.; Purcell, W.M.; Jackson, S.K.; Coakley, W.T. Functional three-dimensional HepG2 aggregate cultures generated from an ultrasound trap: Comparison with HepG2 spheroids. J. Cell. Biochem. 2007, 102, 1180–1189. [Google Scholar] [CrossRef]
- Jafari, J.; Han, X.L.; Palmer, J.; Tran, P.A.; O’Connor, A.J. Remote Control in Formation of 3D Multicellular Assemblies Using Magnetic Forces. ACS Biomater. Sci. Eng. 2019, 5, 2532–2542. [Google Scholar] [CrossRef]
- Ryu, N.E.; Lee, S.H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Hur, W. Scaffold-free formation of a millimeter-scale multicellular spheroid with an internal cavity from magnetically levitated 3T3 cells that ingested iron oxide-containing microspheres. Biotechnol. Bioeng. 2014, 111, 1038–1047. [Google Scholar] [CrossRef]
- Lewis, E.E.; Wheadon, H.; Lewis, N.; Yang, J.; Mullin, M.; Hursthouse, A.; Stirling, D.; Dalby, M.J.; Berry, C.C. A Quiescent, Regeneration-Responsive Tissue Engineered Mesenchymal Stem Cell Bone Marrow Niche Model via Magnetic Levitation. Acs Nano 2016, 10, 8346–8354. [Google Scholar] [CrossRef] [Green Version]
- Metzger, T.A.; Shudick, J.M.; Seekell, R.; Zhu, Y.; Niebur, G.L. Rheological behavior of fresh bone marrow and the effects of storage. J. Mech. Behav. Biomed. Mater. 2014, 40, 307–313. [Google Scholar] [CrossRef]
- Labusca, L.; Herea, D.D.; Minuti, A.E.; Stavila, C.; Danceanu, C.; Grigoras, M.; Ababei, G.; Chiriac, H.; Lupu, N. Magnetic nanoparticle loaded human adipose derived mesenchymal cells spheroids in levitated culture. J. Biomed. Mater. Res. Part B Appl. Biomater. 2020. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, T.; Yamazoe, H.; Mochizuki, N.; Khademhosseini, A.; Suzuki, H.; Fukuda, J. Preparation of arrays of cell spheroids and spheroid-monolayer cocultures within a microfluidic device. J. Biosci. Bioeng. 2010, 110, 572–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, E.L.; Lu, H. Three-dimensional models for studying development and disease: Moving on from organisms to organs-on-a-chip and organoids. Integr. Biol. Quant. Biosci. Nano Macro 2016, 8, 672–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agastin, S.; Giang, U.B.; Geng, Y.; Delouise, L.A.; King, M.R. Continuously perfused microbubble array for 3D tumor spheroid model. Biomicrofluidics 2011, 5, 24110. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.G.; Lee, H.; Cho, D.W. 3D Printing of Organs-On-Chips. Bioengineering 2017, 4, 10. [Google Scholar] [CrossRef]
- Park, J.; Lee, B.K.; Jeong, G.S.; Hyun, J.K.; Lee, C.J.; Lee, S.-H. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab. A Chip 2015, 15, 141–150. [Google Scholar] [CrossRef]
- Rutkowski, J.M.; Swartz, M.A. A driving force for change: Interstitial flow as a morphoregulator. Trends Cell Biol. 2007, 17, 44–50. [Google Scholar] [CrossRef]
- Abbott, N.J. Evidence for bulk flow of brain interstitial fluid: Significance for physiology and pathology. Neurochem. Int. 2004, 45, 545–552. [Google Scholar] [CrossRef]
- Zhuang, P.; Sun, A.X.; An, J.; Chua, C.K.; Chew, S.Y. 3D neural tissue models: From spheroids to bioprinting. Biomaterials 2018, 154, 113–133. [Google Scholar] [CrossRef]
- Park, J.; Wetzel, I.; Marriott, I.; Dréau, D.; D’Avanzo, C.; Kim, D.Y.; Tanzi, R.E.; Cho, H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 2018, 21, 941–951. [Google Scholar] [CrossRef]
- Wu, A.; Loutherback, K.; Lambert, G.; Estévez-Salmerón, L.; Tlsty, T.D.; Austin, R.H.; Sturm, J.C. Cell motility and drug gradients in the emergence of resistance to chemotherapy. Proc. Natl. Acad. Sci. USA 2013, 110, 16103–16108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.J.; Valadez, A.V.; Zuo, P.; Nie, Z. Microfluidic 3D cell culture: Potential application for tissue-based bioassays. Bioanalysis 2012, 4, 1509–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Reid, L.; Huang, Y.; Uhl, C.G.; He, R.; Zhou, C.; Liu, Y. Bi-layer blood vessel mimicking microfluidic platform for antitumor drug screening based on co-culturing 3D tumor spheroids and endothelial layers. Biomicrofluidics 2019, 13, 044108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michailidou, M.; Brown, H.K.; Lefley, D.V.; Evans, A.; Cross, S.S.; Coleman, R.E.; Brown, N.J.; Holen, I. Microvascular endothelial cell responses in vitro and in vivo: Modulation by zoledronic acid and paclitaxel? J. Vasc. Res. 2010, 47, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konagaya, S.; Iwata, H. Reproducible preparation of spheroids of pancreatic hormone positive cells from human iPS cells: An in vitro study. Biochim. Et Biophys. Acta 2016, 1860, 2008–2016. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Konagaya, S.; Turner, A.; Noda, Y.; Kitamura, S.; Kotera, H.; Iwata, H. Closed-channel culture system for efficient and reproducible differentiation of human pluripotent stem cells into islet cells. Biochem. Biophys. Res. Commun. 2017, 487, 344–350. [Google Scholar] [CrossRef]
- Hurrell, T.; Ellero, A.A.; Masso, Z.F.; Cromarty, A.D. Characterization and reproducibility of HepG2 hanging drop spheroids toxicology in vitro. Toxicol. Vitr. Int. J. Publ. Assoc. Bibra 2018, 50, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Frey, O.; Misun, P.M.; Fluri, D.A.; Hengstler, J.G.; Hierlemann, A. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat. Commun. 2014, 5, 4250. [Google Scholar] [CrossRef] [Green Version]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef]
- Izadpanah, S.; Shabani, P.; Aghebati-Maleki, A.; Baghbanzadeh, A.; Fotouhi, A.; Bisadi, A.; Aghebati-Maleki, L. Prospects for the involvement of cancer stem cells in the pathogenesis of osteosarcoma. J. Cell. Physiol. 2020, 235, 4167–4182. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Wu, D.; Wang, Y.; Wang, Z.; Zou, C.; Dai, Y.; Ng, C.F.; Teoh, J.Y.; Chan, F.L. Development of a novel and economical agar-based non-adherent three-dimensional culture method for enrichment of cancer stem-like cells. Stem Cell Res. Ther. 2018, 9, 243. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.; Singh, S.K.; Saxena, A.K.; Tiwari, S.; Sharma, L.K.; Tiwari, M. Role of autophagy in regulation of glioma stem cells population during therapeutic stress. J. Stem Cells Regen. Med. 2020, 16, 80–89. [Google Scholar] [CrossRef]
- Tang, Q.L.; Zhao, Z.Q.; Li, J.C.; Liang, Y.; Yin, J.Q.; Zou, C.Y.; Xie, X.B.; Zeng, Y.X.; Shen, J.N.; Kang, T.; et al. Salinomycin inhibits osteosarcoma by targeting its tumor stem cells. Cancer Lett. 2011, 311, 113–121. [Google Scholar] [CrossRef]
- Goričan, L.; Gole, B. Head and Neck Cancer Stem Cell-Enriched Spheroid Model for Anticancer Compound Screening. Cells 2020, 9, 1707. [Google Scholar] [CrossRef]
- Sant, S.; Johnston, P.A. The production of 3D tumor spheroids for cancer drug discovery. Drug Discov. Today Technol. 2017, 23, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Peitzsch, C.; Nathansen, J.; Schniewind, S.I.; Schwarz, F.; Dubrovska, A. Cancer Stem Cells in Head and Neck Squamous Cell Carcinoma: Identification, Characterization and Clinical Implications. Cancers 2019, 11, 616. [Google Scholar] [CrossRef] [Green Version]
- Shan, F.; Close, D.A.; Camarco, D.P.; Johnston, P.A. High-Content Screening Comparison of Cancer Drug Accumulation and Distribution in Two-Dimensional and Three-Dimensional Culture Models of Head and Neck Cancer. Assay Drug Dev. Technol. 2018, 16, 27–50. [Google Scholar] [CrossRef]
- Hong, Y.J.; Do, J.T. Neural Lineage Differentiation From Pluripotent Stem Cells to Mimic Human Brain Tissues. Front. Bioeng. Biotechnol. 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Finkbeiner, S.R.; Zeng, X.L.; Utama, B.; Atmar, R.L.; Shroyer, N.F.; Estes, M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 2012, 3, e00159-00112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekkers, J.F.; Wiegerinckbv, C.L.; de Jonge, H.R.; Bronsveld, I.; Janssens, H.M.; de Winter-de Groot, K.M.; Brandsma, A.M.; de Jong, N.W.; Bijvelds, M.J.; Scholte, B.J.; et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013, 19, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Hunziker, W. Engineering Microfluidic Organoid-on-a-Chip Platforms. Micromachines 2019, 10, 165. [Google Scholar] [CrossRef] [Green Version]
- Park, S.E.; Georgescu, A. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Chen, H.Y.; Kaya, K.D.; Dong, L.; Swaroop, A. Three-dimensional retinal organoids from mouse pluripotent stem cells mimic in vivo development with enhanced stratification and rod photoreceptor differentiation. Mol. Vis. 2016, 22, 1077–1094. [Google Scholar]
- Lakowski, J.; Welby, E.; Budinger, D.; Di Marco, F.; Di Foggia, V.; Bainbridge, J.W.B.; Wallace, K.; Gamm, D.M.; Ali, R.R.; Sowden, J.C. Isolation of Human Photoreceptor Precursors via a Cell Surface Marker Panel from Stem Cell-Derived Retinal Organoids and Fetal Retinae. Stem Cells 2018, 36, 709–722. [Google Scholar] [CrossRef] [Green Version]
- Lidgerwood, G.E.; Hewitt, A.W.; Pébay, A. Human pluripotent stem cells for the modelling of diseases of the retina and optic nerve: Toward a retina in a dish. Curr. Opin. Pharmacol. 2019, 48, 114–119. [Google Scholar] [CrossRef]
- Groveman, B.R.; Walters, R.; Haigh, C.L. Using our mini-brains: Cerebral organoids as an improved cellular model for human prion disease. Neural Regen. Res. 2020, 15, 1019–1020. [Google Scholar] [CrossRef]
- Faravelli, I.; Costamagna, G.; Tamanini, S.; Corti, S. Back to the origins: Human brain organoids to investigate neurodegeneration. Brain Res. 2020, 1727, 146561. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renner, M.; Lancaster, M.A.; Bian, S.; Choi, H.; Ku, T.; Peer, A.; Chung, K.; Knoblich, J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017, 36, 1316–1329. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kim, W.; Lim, S.; Jeon, J.S. Vasculature-On-A-Chip for In Vitro Disease Models. Bioengineering 2017, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Kam, C.; Shuler, M.L. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab. A Chip 2010, 10, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, K.M.; Saeed, O.; Zaidi, A.; Sanz, J.; Nielsen, J.C.; Hsu, D.T.; Jorde, U.P. 3D Printing to Guide Ventricular Assist Device Placement in Adults With Congenital Heart Disease and Heart Failure. JACC Heart Fail. 2016, 4, 301–311. [Google Scholar] [CrossRef]
- Schievano, S.; Migliavacca, F.; Coats, L.; Khambadkone, S.; Carminati, M.; Wilson, N.; Deanfield, J.E.; Bonhoeffer, P.; Taylor, A.M. Percutaneous Pulmonary Valve Implantation Based on Rapid Prototyping of Right Ventricular Outflow Tract and Pulmonary Trunk from MR Data. Radiolpgy 2007, 242, 490–497. [Google Scholar] [CrossRef]
- Wake, N.; Chandarana, H.; Huang, W.C.; Taneja, S.S.; Rosenkrantz, A.B. Application of anatomically accurate, patient-specific 3D printed models from MRI data in urological oncology. Clin. Radiol. 2016, 71, 610–614. [Google Scholar] [CrossRef]
- Kusaka, M.; Sugimoto, M.; Fukami, N.; Sasaki, H.; Takenaka, M.; Anraku, T.; Ito, T.; Kenmochi, T.; Shiroki, R.; Hoshinaga, K. Initial Experience With a Tailor-made Simulation and Navigation Program Using a 3-D Printer Model of Kidney Transplantation Surgery. Transplant. Proc. 2015, 47, 596–599. [Google Scholar] [CrossRef]
- Erbano, B.O.; Opolski, A.C.; Olandoski, M.; Foggiatto, J.A.; Kubrusly, L.F.; Dietz, U.A.; Zini, C.; Marinho, M.M.; Leal, A.G.; Ramina, R. Rapid prototyping of three-dimensional biomodels as an adjuvant in the surgical planning for intracranial aneurysms. Acta Cir. Bras. 2013, 28, 756–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.H.; Kang, J.-W.; Kim, N.; Song, J.-K.; Lee, J.-W.; Lim, T.-H. Myocardial 3-Dimensional Printing for Septal Myectomy Guidance in a Patient With Obstructive Hypertrophic Cardiomyopathy. Circulationaha 2015, 132, 300–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurenov, S.N.; Ionita, C.; Sammons, D.; Demmy, T.L. Three-dimensional printing to facilitate anatomic study, device development, simulation, and planning in thoracic surgery. J. Thorac. Cardiovasc. Surg. 2015, 149, 973–979.e971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderburgh, J.; Sterling, J.A.; Guelcher, S.A. 3D Printing of Tissue Engineered Constructs for In Vitro Modeling of Disease Progression and Drug Screening. Ann. Biomed. Eng. 2017, 45, 164–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Choudhury, D. Microfluidic bioprinting for organ-on-a-chip models. Drug Discov. Today 2019, 24, 1248–1257. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Davoudi, F.; Walch, P.; Manbachi, A.; Luo, X.; Dell’Erba, V.; Miri, A.K.; Albadawi, H.; Arneri, A.; Li, X.; et al. Bioprinted thrombosis-on-a-chip. Lab. A Chip 2016, 16, 4097–4105. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Lee, S.J.; Cheng, H.-J.; Yoo, J.J.; Atala, A. 3D bioprinted functional and contractile cardiac tissue constructs. Acta Biomater. 2018, 70, 48–56. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.-R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Park, J.Y.; Ryu, H.; Lee, B.; Ha, D.-H.; Ahn, M.; Kim, S.; Kim, J.Y.; Jeon, N.L.; Cho, D.-W. Development of a functional airway-on-a-chip by 3D cell printing. Biofabrication 2018, 11, 015002. [Google Scholar] [CrossRef]
- Seo, J.; Byun, W.Y.; Alisafaei, F.; Georgescu, A.; Yi, Y.-S.; Massaro-Giordano, M.; Shenoy, V.B.; Lee, V.; Bunya, V.Y.; Huh, D. Multiscale reverse engineering of the human ocular surface. Nat. Med. 2019, 25, 1310–1318. [Google Scholar] [CrossRef]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Homan, K.A.; Kolesky, D.B.; Skylar-Scott, M.A.; Herrmann, J.; Obuobi, H.; Moisan, A.; Lewis, J.A. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci. Rep. 2016, 6, 34845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, D.B.; Frismantas, V.; Milton, Y.; David, R.; Pop-Damkov, P.; Ferguson, D.; MacDonald, A.; Vargel Bölükbaşı, Ö. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat. Biomed. Eng. 2020, 4, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Chitty, J.L.; Skhinas, J.N. The Mini-Organo: A rapid high-throughput 3D coculture organotypic assay for oncology screening and drug development. Cancer Rep. 2020, 3, e1209. [Google Scholar] [CrossRef] [Green Version]
- Marturano-Kruik, A.; Nava, M.M.; Yeager, K.; Chramiec, A.; Hao, L.; Robinson, S.; Guo, E.; Raimondi, M.T. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA 2018, 115, 1256–1261. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Witkowski, M.T. Leukemia-on-a-chip: Dissecting the chemoresistance mechanisms in B cell acute lymphoblastic leukemia bone marrow niche. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro-Oncol. 2016, 18, v1–v75. [Google Scholar] [CrossRef] [Green Version]
- Yi, H.G.; Jeong, Y.H.; Kim, Y.; Choi, Y.J.; Moon, H.E.; Park, S.H. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef]
- Li, J.; Parra-Cantu, C.; Wang, Z.; Zhang, Y.S. Improving Bioprinted Volumetric Tumor Microenvironments In Vitro. Trends Cancer 2020. [Google Scholar] [CrossRef]
- Cui, X.; Ma, C. Dissecting the immunosuppressive tumor microenvironments in Glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. ELife 2020, 9. [Google Scholar] [CrossRef]
- Brancato, V.; Oliveira, J.M.; Correlo, V.M.; Reis, R.L.; Kundu, S.C. Could 3D models of cancer enhance drug screening? Biomaterials 2020, 232, 119744. [Google Scholar] [CrossRef] [PubMed]
- Eduati, F.; Utharala, R.; Madhavan, D.; Neumann, U.P.; Longerich, T.; Cramer, T.; Saez-Rodriguez, J.; Merten, C.A. A microfluidics platform for combinatorial drug screening on cancer biopsies. Nat. Commun. 2018, 9, 2434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhury, D.; Yee, M.; Sheng, Z.L.J.; Amirul, A.; Naing, M.W. Decellularization systems and devices: State-of-the-art. Acta Biomater. 2020, 115, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Jun, Y.; Qin, J.; Lee, S.H. Electrospinning versus microfluidic spinning of functional fibers for biomedical applications. Biomaterials 2017, 114, 121–143. [Google Scholar] [CrossRef] [PubMed]
- Eatemadi, A.; Daraee, H.; Zarghami, N.; Yar, H.M.; Akbarzadeh, A. Nanofiber: Synthesis and biomedical applications. Artif. Cells Nanomed. Biotechnol. 2016, 44, 111–121. [Google Scholar] [CrossRef]
- Farhangdoust, S.; Zamanian, A.; Yasaei, M.; Khorami, M. The effect of processing parameters and solid concentration on the mechanical and microstructural properties of freeze-casted macroporous hydroxyapatite scaffolds. Mater. Sci. Eng. C Mater. Biol. Appl. 2013, 33, 453–460. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, S.K.; Bardia, A.; Lakkireddy, C.; Nagarapu, R.; Habeeb, M.A.; Khan, A.A. Bioengineered humanized livers as better three-dimensional drug testing model system. World J. Hepatol. 2018, 10, 22–33. [Google Scholar] [CrossRef]
- Taylor, D.A.; Sampaio, L.C.; Ferdous, Z.; Gobin, A.S.; Taite, L.J. Decellularized matrices in regenerative medicine. Acta Biomater. 2018, 74, 74–89. [Google Scholar] [CrossRef]
- Uday Chandrika, K.; Tripathi, R.; Kameshwari, Y.; Rangaraj, N.; Mahesh Kumar, J.; Singh, S. Refunctionalization of Decellularized Organ Scaffold of Pancreas by Recellularization: Whole Organ Regeneration into Functional Pancreas. Tissue Eng. Regen. Med. 2020. [Google Scholar] [CrossRef]
- Lehmann, J.; Nürnberger, S.; Narcisi, R.; Stok, K.S.; van der Eerden, B.C.J.; Koevoet, W.; Kops, N.; Ten Berge, D.; van Osch, G.J. Recellularization of auricular cartilage via elastase-generated channels. Biofabrication 2019, 11, 035012. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.R.; Rudert, M.; Herrmann, M. Decellularized human bone as a 3D model to study skeletal progenitor cells in a natural environment. Methods Cell Biol. 2020, 157, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, J.; Liu, C.; Zhang, S.; Gao, F.; Guo, W.; Sun, X.; Zhang, C.; Li, H.; Rao, Z.; et al. Understanding the role of tissue-specific decellularized spinal cord matrix hydrogel for neural stem/progenitor cell microenvironment reconstruction and spinal cord injury. Biomaterials 2020, 268, 120596. [Google Scholar] [CrossRef]
- Sobreiro-Almeida, R.; Melica, M.E.; Lasagni, L.; Osório, H.; Romagnani, P.; Neves, N.M. Particulate kidney extracellular matrix: Bioactivity and proteomic analysis of a novel scaffold from porcine origin. Biomater. Sci. 2021, 9, 186–198. [Google Scholar] [CrossRef]
- Alabi, B.R.; LaRanger, R.; Shay, J.W. Decellularized mice colons as models to study the contribution of the extracellular matrix to cell behavior and colon cancer progression. Acta Biomater. 2019, 100, 213–222. [Google Scholar] [CrossRef]
- Lü, W.D.; Zhang, L.; Wu, C.L.; Liu, Z.G.; Lei, G.Y.; Liu, J.; Gao, W.; Hu, Y.R. Development of an acellular tumor extracellular matrix as a three-dimensional scaffold for tumor engineering. PLoS ONE 2014, 9, e103672. [Google Scholar] [CrossRef]
- Gaggi, G.; Di Credico, A.; Izzicupo, P.; Sancilio, S.; Di Mauro, M.; Iannetti, G.; Dolci, S.; Amabile, G.; Di Baldassarre, A.; Ghinassi, B. Decellularized Extracellular Matrices and Cardiac Differentiation: Study on Human Amniotic Fluid-Stem Cells. Int. J. Mol. Sci. 2020, 21, 6317. [Google Scholar] [CrossRef]
- Choudhury, D.; Tun, H.W.; Wang, T.; Naing, M.W. Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing? Trends Biotechnol. 2018, 36, 787–805. [Google Scholar] [CrossRef]
- Saldin, L.T.; Cramer, M.C.; Velankar, S.S.; White, L.J.; Badylak, S.F. Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomater. 2017, 49, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Pouliot, R.A.; Link, P.A.; Mikhaiel, N.S.; Schneck, M.B.; Valentine, M.S.; Kamga Gninzeko, F.J.; Herbert, J.A.; Sakagami, M.; Heise, R.L. Development and characterization of a naturally derived lung extracellular matrix hydrogel. J. Biomed. Mater. Res. Part. A 2016, 104, 1922–1935. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Pr, A.K.; Yoo, J.J.; Zahran, F.; Atala, A.; Lee, S.J. A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation. Adv. Healthc. Mater. 2019, 8, e1800992. [Google Scholar] [CrossRef] [PubMed]
- DeQuach, J.A.; Yuan, S.H.; Goldstein, L.S.; Christman, K.L. Decellularized porcine brain matrix for cell culture and tissue engineering scaffolds. Tissue Eng. Part. A 2011, 17, 2583–2592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medberry, C.J.; Crapo, P.M.; Siu, B.F.; Carruthers, C.A.; Wolf, M.T.; Nagarkar, S.P.; Agrawal, V.; Jones, K.E.; Kelly, J.; Johnson, S.A.; et al. Hydrogels derived from central nervous system extracellular matrix. Biomaterials 2013, 34, 1033–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paduano, F.; Marrelli, M.; White, L.J.; Shakesheff, K.M.; Tatullo, M. Odontogenic Differentiation of Human Dental Pulp Stem Cells on Hydrogel Scaffolds Derived from Decellularized Bone Extracellular Matrix and Collagen Type I. PLoS ONE 2016, 11, e0148225. [Google Scholar] [CrossRef] [Green Version]
- Keane, T.J.; Swinehart, I.T.; Badylak, S.F. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods (San Diegocalif.) 2015, 84, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Kočí, Z.; Výborný, K.; Dubišová, J.; Vacková, I.; Jäger, A.; Lunov, O.; Jiráková, K.; Kubinová, Š. Extracellular Matrix Hydrogel Derived from Human Umbilical Cord as a Scaffold for Neural Tissue Repair and Its Comparison with Extracellular Matrix from Porcine Tissues. Tissue Eng. Part. C Methods 2017, 23, 333–345. [Google Scholar] [CrossRef]
- Yao, Q.; Zheng, Y.W.; Lan, Q.H.; Kou, L.; Xu, H.L.; Zhao, Y.Z. Recent development and biomedical applications of decellularized extracellular matrix biomaterials. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 104, 109942. [Google Scholar] [CrossRef]
- Kc, P.; Hong, Y.; Zhang, G. Cardiac tissue-derived extracellular matrix scaffolds for myocardial repair: Advantages and challenges. Regen. Biomater. 2019, 6, 185–199. [Google Scholar] [CrossRef]
- Tebyanian, H.; Karami, A.; Motavallian, E.; Samadikuchaksaraei, A.; Arjmand, B.; Nourani, M.R. Rat lung decellularization using chemical detergents for lung tissue engineering. Biotech. Histochem. 2019, 94, 214–222. [Google Scholar] [CrossRef]
- Ahmadi-Aghkand, F.; Gholizadeh-Ghaleh Aziz, S.; Panahi, Y.; Daraee, H.; Gorjikhah, F.; Gholizadeh-Ghaleh Aziz, S.; Hsanzadeh, A.; Akbarzadeh, A. Recent prospective of nanofiber scaffolds fabrication approaches for skin regeneration. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Daud, M.F.; Pawar, K.C.; Claeyssens, F.; Ryan, A.J.; Haycock, J.W. An aligned 3D neuronal-glial co-culture model for peripheral nerve studies. Biomaterials 2012, 33, 5901–5913. [Google Scholar] [CrossRef] [PubMed]
- Malheiro, A.; Morgan, F.; Baker, M.; Moroni, L.; Wieringa, P. A three-dimensional biomimetic peripheral nerve model for drug testing and disease modelling. Biomaterials 2020, 257, 120230. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; Wieringa, P.; Moroni, L.; Navarro, X.; Valle, J.D. PEOT/PBT Guides Enhance Nerve Regeneration in Long Gap Defects. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef]
- Halabian, R.; Moridi, K.; Korani, M.; Ghollasi, M. Composite Nanoscaffolds Modified with Bio-ceramic Nanoparticles (Zn2SiO4) Prompted Osteogenic Differentiation of Human Induced Pluripotent Stem Cells. Int. J. Mol. Cell. Med. 2019, 8, 24–38. [Google Scholar] [CrossRef]
- Naudot, M.; Garcia, A.G.; Jankovsky, N.; Barre, A.; Zabijak, L.; Azdad, S.Z.; Collet, L.; Bedoui, F.; Hébraud, A.; Schlatter, G.; et al. The combination of a poly-caprolactone/nano-hydroxyapatite honeycomb scaffold and mesenchymal stem cells promotes bone regeneration in rat calvarial defects. J. Tissue Eng. Regen. Med. 2020, 14, 1570–1580. [Google Scholar] [CrossRef]
- Sridharan, D.; Palaniappan, A.; Blackstone, B.N.; Powell, H.M.; Khan, M. Electrospun Aligned Coaxial Nanofibrous Scaffold for Cardiac Repair. Methods Mol. Biol. (Cliftonn. J.) 2021, 2193, 129–140. [Google Scholar] [CrossRef]
- Anitha, R.; Vaikkath, D.; Shenoy, S.J.; Nair, P.D. Tissue-engineered islet-like cell clusters generated from adipose tissue-derived stem cells on three-dimensional electrospun scaffolds can reverse diabetes in an experimental rat model and the role of porosity of scaffolds on cluster differentiation. J. Biomed. Mater. Res. Part. A 2020, 108, 749–759. [Google Scholar] [CrossRef]
- Liu, W.; Dai, D. Cellular responses to flow diverters in a tissue-engineered aneurysm model. J. NeuroInterv. Surg. 2020. [Google Scholar] [CrossRef]
- Raeisdasteh Hokmabad, V.; Davaran, S.; Ramazani, A.; Salehi, R. Design and fabrication of porous biodegradable scaffolds: A strategy for tissue engineering. J. Biomater. Sci. Polym. Ed. 2017, 28, 1797–1825. [Google Scholar] [CrossRef]
- Wubneh, A.; Tsekoura, E.K.; Ayranci, C.; Uludağ, H. Current state of fabrication technologies and materials for bone tissue engineering. Acta Biomater. 2018, 80, 1–30. [Google Scholar] [CrossRef]
- Conde-González, A.; Dutta, D.; Wallace, R.; Callanan, A.; Bradley, M. Rapid fabrication and screening of tailored functional 3D biomaterials. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 108, 110489. [Google Scholar] [CrossRef]
- Nematollahi, Z.; Tafazzoli-Shadpour, M.; Zamanian, A.; Seyedsalehi, A.; Mohammad-Behgam, S.; Ghorbani, F.; Mirahmadi, F. Fabrication of Chitosan Silk-based Tracheal Scaffold Using Freeze-Casting Method. Iran. Biomed. J. 2017, 21, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.T.; Gullane, P.J. Current concepts in tracheal reconstruction. Curr. Opin. Otolaryngol. Head Neck Surg. 2012, 20, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Mirahmadi, F.; Tafazzoli-Shadpour, M.; Shokrgozar, M.A.; Bonakdar, S. Enhanced mechanical properties of thermosensitive chitosan hydrogel by silk fibers for cartilage tissue engineering. Mater. Sci. Eng. C Mater. Biol. Appl. 2013, 33, 4786–4794. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, J.; Bhatnagar, I.; Kim, S.K. Chitosan-alginate biocomposite containing fucoidan for bone tissue engineering. Mar. Drugs 2014, 12, 300–316. [Google Scholar] [CrossRef]
- Hsu, S.H.; Whu, S.W.; Hsieh, S.C.; Tsai, C.L.; Chen, D.C.; Tan, T.S. Evaluation of chitosan-alginate-hyaluronate complexes modified by an RGD-containing protein as tissue-engineering scaffolds for cartilage regeneration. Artif. Organs 2004, 28, 693–703. [Google Scholar] [CrossRef]
- Li, Z.; Leung, M.; Hopper, R.; Ellenbogen, R.; Zhang, M. Feeder-free self-renewal of human embryonic stem cells in 3D porous natural polymer scaffolds. Biomaterials 2010, 31, 404–412. [Google Scholar] [CrossRef]
- Florczyk, S.J.; Kim, D.J.; Wood, D.L.; Zhang, M. Influence of processing parameters on pore structure of 3D porous chitosan-alginate polyelectrolyte complex scaffolds. J. Biomed. Mater. Res. Part. A 2011, 98, 614–620. [Google Scholar] [CrossRef]
- Irvine, S.A.; Venkatraman, S.S. Bioprinting and Differentiation of Stem Cells. Molecules 2016, 21, 1188. [Google Scholar] [CrossRef]
- Gu, Q.; Tomaskovic-Crook, E.; Lozano, R.; Chen, Y.; Kapsa, R.M.; Zhou, Q.; Wallace, G.G.; Crook, J.M. Functional 3D Neural Mini-Tissues from Printed Gel-Based Bioink and Human Neural Stem Cells. Adv. Healthc. Mater. 2016, 5, 1429–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.B.; Polio, S.; Lee, W.; Dai, G.; Menon, L.; Carroll, R.S.; Yoo, S.S. Bio-printing of collagen and VEGF-releasing fibrin gel scaffolds for neural stem cell culture. Exp. Neurol. 2010, 223, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Sarker, M.D.; Naghieh, S.; Sharma, N.K.; Ning, L.; Chen, X. Bioprinting of Vascularized Tissue Scaffolds: Influence of Biopolymer, Cells, Growth Factors, and Gene Delivery. J. Healthc. Eng. 2019, 2019, 9156921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedir, T.; Ulag, S.; Ustundag, C.B.; Gunduz, O. 3D bioprinting applications in neural tissue engineering for spinal cord injury repair. Mater. Sci Eng. C Mater. Biol. Appl. 2020, 110, 110741. [Google Scholar] [CrossRef]
- Sagner, A.; Briscoe, J. Establishing neuronal diversity in the spinal cord: A time and a place. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Joung, D.; Truong, V.; Neitzke, C.C.; Guo, S.Z.; Walsh, P.J.; Monat, J.R.; Meng, F.; Park, S.H.; Dutton, J.R.; Parr, A.M.; et al. 3D Printed Stem-Cell Derived Neural Progenitors Generate Spinal Cord Scaffolds. Adv. Funct. Mater. 2018, 28. [Google Scholar] [CrossRef]
- Pawelec, K.M.; Koffler, J.; Shahriari, D.; Galvan, A.; Tuszynski, M.H.; Sakamoto, J. Microstructure and in vivo characterization of multi-channel nerve guidance scaffolds. Biomed. Mater. 2018, 13, 044104. [Google Scholar] [CrossRef]
- Krych, A.J.; Rooney, G.E.; Chen, B.; Schermerhorn, T.C.; Ameenuddin, S.; Gross, L.; Moore, M.J.; Currier, B.L.; Spinner, R.J.; Friedman, J.A.; et al. Relationship between scaffold channel diameter and number of regenerating axons in the transected rat spinal cord. Acta Biomater. 2009, 5, 2551–2559. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.N.; Lancaster, K.Z.; Zhen, G.; He, J.; Gupta, M.K.; Kong, Y.L.; Engel, E.A.; Krick, K.D.; Ju, A.; Meng, F.; et al. 3D Printed Anatomical Nerve Regeneration Pathways. Adv. Funct. Mater. 2015, 25, 6205–6217. [Google Scholar] [CrossRef]
- Gupta, M.K.; Meng, F.; Johnson, B.N.; Kong, Y.L.; Tian, L.; Yeh, Y.W.; Masters, N.; Singamaneni, S.; McAlpine, M.C. 3D Printed Programmable Release Capsules. Nano Lett. 2015, 15, 5321–5329. [Google Scholar] [CrossRef] [Green Version]
- Qiu, K.; Zhao, Z.; Haghiashtiani, G.; Guo, S.Z.; He, M.; Su, R.; Zhu, Z.; Bhuiyan, D.B.; Murugan, P.; Meng, F.; et al. 3D Printed Organ Models with Physical Properties of Tissue and Integrated Sensors. Adv. Mater. Technol. 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betsch, M.; Cristian, C.; Lin, Y.Y.; Blaeser, A.; Schöneberg, J.; Vogt, M.; Buhl, E.M.; Fischer, H.; Duarte Campos, D.F. Incorporating 4D into Bioprinting: Real-Time Magnetically Directed Collagen Fiber Alignment for Generating Complex Multilayered Tissues. Adv. Health Mater. 2018, 7, e1800894. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, C.; Fosang, A.; Donati, D.M.; Wallace, G.G.; Choong, P.F. 3D Bioprinting of Cartilage for Orthopedic Surgeons: Reading between the Lines. Front. Surg. 2015, 2, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte Campos, D.F.; Drescher, W.; Rath, B.; Tingart, M.; Fischer, H. Supporting Biomaterials for Articular Cartilage Repair. Cartilage 2012, 3, 205–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioannidis, K.; Danalatos, R.I.; Champeris Tsaniras, S.; Kaplani, K.; Lokka, G.; Kanellou, A.; Papachristou, D.J.; Bokias, G.; Lygerou, Z.; Taraviras, S. A Custom Ultra-Low-Cost 3D Bioprinter Supports Cell Growth and Differentiation. Front. Bioeng. Biotechnol. 2020, 8, 580889. [Google Scholar] [CrossRef] [PubMed]
- Ashammakhi, N.; Ahadian, S.; Zengjie, F.; Suthiwanich, K.; Lorestani, F.; Orive, G.; Ostrovidov, S.; Khademhosseini, A. Advances and Future Perspectives in 4D Bioprinting. Biotechnol. J. 2018, 13, e1800148. [Google Scholar] [CrossRef]
- Thomas, A.C.; Campbell, G.R.; Campbell, J.H. Advances in vascular tissue engineering. Cardiovasc. Pathol. Off. J. Soc. Cardiovasc. Pathol. 2003, 12, 271–276. [Google Scholar] [CrossRef]
- Wu, P.; Wang, L.; Li, W.; Zhang, Y.; Wu, Y.; Zhi, D.; Wang, H.; Wang, L.; Kong, D.; Zhu, M. Construction of vascular graft with circumferentially oriented microchannels for improving artery regeneration. Biomaterials 2020, 242, 119922. [Google Scholar] [CrossRef]
- Zhu, M.; Li, W.; Dong, X.; Yuan, X.; Midgley, A.C.; Chang, H.; Wang, Y.; Wang, H.; Wang, K.; Ma, P.X.; et al. In vivo engineered extracellular matrix scaffolds with instructive niches for oriented tissue regeneration. Nat. Commun. 2019, 10, 4620. [Google Scholar] [CrossRef] [Green Version]
- Pennings, I.; van Haaften, E.E.; Jungst, T.; Bulsink, J.A.; Rosenberg, A.; Groll, J.; Bouten, C.V.C.; Kurniawan, N.A.; Smits, A.; Gawlitta, D. Layer-specific cell differentiation in bi-layered vascular grafts under flow perfusion. Biofabrication 2019, 12, 015009. [Google Scholar] [CrossRef]
- Jun, Y.; Kang, E.; Chae, S.; Lee, S.H. Microfluidic spinning of micro- and nano-scale fibers for tissue engineering. Lab. A Chip 2014, 14, 2145–2160. [Google Scholar] [CrossRef] [PubMed]
- Du, X.Y.; Li, Q.; Wu, G.; Chen, S. Multifunctional Micro/Nanoscale Fibers Based on Microfluidic Spinning Technology. Adv. Mater. 2019, 31, e1903733. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Sun, T.; Shi, Q.; Liang, Q.; Yao, Y.; Wang, H.; Sun, J.; Huang, Q.; Fukuda, T. Fabrication of vascular smooth muscle-like tissues based on self-organization of circumferentially aligned cells in microengineered hydrogels. Lab. A Chip 2020, 20, 3120–3131. [Google Scholar] [CrossRef]
- Callens, S.J.P.; Uyttendaele, R.J.C.; Fratila-Apachitei, L.E.; Zadpoor, A.A. Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials 2020, 232, 119739. [Google Scholar] [CrossRef]
- Cui, T.; Yu, J.; Li, Q.; Wang, C.F. Large-Scale Fabrication of Robust Artificial Skins from a Biodegradable Sealant-Loaded Nanofiber Scaffold to Skin Tissue via Microfluidic Blow-Spinning. Adv. Mater. 2020, 32, e2000982. [Google Scholar] [CrossRef]
- Lee, K.H.; Shin, S.J.; Kim, C.B.; Kim, J.K.; Cho, Y.W.; Chung, B.G.; Lee, S.H. Microfluidic synthesis of pure chitosan microfibers for bio-artificial liver chip. Lab. A Chip 2010, 10, 1328–1334. [Google Scholar] [CrossRef]
- Lee, K.H.; Shin, S.J.; Park, Y.; Lee, S.H. Synthesis of cell-laden alginate hollow fibers using microfluidic chips and microvascularized tissue-engineering applications. Small 2009, 5, 1264–1268. [Google Scholar] [CrossRef]
- Park, D.; Park, J.; Jang, H.; Cheng, J.; Hyun Kim, S.; Lee, S.H. Simultaneous microfluidic spinning of multiple strands of submicron fiber for the production of free-standing porous membranes for biological application. Biofabrication 2017, 9, 025026. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Khademhosseini, A. Advances in engineering hydrogels. Science 2017, 356. [Google Scholar] [CrossRef]
- Acome, E.; Mitchell, S.K. Hydraulically amplified self-healing electrostatic actuators with muscle-like performance. Science 2018, 359, 61–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Li, Y.; Li, W.; Xiao, C.; Liu, D.; Dong, C.; Zhang, M.; Mäkilä, E.; Kemell, M.; Salonen, J.; et al. Multifunctional Nanohybrid Based on Porous Silicon Nanoparticles, Gold Nanoparticles, and Acetalated Dextran for Liver Regeneration and Acute Liver Failure Theranostics. Adv. Mater. 2018, 30, e1703393. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Liu, M.; Ishida, Y.; Ebina, Y.; Osada, M.; Sasaki, T.; Hikima, T.; Takata, M.; Aida, T. Thermoresponsive actuation enabled by permittivity switching in an electrostatically anisotropic hydrogel. Nat. Mater. 2015, 14, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Hines, L.; Petersen, K.; Lum, G.Z.; Sitti, M. Soft Actuators for Small-Scale Robotics. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef]
- Xu, L.; Mou, F.; Gong, H.; Luo, M.; Guan, J. Light-driven micro/nanomotors: From fundamentals to applications. Chem. Soc. Rev. 2017, 46, 6905–6926. [Google Scholar] [CrossRef]
- Ricotti, L.; Trimmer, B. Biohybrid actuators for robotics: A review of devices actuated by living cells. Sci. Robot. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Zhou, Q. Multifunctional biohybrid magnetite microrobots for imaging-guided therapy. Sci. Robot. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Park, B.W.; Zhuang, J.; Yasa, O. Multifunctional Bacteria-Driven Microswimmers for Targeted Active Drug Delivery. ACS Nano 2017, 11, 8910–8923. [Google Scholar] [CrossRef]
- Sun, L.; Chen, Z.; Bian, F.; Zhao, Y. Bioinspired Soft Robotic Caterpillar with Cardiomyocyte Drivers. Adv. Funct. Mater. 2020, 30, 1907820. [Google Scholar] [CrossRef]
- Tibbitt, M.W.; Anseth, K.S. Dynamic microenvironments: The fourth dimension. Sci. Transl. Med. 2012, 4, 160ps124. [Google Scholar] [CrossRef]

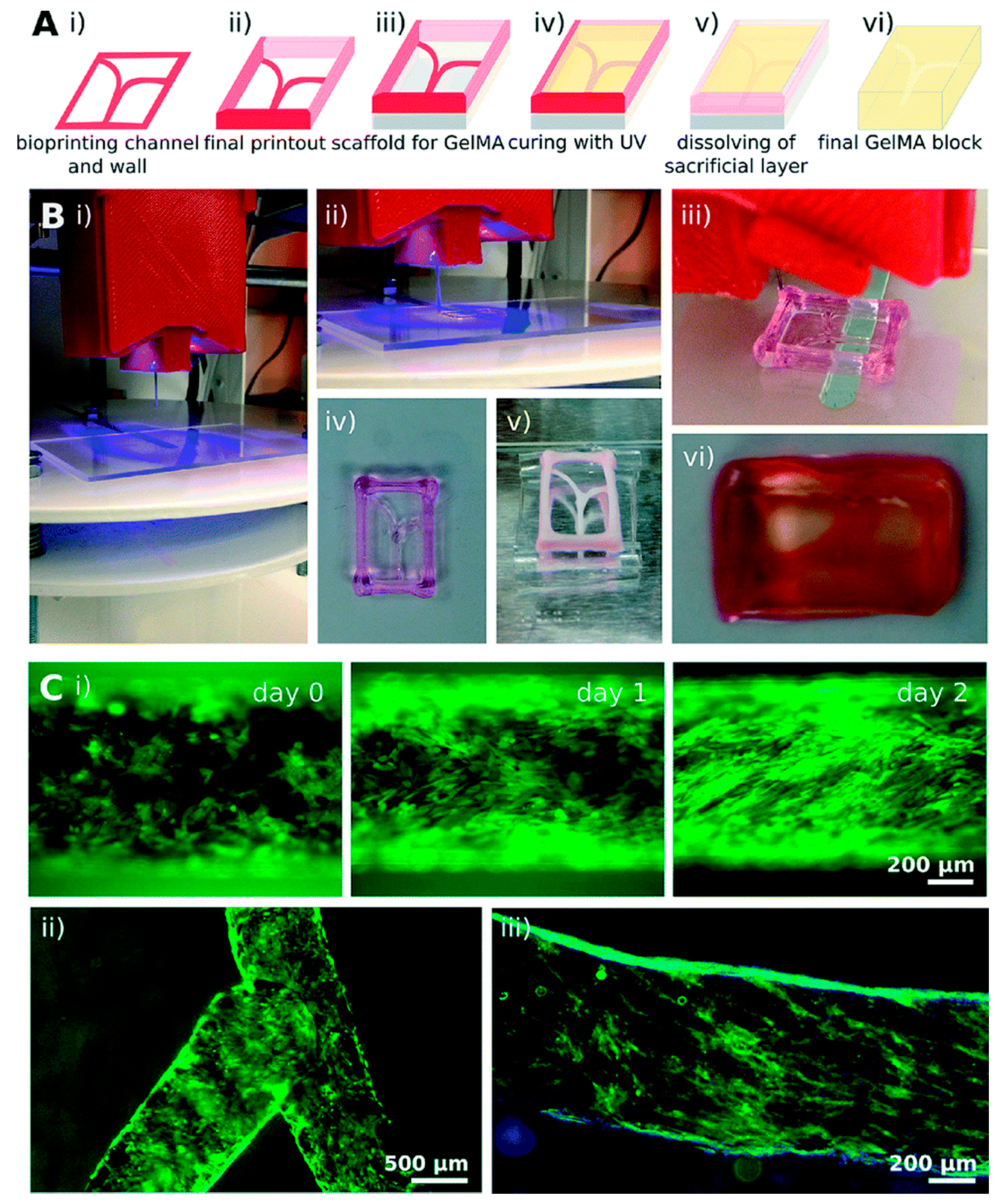
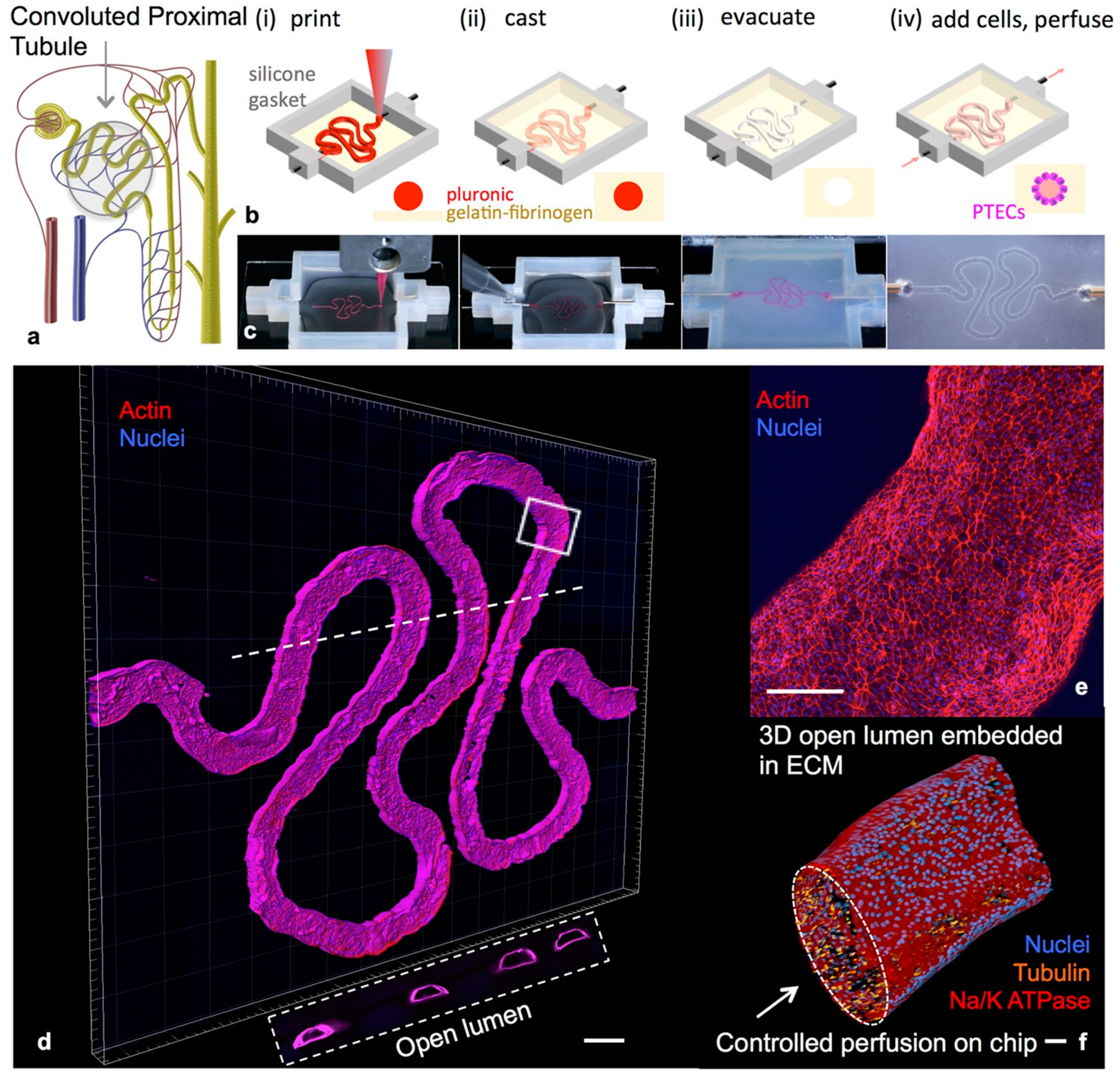
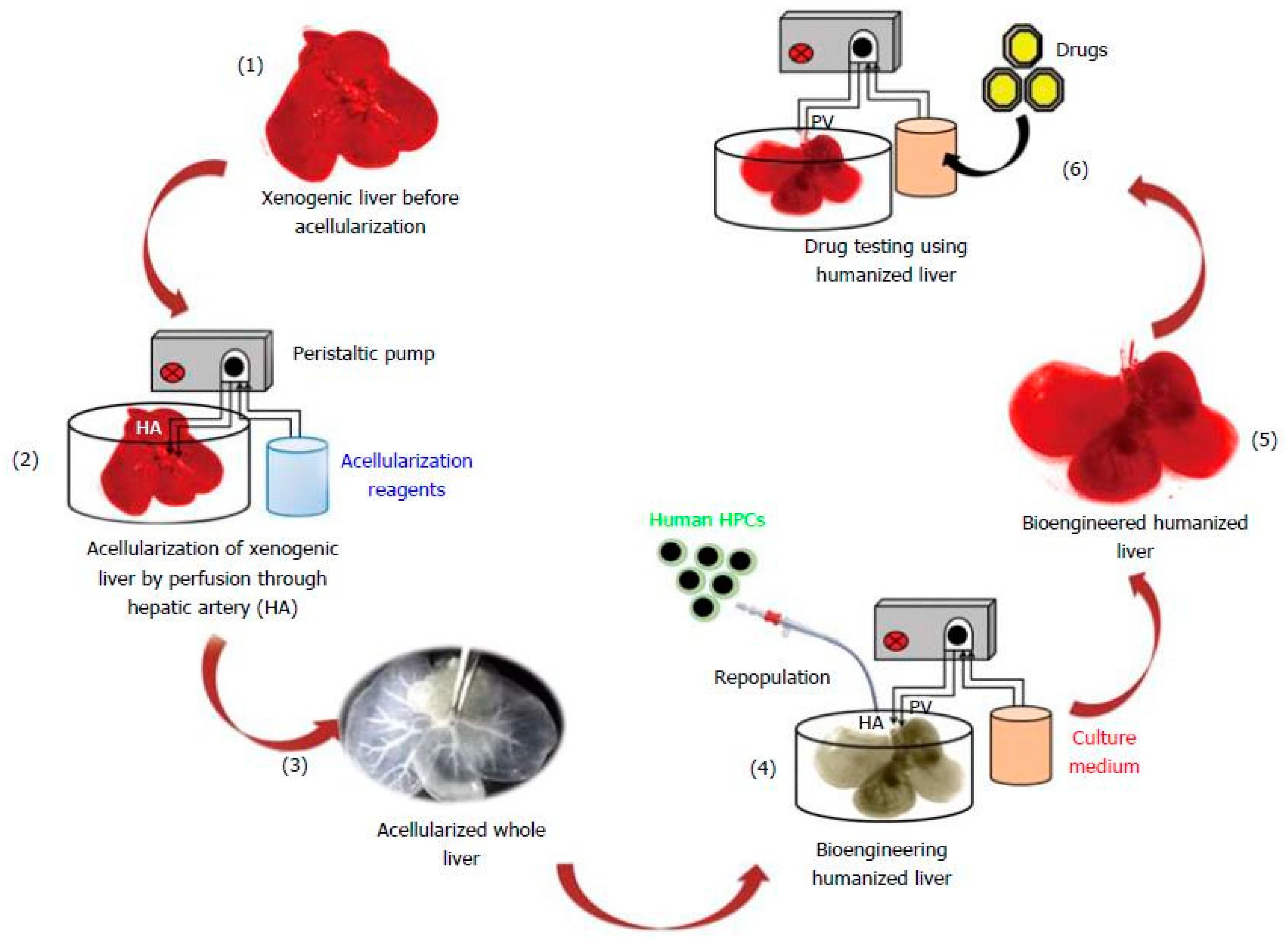

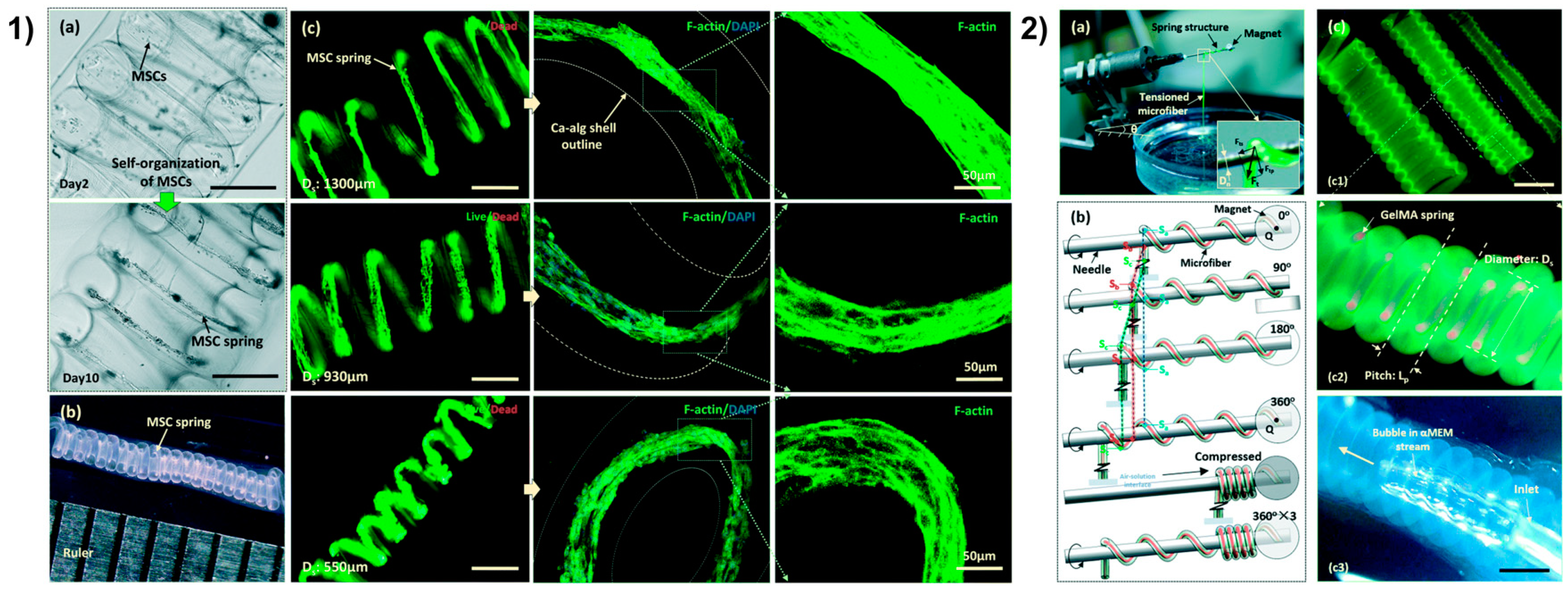
| Organ | Cells Population | Biomaterial(s) | Purpose of the Research | References |
|---|---|---|---|---|
| Heart | Primary cardiomyocytes | Polycaprolactone (PCL), fibrin | Contractive cardiac tissue model | [109] |
| Vessels | HUVECs; human dermal fibroblasts | GelMA | Thrombosis model | [108] |
| Heart | HUVECs, hiPSCs | GelMA, alginate | Drug screening | [110] |
| Lung | Human dermal microvascular endothelial cells, human lung fibroblast | Collagen type I, PCL | Airway epithelium model | [111] |
| Eye | Human corneal and conjunctival cells | GelMA | Dry-eye disease model, drug screening | [112] |
| GI tract | Human epithelial colorectal cell line Caco-2, non-cancerous colonic cell line CCD-18Co | Polycarbonate | Gastrointestinal microbiome model | [113] |
| Kidney | Proximal tubule epithelial cells, renal cancer cell line A498 | Gelatin, fibrin, Pluronic® F127 | Renal proximal tubules model | [114] |
| Bone | Human CD34+, nBM-MSCs, HUVECs | Fibrin | Investigation of myelothroid toxiciy | [115] |
| Bone | MDA-MB-231 breast cancer cells, bone marrow stem cells, endothelial cells | Decellularized bone matrix | Investigation of metastatic colonization | [117] |
| Bone | B cell acute lymphoblastic leukemia cell lines | PDMS | Leukemia model | [118] |
| Brain | Patient-derived cancer cells, HUVECs | Decellularized pig brain matrix | Glioblastoma model | [120] |
| Brain | Human brain microvascular endothelial cells, tumor associated macrophages, patient-derived cancer cells, human CD8+ T-cells | Hyaluronan | Glioblastoma model, chemotherapy testing | [122] |
| Advantages | Disadvantages |
|---|---|
| MULTICELLULAR SPHEROIDS | |
| 3D cell distribution, control on cell arrangement [22,45,46,47,48,53] High reproducibility [15] Cost-effectiveness [15] Few reagents [15] Easy high throughput production and scaling up [55,56,69] CSCs enrichment-method [72,73,74,78] | Absence of extracellular matrix [17] Variable size and shape [17,18,34,35] Poor control on cell functions within the spheroid [17] Inhomogeneous distribution of nutrients and gas [17] Compact cell arrangement [17] |
| ORGANOIDS | |
| Presence of basilar anatomic microstructure and cells functions [82,83,84,85,88,89] Possibility to combine cell layers of tissue-specific cell types [82,93,94,95] High cell density of systems culture [87] | Small size [86] Inadequate nutrients, factor gradients and gases supply to cells [86,87] Improper removal of cells waste products [86,87] Poor reproducibility [86] Lack of vascularization [96] |
| ORGAN-ON-A-CHIP | |
| Presence of cell–cell interactions [86,97] Presence of spatio-temporal gradients of chemicals [86,97,120,121] Proper mechanical strain [86,97] Presence of vasculature [107,108,109,110,112,113,114,115,117,118] High cell density of systems culture [87] | Poor reproducibility [97] High costs and time-consuming methodology [97] |
| NANOSTRUCTURED BIOMATERIALS | |
| Presence of 3D extracellular matrix [123,125,126] 3D cell distribution and arrangement [123,125,126,131,183,184,185,186,192,193,194] Tailoring of physico/chemical features (from nano to macro) [130,131,155,166,167,175] | Variable scaling up [127,152,153,164,165] Variable expensiveness in dependence on the technique [127,152,153,189] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bassi, G.; Grimaudo, M.A.; Panseri, S.; Montesi, M. Advanced Multi-Dimensional Cellular Models as Emerging Reality to Reproduce In Vitro the Human Body Complexity. Int. J. Mol. Sci. 2021, 22, 1195. https://doi.org/10.3390/ijms22031195
Bassi G, Grimaudo MA, Panseri S, Montesi M. Advanced Multi-Dimensional Cellular Models as Emerging Reality to Reproduce In Vitro the Human Body Complexity. International Journal of Molecular Sciences. 2021; 22(3):1195. https://doi.org/10.3390/ijms22031195
Chicago/Turabian StyleBassi, Giada, Maria Aurora Grimaudo, Silvia Panseri, and Monica Montesi. 2021. "Advanced Multi-Dimensional Cellular Models as Emerging Reality to Reproduce In Vitro the Human Body Complexity" International Journal of Molecular Sciences 22, no. 3: 1195. https://doi.org/10.3390/ijms22031195
APA StyleBassi, G., Grimaudo, M. A., Panseri, S., & Montesi, M. (2021). Advanced Multi-Dimensional Cellular Models as Emerging Reality to Reproduce In Vitro the Human Body Complexity. International Journal of Molecular Sciences, 22(3), 1195. https://doi.org/10.3390/ijms22031195






