Moonlighting Proteins Shine New Light on Molecular Signaling Niches
Abstract
1. Introduction
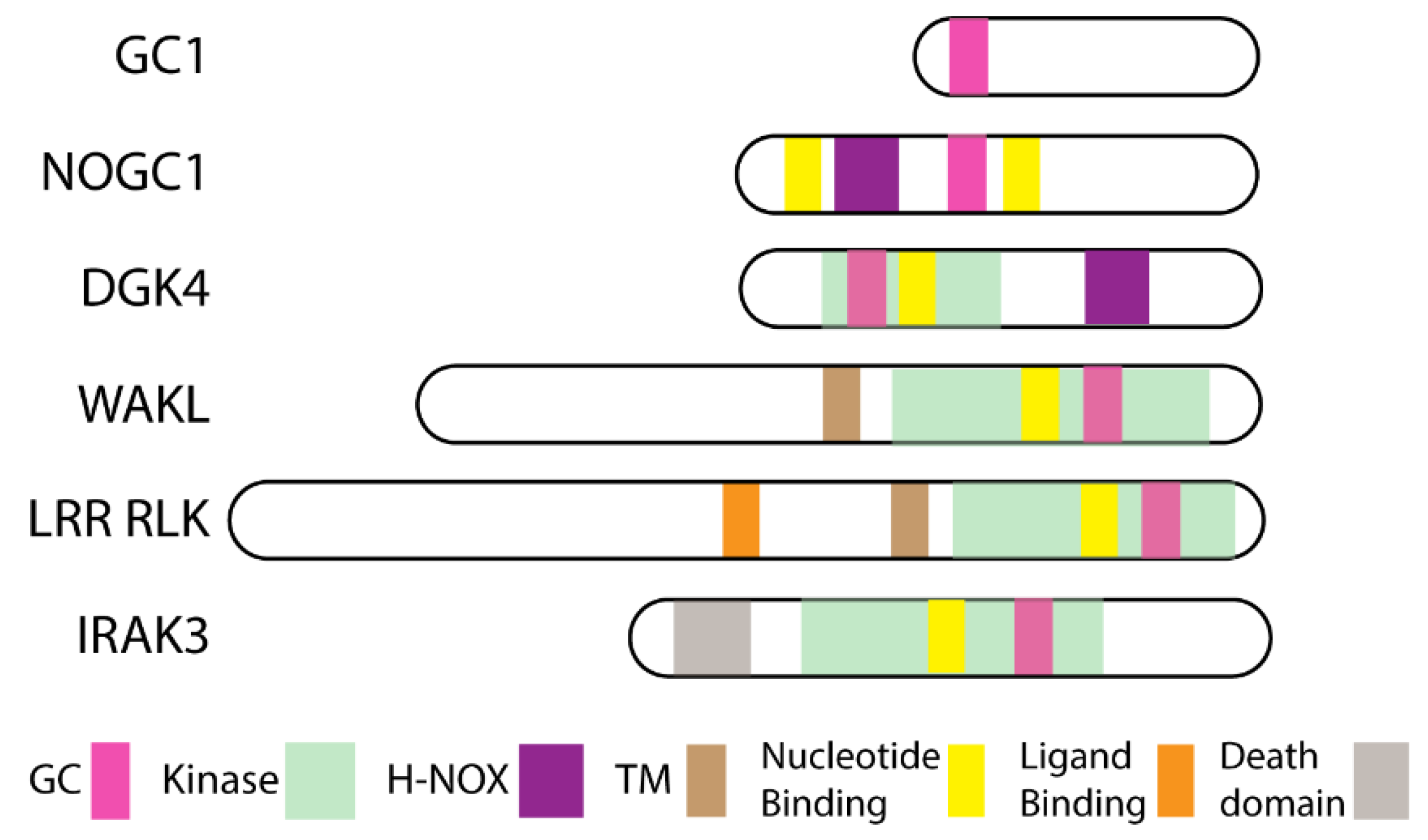
2. Moonlighting Kinases
2.1. Wall Associated Kinase Like (WAKL)
2.2. Leucine Rich Repeat Receptor Like Kinases (LRR RLK)
2.3. Nitric Oxide (NO)-Responsive Moonlighting Proteins
3. Moonlighting Kinase Guanylate Cyclase Centers
4. Nanodomains Surrounding Moonlighting Kinases
5. Degradation of Moonlighting Proteins
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gentili, P.L. The fuzziness of the molecular world and its perspectives. Molecules 2018, 23, 2074. [Google Scholar] [CrossRef] [PubMed]
- Baverstock, K. Crick’s sequence hypothesis—A review. Commun. Integr. Biol. 2019, 12, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jeffery, C.J. Moonlighting proteins in the fuzzy logic of cellular metabolism. Molecules 2020, 25, 3440. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Anand, R.; Agrawal, M.; Mattaparthi, V.S.K.; Swaminathan, R.; Santra, S.B. Consequences of heterogeneous crowding on an enzymatic reaction: A residence time Monte Carlo approach. ACS Omega 2019, 4, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Luby-Phelps, K. The physical chemistry of cytoplasm and its influence on cell function: An update. Mol. Biol. Cell 2013, 24, 2593–2596. [Google Scholar] [CrossRef]
- Goldstein, R.E.; van de Meert, J.-W. A physical perspective on cytoplasmic streaming. Interface Focus 2015, 5, 20150030. [Google Scholar] [CrossRef]
- Vestergaard, C.L.; Flyvbjerg, H.; Møller, I.M. Intracellular signaling by diffusion: Can waves of hydrogen peroxide transmit intracellular information in plant cells? Front. Plant. Sci. 2012, 3, 295. [Google Scholar] [CrossRef]
- Illukkumbura, R.; Bland, T.; Goehring, N.W. Patterning and polarization of cells by intracellular flows. Curr. Opin. Cell Biol. 2020, 62, 123–134. [Google Scholar] [CrossRef]
- Needleman, D.; Shelley, M. The stormy fluid dynamics of the living cell. Physics Today 2019, 72, 32–38. [Google Scholar] [CrossRef]
- Geremia, S.; Campagnolo, M.; Demitri, N.; Johnson, L.N. Simulation of diffusion time of small molecules in protein crystals. Structure 2006, 14, 393–400. [Google Scholar] [CrossRef]
- Mori, K.; Kuhn, B. Imaging Ca2+ concentration and pH in nanopores/channels of protein crystals. J. Phys. Chem. B 2018, 122, 9646–9653. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Moonlighting proteins. Trends Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef]
- Jeffery, C.J. Moonlighting proteins—An update. Mol. Biosyst. 2009, 5, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. An enzyme in the test tube, and a transcription factor in the cell: Moonlighting proteins and cellular factors that affect their behavior. Protein Sci. 2019, 28, 1233–1238. [Google Scholar] [CrossRef]
- Monaghan, R.M.; Whitmarsh, A.J. Mitochondrial proteins moonlighting in the nucleus. Trends Biochem. Sci. 2015, 40, 728–735. [Google Scholar] [CrossRef]
- Piatigorsky, J.; Brien, W.E.; Norman, B.L.; Kalumuck, K.; Wistow, G.J.; Borras, T.; Nickerson, J.M.; Wawrousek, E.F. Gene sharing by delta-crystallin and argininosuccinate lyase. Proc. Natl. Acad. Sci. USA 1988, 85, 3479. [Google Scholar] [CrossRef]
- Chen, C.; Zabad, S.; Liu, H.; Wang, W.; Jeffery, C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Res. 2018, 46, D640–D644. [Google Scholar] [CrossRef]
- Khan, I.K.; Bhuiyan, M.; Kihara, D. DextMP: Deep dive into text for predicting moonlighting proteins. Bioinformatics 2017, 33, i83–i91. [Google Scholar] [CrossRef]
- Khan, I.K.; Kihara, D. Computational characterization of moonlighting proteins. Biochem. Soc. Trans. 2014, 42, 1780–1785. [Google Scholar] [CrossRef]
- Mani, M.; Chen, C.; Amblee, V.; Liu, H.; Mathur, T.; Zwicke, G.; Zabad, S.; Patel, B.; Thakkar, J.; Jeffery, C.J. MoonProt: A database for proteins that are known to moonlight. Nucleic Acids Res. 2015, 43, D277–D282. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Qian, Z.; Li, T.; Zhou, Y.; Wong, A. PlantMP: A database for moonlighting plant proteins. Database 2019, 2019, baz050. [Google Scholar] [CrossRef] [PubMed]
- Irving, H.R.; Cahill, D.M.; Gehring, C. Moonlighting proteins and their role in the control of signalling microenvironments, as exemplified by cGMP and phytosulfokine receptor 1 (PSKR1). Front. Plant. Sci. 2018, 9, 415. [Google Scholar] [CrossRef] [PubMed]
- Irving, H.R.; Kwezi, L.; Wheeler, J.I.; Gehring, C. Moonlighting kinases with guanylate cyclase activity can tune regulatory signal networks. Plant. Signal. Behav. 2012, 7, 201–204. [Google Scholar] [CrossRef]
- Gehring, C.; Turek, I.S. Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front. Plant. Sci. 2017, 8, 1704. [Google Scholar] [CrossRef]
- Marondedze, C.; Wong, A.; Thomas, L.; Irving, H.; Gehring, C. Cyclic Nucleotide Monophosphates in Plants and Plant Signaling. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Newton, R.P.; Kingston, E.E.; Evans, D.E.; Younis, L.M.; Brown, E.G. Occurrence of guanosine 3′,5′-cyclic monophosphate (Cyclic GMP) and associated enzyme systems in Phaseolus vulgaris. Phytochemistry 1984, 23, 1367–1372. [Google Scholar]
- Newton, R.P.; Smith, C.J. Cyclic nucleotides. Phytochemistry 2004, 65, 2423–2437. [Google Scholar] [CrossRef]
- Cousson, A. Pharmacological evidence for the implication of both cyclic GMP-dependent and -independent transduction pathways within auxin-induced stomatal opening in Commelina communis (L.). Plant Sci. 2001, 161, 249–258. [Google Scholar] [CrossRef]
- Donaldson, L.; Ludidi, N.; Knight, M.R.; Gehring, C.; Denby, K. Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS Lett. 2004, 569, 317–320. [Google Scholar] [CrossRef]
- Neuhaus, G.; Bowler, C.; Hiratsuka, K.; Yamagata, H.; Chua, N.H. Phytochrome-regulated repression of gene expression requires calcium and cGMP. EMBO J. 1997, 16, 2554–2564. [Google Scholar] [CrossRef]
- Penson, S.P.; Schuurink, R.C.; Fath, A.; Gubler, F.; Jacobsen, J.V.; Jones, R.L. cGMP is required for giberellic acid-induced gene expression in barley aleurone. Plant Cell 1996, 8, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Pharmawati, M.; Gehring, C.A.; Irving, H.R. An immunoaffinity purified plant natriuretic peptide analogue modulates cGMP level in the Zea mays root stele. Plant Sci. 1998, 137, 107–115. [Google Scholar] [CrossRef]
- Pharmawati, M.; Maryani, M.M.; Nikolakopoulos, T.; Gehring, C.A.; Irving, H.R. Cyclic GMP modulates stomatal opening induced by natriuretic peptides and immunoreactive analogues. Plant Physiol. Biochem. 2001, 39, 385–394. [Google Scholar] [CrossRef]
- Ludidi, N.N.; Gehring, C. Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 6490–6494. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, J.; Meima, M.; Schaap, P.; Van Haastert, P.J.M. The Dictyostelium homologue of mammalian soluble adenylyl cyclase encodes a guanylyl cyclase. EMBO J. 2001, 20, 4341–4348. [Google Scholar] [CrossRef]
- Kwezi, L.; Meier, S.; Mungur, L.; Ruzvidzo, O.; Irving, H.; Gehring, C. The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS ONE 2007, 2, e449. [Google Scholar] [CrossRef]
- Wong, A.; Gehring, C.; Irving, H.R. Conserved functional motifs and homology modeling to predict hidden moonlighting functional sites. Front. Bioengin. Biotech. 2015, 3, 82. [Google Scholar] [CrossRef]
- Schaap, P. Guanylyl cyclases across the tree of life. Front. Biosci. 2005, 10, 1485–1498. [Google Scholar] [CrossRef]
- Liu, Y.; Ruoho, A.E.; Rao, V.D.; Hurley, J.H. Catalytic mechanism of the adenylyl and guanylyl cyclases: Modeling and mutational analysis. Proc. Natl. Acad. Sci. USA 1997, 94, 13414–13419. [Google Scholar] [CrossRef]
- McCue, L.A.; McDonough, K.A.; Lawrence, C.E. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 2000, 10, 204–219. [Google Scholar] [CrossRef]
- Tucker, C.L.; Hurley, J.H.; Miller, T.R.; Hurley, J.B. Two amino acid substitutions convert a guanylyl cyclase, RetGC-1, into an adenylyl cyclase. Proc. Natl. Acad. Sci. USA 1998, 95, 5993–5997. [Google Scholar] [CrossRef] [PubMed]
- Świeżawska, B.; Jaworski, K.; Szewczuk, P.; Pawełek, A.; Szmidt-Jaworska, A. Identification of a Hippeastrum hybridum guanylyl cyclase responsive to wounding and pathogen infection. J. Plant Physiol. 2015, 189, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Szmidt-Jaworska, A.; Jaworski, K.; Pawelek, A.; Kocewicz, J. Molecular cloning and characterization of a guanylyl cyclase, PNGC-1, involved in light signaling in Pharbitis nil. J. Plant Growth Reg. 2009, 28, 367–380. [Google Scholar] [CrossRef]
- Yuan, J.; Liakat Ali, M.; Taylor, J.; Liu, J.; Sun, G.; Liu, W.; Masilimany, P.; Gulati-Sakhuja, A.; Pauls, K.P. A guanylyl cyclase-like gene is associated with Gibberella ear rot resistance in maize (Zea mays L.). Theoret. Appl. Genet. 2008, 116, 465–479. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.X.; Li, W.-H. Comparative Analysis of the Receptor-Like Kinase Family in Arabidopsis and Rice. Plant Cell 2004, 16, 1220. [Google Scholar] [CrossRef]
- He, Z.-H.; Fujiki, M.; Kohorn, B.D. A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 1996, 271, 19789–19793. [Google Scholar] [CrossRef]
- Kohorn, B.D.; Kohorn, S.L. The cell wall-associated kinases, WAK, as pectin receptors. Front. Plant Sci. 2012, 3, 88. [Google Scholar] [CrossRef]
- Verica, J.A.; Chae, L.; Tong, H.; Ingmire, P.; He, Z.-H. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003, 133, 1732–1746. [Google Scholar] [CrossRef]
- Verica, J.A.; He, Z.H. The cell wall-associated kinase (WAK) and WAK-Like kinase gene family. Plant Physiol. 2002, 129, 455–459. [Google Scholar] [CrossRef]
- Kohorn, B.D. Cell wall-associated kinases and pectin perception. J. Exp. Bot. 2016, 67, 489–494. [Google Scholar] [CrossRef]
- Meier, S.; Ruzvidzo, O.; Morse, M.; Donaldson, L.; Kwezi, L.; Gehring, C. The Arabidopsis Wall Associated Kinase-Like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS ONE 2010, 5, e8904. [Google Scholar] [CrossRef]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef] [PubMed]
- Klessig, D.F.; Durner, J.; Noad, R.; Navarre, D.A.; Wendehenne, D.; Kumar, D.; Zhou, J.M.; Shah, J.; Zhang, S.; Kachroo, P.; et al. Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 2000, 97, 8849. [Google Scholar] [CrossRef]
- Pasqualini, S.; Meier, S.; Gehring, C.; Madeo, L.; Fornaciari, M.; Romano, B.; Ederli, L. Ozone and NO induce cGMP-dependent and -independent transcription of defence genes in tobacco. New Phyt. 2009, 181, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Bot, P.; Mun, B.-G.; Imran, Q.M.; Hussain, A.; Lee, S.-U.; Loake, G.; Yun, B.-W. Differential expression of AtWAKL10 response to nitric oxide suggests a putative role in biotic and abiotic stress responses. Peer J. 2019, 7, e7383. [Google Scholar] [CrossRef] [PubMed]
- Larkan, N.J.; Ma, L.; Haddadi, P.; Buchwaldt, M.; Parkin, I.A.P.; Djavaheri, M.; Borhan, M.H. The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 2020, 104, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Saintenac, C.; Lee, W.S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C.; et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Malukani, K.K.; Ranjan, A.; Hota, S.J.; Patel, H.K.; Sonti, R.V. Dual activities of receptor-like kinase OsWAKL21.2 induce immune responses. Plant Physiol. 2020, 183, 1345. [Google Scholar] [CrossRef]
- Wong, A.; Gehring, C. The Arabidopsis thaliana proteome harbors undiscovered multi-domain molecules with functional guanylyl cyclase catalytic centers. Cell Commun. Signal. 2013, 11, 48. [Google Scholar] [CrossRef]
- Wong, A.; Tian, X.; Gehring, C.; Marondedze, C. Discovery of novel functional centers with rationally designed amino acid motifs. Comput. Struct. Biotech. J. 2018, 16, 70–76. [Google Scholar] [CrossRef]
- Hosseini, S.; Schmidt, E.D.L.; Bakker, F.T. Leucine-rich repeat receptor-like kinase II phylogenetics reveals five main clades throughout plant kingdom. Plant J. 2020, 103, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-L.; Du, L.; Huang, Y.; Gao, S.-M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, e47. [Google Scholar] [CrossRef] [PubMed]
- Man, J.; Gallagher, J.P.; Bartlett, M. Structural evolution drives diversification of the large LRR-RLK gene family. New Phyt. 2020, 226, 1492–1505. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, 2001, re22. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C. Receptor-like protein kinase genes in Arabidopsis thaliana. Plant J. 1993, 3, 451–458. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Zou, C.; Hanada, K.; Shiu, S.-H. Evolutionary history and stress regulation of plant receptor-like kinase/Pelle genes. Plant Physiol. 2009, 150, 12–26. [Google Scholar] [CrossRef]
- Gosu, V.; Basith, S.; Durai, P.; Choi, S. Molecular evolution and structural features of IRAK family members. PLoS ONE 2012, 7, e49771. [Google Scholar] [CrossRef]
- aan den Toorn, M.; Albrecht, C.; de Vries, S. On the origin of SERKs: Bioinformatics analysis of the somatic embryogenesis receptor kinases. Mol. Plant 2015, 8, 762–782. [Google Scholar] [CrossRef]
- He, Y.; Zhou, J.; Shan, L.; Meng, X. Plant cell surface receptor-mediated signaling—A common theme amid diversity. J. Cell Sci. 2018, 131, jcs209353. [Google Scholar] [CrossRef]
- Jose, J.; Ghantasala, S.; Choudhury, S.R. Arabidopsis transmembrane receptor-like kinases (RLKs): A Bridge between extracellular signal and intracellular regulatory machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Lou, Y. PRRs and NB-LRRs: From Signal Perception to Activation of Plant Innate Immunity. Int. J. Mol. Sci. 2019, 20, 1882. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Morita, A.; Sakagami, Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 2002, 296, 1470–1472. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y.; Sakagami, Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinales L. Proc. Natl. Acad. Sci. USA 1996, 93, 7623–7627. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.I.; Irving, H.R. Evolutionary advantages of secreted peptide signalling molecules. Funct. Plant Biol. 2010, 37, 382–394. [Google Scholar] [CrossRef]
- Chakraborty, S.; Nguyen, B.; Wasti, S.D.; Guozhou, X. Plant Leucine-Rich Repeat Receptor Kinase (LRR-RK): Structure, ligand perception, and activation mechanism. Molecules 2019, 24, 3081. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, U.; Lau, K.; Hothorn, M. The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 2017, 68, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.I.; Wong, A.; Marondedze, C.; Groen, A.J.; Kwezi, L.; Freihat, L.; Vyas, J.; Raji, M.R.; Irving, H.R.; Gehring, C. The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling. Plant J. 2017, 91, 590–600. [Google Scholar] [CrossRef]
- Kwezi, L.; Ruzvidzo, O.; Wheeler, J.I.; Govender, K.; Iacuone, S.; Thompson, P.E.; Gehring, C.; Irving, H.R. The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependant signaling in plants. J. Biol. Chem. 2011, 286, 22580–22588. [Google Scholar] [CrossRef]
- Muleya, V.; Marondedze, C.; Wheeler, J.I.; Thomas, L.; Mok, Y.F.; Griffin, M.W.D.; Manallack, D.T.; Kwezi, L.; Lilley, K.S.; Gehring, C.; et al. Phosphorylation of the dimeric cytoplasmic domain of the phytosulfokine receptor, PSKR1. Biochem. J. 2016, 473, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Muleya, V.; Wheeler, J.I.; Ruzvidzo, O.; Freihat, L.; Manallack, D.T.; Gehring, C.; Irving, H.R. Calcium is the switch in the moonlighting dual function of the ligand-activated receptor kinase phytosulfokine receptor 1. Cell Commun. Signal. 2014, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Verma, R.; Gehring, C.; Yamaguchi, Y.; Zhao, Y.; Ryan, C.A.; Berkowitz, G.A. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA 2010, 107, 21193–21198. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Wang, X.-Y.; Xu, Y.-P.; He, Y.-H.; Cai, X.-Z. Characterization of tomato protein kinases embedding guanylate cyclase catalytic center motif. Sci. Rep. 2020, 10, 4078. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Dufayard, J.-F.; Bettembourg, M.; Fischer, I.; Droc, G.; Guiderdoni, E.; Périn, C.; Chantret, N.; Diévart, A. New Insights on Leucine-Rich Repeats Receptor-Like Kinase Orthologous Relationships in Angiosperms. Front. Plant Sci. 2017, 8, 381. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Clouse, S.D. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 2011, 23, 1219–1230. [Google Scholar] [CrossRef]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–471. [Google Scholar] [CrossRef]
- Sun, Y.; Han, Z.; Tang, J.; Hu, Z.; Chai, C.; Zhou, B.; Chai, J. Structure reveals that BAK1 as a co-receptor recognises the BRI1-bound brassinolide. Cell Res. 2013, 23, 1326–1329. [Google Scholar] [CrossRef]
- Bücherl, C.A.; van Esse, G.W.; Kruis, A.; Luchtenberg, J.; Westphal, A.H.; Aker, J.; van Hoek, A.; Albrecht, C.; Borst, J.W.; de Vries, S.C. Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant. Physiol. 2013, 162, 1911–1925. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.-H.; Ray, W.K.; Huber, S.C.; Asara, J.M.; Gage, D.A.; Clouse, S.D. Recombinant Brassinosteroid Insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000, 124, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Sreeramulu, S.; Mostizky, Y.; Sunitha, S.; Shani, E.; Nahum, H.; Salomon, D.; Hayun, L.B.; Gruetter, C.; Rauh, D.; Ori, N.; et al. BSKs are partially redundant positive regulators of brassinosteriod signaling in Arabidopsis. Plant J. 2013, 74, 905–919. [Google Scholar] [CrossRef]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef] [PubMed]
- van Esse, G.W.; van Mourik, S.; Stigter, H.; ten Hove, C.A.; Molenaar, J.; de Vries, S.C. A mathematical model for BRASSINOSTEROID INSENSITIVE1-mediated signaling in root growth and hypocotyl elongation. Plant Physiol. 2012, 160, 523–532. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, Y.; Qi, Z.; Berkowitz, G.A. Teaching an old hormone new tricks: Cytosolic Ca2+ elevation involvement in plant brassinosteroid signal transduction cascades. Plant Physiol. 2013, 163, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Isner, J.C.; Nuhse, T.; Maathuis, F.J. The cyclic nucleotide cGMP is involved in plant hormone signalling and alters phosphorylation of Arabidopsis thaliana root proteins. J. Exp. Bot. 2012, 63, 3199–3205. [Google Scholar] [CrossRef]
- Nausch, L.W.; Ledoux, J.; Bonev, A.D.; Nelson, M.T.; Dostmann, W.R. Differential patterning of cGMP in vascular smooth muscle cells revealed by single GFP-linked biosensors. Proc. Natl. Acad. Sci. USA 2009, 105, 365–370. [Google Scholar] [CrossRef]
- Isner, J.-C.; Maathuis, F.J.M. Measurement of cellular cGMP in plant cells and tissues using the endogenous fluorescent reporter FlincG. Plant J. 2011, 65, 329–334. [Google Scholar] [CrossRef]
- Marondedze, C.; Groen, A.; Thomas, L.; Lilley, K.S.; Gehring, C. A quantitative phosphoproteome analysis of cGMP-dependent cellular responses in Arabidopsis thaliana. Mol. Plant 2016, 9. [Google Scholar] [CrossRef]
- Bojar, D.; Martinez, J.; Santiago, J.; Rybin, V.; Bayliss, R.; Hothorn, M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014, 78, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; van der Does, D.; Ladwig, F.; Sticht, C.; Kolbeck, A.; Schürholz, A.-K.; Augustin, S.; Keinath, N.; Rausch, T.; Greiner, S.; et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 15261–15266. [Google Scholar] [CrossRef]
- Holzwart, E.; Huerta, A.I.; Glöckner, N.; Garnelo Gómez, B.; Wanke, F.; Augustin, S.; Askani, J.C.; Schürholz, A.-K.; Harter, K.; Wolf, S. BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc. Natl. Acad. Sci. USA 2018, 115, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Holzwart, E.; Wanke, F.; Glöckner, N.; Höfte, H.; Harter, K.; Wolf, S. A mutant allele uncouples the brassinosteroid-dependent and independent functions of BRASSINOSTEROID INSENSITIVE 1. Plant Physiol. 2020, 182, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Amano, Y.; Tsubouchi, H.; Shinohara, H.; Ogawa, M.; Matsubayashi, Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18333–18338. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Kihara, H.; Niwa, M.; Sakagami, Y. Disruption and overexpression of Arabidopsis phytosulokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006, 142, 45–53. [Google Scholar] [CrossRef]
- Sauter, M. Phytosulfokine peptide signaling. J. Exp. Bot. 2015, 66, 5161–5169. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Han, Z.; Zhang, H.; Wang, T.; Lin, G.; Chang, J.; Yang, W.; Chai, J. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 2015, 525, 265–268. [Google Scholar] [CrossRef]
- Ladwig, F.; Dahlke, R.I.; Stuhrwohdldt, N.; Hartmann, J.; Harter, K.; Sauter, M. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H-ATPase, and BAK1. Plant Cell 2015, 27, 1718–1729. [Google Scholar] [CrossRef]
- Hartmann, J.; Linke, D.; Bönniger, C.; Tholey, A.; Sauter, M. Conserved phosphorylation sites in the activation loop of Arabidopsis phytosulokine receptor PSKR1 differentially affect kinase and receptor activity. Biochem. J. 2015, 472, 379–391. [Google Scholar] [CrossRef]
- Kaufmann, C.; Motzkus, M.; Sauter, M. Phosphorylation of the phytosulokine peptide receptor PSKR1 controls receptor activity. J. Exp. Bot. 2017, 68, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, Z.; Lei, C.; Zheng, C.; Wang, J.; Shao, S.; Li, X.; Xia, X.; Cai, X.; Zhou, J.; et al. A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. Plant Cell 2018, 30, 652–667. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Liu, J.-X.; Howell, S.H. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilsin serine protease in Arabidopsis. Plant J. 2008, 56, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, D.; Tsuda, K.; Katagiri, F. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant. J. 2012, 71, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Loivamaki, M.; Stührwohldt, N.; Deeken, R.; Steffens, B.; Roitsch, T.; Hedrich, R.; Sauter, M. A role for PSK signaling in wounding and microbial interactions in Arabidopsis. Physiol. Plant. 2010, 139, 348–537. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Pearce, G.; Ryan, C.A. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 2006, 103, 10104–10109. [Google Scholar] [CrossRef]
- Bartels, S.; Lori, M.; Mbengue, M.; van Verk, M.; Klauser, D.; Hander, T.; Böni, R.; Robatzek, S.; Boller, T. The family of Peps and their precursors in Arabidopsis: Differential expression and localization but similar induction of pattern-triggered immune responses. J. Exp. Bot. 2013, 64, 5309–5321. [Google Scholar] [CrossRef]
- Huffaker, A.; Ryan, C.A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 10732. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Huffaker, A.; Bryan, A.C.; Tax, F.E.; Ryan, C.A. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 2010, 22, 508–522. [Google Scholar] [CrossRef]
- Hander, T.; Fernandez-Fernandez, A.D.; Kumpf, R.P.; Willems, P.; Schatowitz, H.; Rombaut, D.; Staes, A.; Nolf, J.; Pottie, R.; Yao, P.; et al. Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 2019, 363, eaar7486. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [PubMed]
- Klauser, D.; Desurmont, G.A.; Glauser, G.; Vallat, A.; Flury, P.; Boller, T.; Turlings, T.C.J.; Bartels, S. The Arabidopsis Pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 2015, 66, 5327–5336. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, Y.; Yang, F.; Zhang, Y.; Chen, S.; Xie, Q.; Tian, X.; Zhou, J.M. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 6205–6210. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Walker, R.K.; Zhao, Y.; Berkowitz, G.A. Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. USA 2012, 109, 19852–19857. [Google Scholar] [CrossRef]
- Tintor, N.; Ross, A.; Kanehara, K.; Yamada, K.; Fan, L.; Kemmerling, B.; Nürnberger, T.; Tsuda, K.; Saijo, Y. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 6211. [Google Scholar] [CrossRef]
- Yamada, K.; Yamashita-Yamada, M.; Hirase, T.; Fujiwara, T.; Tsuda, K.; Hiruma, K.; Saijo, Y. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 2016, 35, 46–61. [Google Scholar] [CrossRef]
- Krol, E.; Mentzel, T.; Chinchilla, D.; Boller, T.; Felix, G.; Kemmerling, B.; Postel, S.; Arents, M.; Jeworutzki, E.; Al-Rasheid, K.A.S.; et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 2010, 285, 13471–13479. [Google Scholar] [CrossRef]
- Postel, S.; Küfner, I.; Beuter, C.; Mazzotta, S.; Schwedt, A.; Borlotti, A.; Halter, T.; Kemmerling, B.; Nürnberger, T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 2010, 89, 169–174. [Google Scholar] [CrossRef]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 2010, 285, 9444–9451. [Google Scholar] [CrossRef]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tör, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef]
- Ma, X.; Claus, L.A.N.; Leslie, M.E.; Tao, K.; Wu, Z.; Liu, J.; Yu, X.; Li, B.; Zhou, J.; Savatin, D.V.; et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 2020, 581, 199–203. [Google Scholar] [CrossRef]
- Jing, Y.; Shen, N.; Zheng, X.; Fu, A.; Zhao, F.; Lan, W.; Luan, S. Danger-Associated Peptide regulates root immune responses and root growth by affecting ROS formation in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4590. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Zheng, X.; Zhang, D.; Shen, N.; Wang, Y.; Yang, L.; Fu, A.; Shi, J.; Zhao, F.; Lan, W.; et al. Danger-Associated Peptides interact with PIN-dependent local auxin distribution to inhibit root growth in Arabidopsis. Plant Cell 2019, 31, 1767. [Google Scholar] [CrossRef] [PubMed]
- Nakaminami, K.; Okamoto, M.; Higuchi-Takeuchi, M.; Yoshizumi, T.; Yamaguchi, Y.; Fukao, Y.; Shimizu, M.; Ohashi, C.; Tanaka, M.; Matsui, M.; et al. AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl. Acad. Sci. USA 2018, 115, 5810. [Google Scholar] [CrossRef] [PubMed]
- Świeżawska, B.; Jaworski, K.; Duszyn, M.; Pawelek, A.; Szmidt-Jaworska, A. The Hippeastrum hybridum PepR1 gene (HpPepR1) encodes a functional guanylyl cyclase and is involved in early response to fungal infection. J. Plant Physiol. 2017, 216, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.K.; Prajapati, R.; Krishna, D.; Divakaran, K.; Pandey, Y.; Reichelt, M.; Mathew, M.K.; Boland, W.; Mithöfer, A.; Vadassery, J. The Ca2+ channel CNGC19 regulates Arabidopsis defense against Spodoptera herbivory. Plant Cell 2019, 31, 1539–1562. [Google Scholar] [CrossRef] [PubMed]
- Jogawat, A.; Meena, M.K.; Kundu, A.; Varma, M.; Vadassery, J. Calcium channel CNGC19 mediates basal defense signaling to regulate colonization by Piriformospora indica in Arabidopsis roots. J. Exp. Bot. 2020, 71, 2752–2768. [Google Scholar] [CrossRef]
- Chou, H.; Zhu, Y.; Ma, Y.; Berkowitz, G.A. The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca2+ as a secondary cytosolic messenger. Plant J. 2016, 85, 494–506. [Google Scholar] [CrossRef]
- Domingos, P.; Prado, A.M.; Wong, A.; Gehring, C.; Feijó, J.A. Nitric oxide: A multitasked signalling gas in plants. Mol. Plant 2015, 8, 506–520. [Google Scholar] [CrossRef]
- Mulaudzi, T.; Ludidi, N.; Ruzvidzo, O.; Morse, M.; Hendricks, N.; Iwuoha, E.; Gehring, C. Identification of a novel Arabidopsis thaliana nitric oxide-binding molecule with guanylate cyclase activity in vitro. FEBS Lett. 2011, 585, 2693–2697. [Google Scholar] [CrossRef]
- Vaz Dias, F.; Serrazina, S.; Vitorino, M.; Marchese, D.; Heilmann, I.; Godinho, M.; Rodrigues, M.; Malhó, R. A role for diacylglycerol kinase 4 in signalling crosstalk during Arabidopsis pollen tube growth. New Phyt. 2019, 222, 1434–1446. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Donaldson, L.; Portes, M.T.; Eppinger, J.; Feijó, J.A.; Gehring, C. Arabidopsis DIACYLGLYCEROL KINASE4 is involved in nitric oxide-dependent pollen tube guidance and fertilization. Development 2020, 147, dev183715. [Google Scholar] [CrossRef]
- Hanks, S.K.; Hunter, T. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain tructure and classification. FASEB J. 1995, 9, 576–596. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef]
- Ahuja, L.G.; Aoto, P.C.; Kornev, A.P.; Veglia, G.; Taylor, S.S. Dynamic allostery-based molecular workings of kinase:peptide complexes. Proc. Natl. Acad. Sci. USA 2019, 116, 15052. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, L.G.; Kornev, A.P.; McClendon, C.L.; Veglia, G.; Taylor, S.S. Mutation of a kinase allosteric node uncouples dynamics linked to phosphotransfer. Proc. Natl. Acad. Sci. USA 2017, 114, E931. [Google Scholar] [CrossRef]
- Ahuja, L.G.; Taylor, S.S.; Kornev, A.P. Tuning the “violin” of protein kinases: The role of dynamics-based allostery. IUBMB Life 2019, 71, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Endicott, J.A.; Noble, M.E.M.; Johnson, L.N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012, 81. [Google Scholar] [CrossRef]
- McClendon, C.L.; Kornev, A.P.; Gilson, M.K.; Taylor, S.S. Dynamic architecture of a protein kinase. Proc. Natl. Acad. Sci. USA 2014, 111, E4623. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821. [Google Scholar] [CrossRef]
- Kanev, G.K.; de Graaf, C.; de Esch, I.J.P.; Leurs, R.; Würdinger, T.; Westerman, B.A.; Kooistra, A.J. The landscape of atypical and eukaryotic protein kinases. Trends Pharmacol. Sci. 2019, 40, 818–832. [Google Scholar] [CrossRef] [PubMed]
- Freihat, L.; Muleya, V.; Manallack, D.T.; Wheeler, J.I.; Irving, H.R. Comparison of moonlighting guanylate cyclases: Roles in signal direction? Biochem. Soc. Trans. 2014, 42, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Kwezi, L.; Wheeler, J.I.; Marondedze, C.; Gehring, C.; Irving, H.R. Intramolecular crosstalk between catalytic activities of receptor kinases. Plant Signal. Behav. 2018, 13, e1430544. [Google Scholar] [CrossRef] [PubMed]
- Boudeau, J.; Miranda-Saavedra, D.; Barton, G.J.; Alessi, D.R. Emerging roles of pseudokinases. Trends Cell Biol. 2006, 16, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Raju, S.; Shaw, A.S. What is the point of pseudokinases? eLife 2015, 4, e07771. [Google Scholar] [CrossRef] [PubMed]
- Rauch, J.; Volinsky, N.; Romano, D.; Kolch, W. The secret life of kinases: Function beyond catalysis. Cell Commun. Signal. 2011, 9, 23. [Google Scholar] [CrossRef]
- Tomoni, A.; Lees, J.; Santana, A.G.; Bolanos-Garcia, V.M.; Bastida, A. Pseudokinases: From allosteric regulation of catalytic domains and the formation of macromolecular assemblies to emerging drug targets. Catalysts 2019, 9, 778. [Google Scholar] [CrossRef]
- Freihat, L.A.; Wheeler, J.I.; Wong, A.; Turek, I.; Manallack, D.T.; Irving, H.R. IRAK3 modulates downstream innate immune signalling through its guanylate cyclase activity. Sci. Rep. 2019, 9, 15468. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hernandez, L.D.; Galan, J.E.; Janeway, C.A.; Medzhitov, R.; Flavell, R.A. IRAK-M is a negative regulator of toll-like receptor signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef]
- Rothschild, D.E.; McDaniel, D.K.; Ringel-Scaia, V.M.; Allen, I.C. Modulating inflammation through the negative regulation of NF-κB signaling. J. Leuk. Biol. 2018, 103, 1131–1150. [Google Scholar] [CrossRef] [PubMed]
- Bücherl, C.A.; Jarsch, I.K.; Schudoma, C.; Segonzac, C.; Mbengue, M.; Robatzek, S.; MacLean, D.; Ott, T.; Zipfel, C. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 2017, 6, e25114. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, H.; Lv, X.; Chen, T.; Li, R.; Xue, Y.; Jiang, J.; Jin, B.; Baluška, F.; Šamaj, J.; et al. Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol. Plant 2015, 8, 1334–1349. [Google Scholar] [CrossRef] [PubMed]
- Jallais, Y.; Ott, T. The nanoscale organization of the plasma membrane and its importance in signaling: A proteolipid perspective. Plant Physiol. 2020, 182, 1682–1696. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, J.; Saunders, R.E.; Warwicker, J. Charge environments around phosphorylation sites in proteins. BMC Struct. Biol. 2008, 8, 19. [Google Scholar] [CrossRef]
- Cohen, P. The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 2000, 25, 596–601. [Google Scholar] [CrossRef]
- Ashton, A.R. Guanylyl cyclase activity in plants? Proc. Natl. Acad. Sci. USA 2011, 108. E96 [(author reply), E97-E98]. [Google Scholar] [CrossRef]
- Berkowitz, G.A.; Gehring, C.; Irving, H.R.; Kwezi, L. Reply to Ashton: The putative guanylyl cyclase domain of AtPepR1 and similar plant receptors. Proc. Natl. Acad. Sci. USA 2011, 108, E97–E98. [Google Scholar] [CrossRef]
- Jackson, P.K. cAMP signaling in nanodomains. Cell 2020, 182, 1379–1381. [Google Scholar] [CrossRef]
- Yuan, P.; Jauregui, E.; Du, L.; Tanaka, K.; Poovaiah, B.W. Calcium signatures and signaling events orchestrate plant-microbe interactions. Curr. Opin. Plant Biol. 2017, 38, 173–183. [Google Scholar] [CrossRef]
- Hussain, J.; Chen, J.; Locato, V.; Sabetta, W.; Behera, S.; Cimini, S.; Griggio, F.; Martinez-Jaime, S.; Graf, A.; Bouneb, M.; et al. Constitutive cyclic GMP accumulation in Arabidopsis thaliana compromises systemic acquired resistance induced by an avirulent pathogen by modulating local signals. Sci. Rep. 2016, 6, 36423. [Google Scholar] [CrossRef]
- Kekenes-Huskey, P.M.; Scott, C.E.; Atalay, S. Quantifying the influence of the crowded cytoplasm on small molecule diffusion. J. Phys. Chem. B 2016, 120, 8696–8706. [Google Scholar] [CrossRef] [PubMed]
- Bock, A.; Annibale, P.; Konrad, C.; Hannawacker, A.; Anton, S.E.; Maiellaro, I.; Zabel, U.; Sivaramakrishnan, S.; Falcke, M.; Lohse, M.J. Optical mapping of cAMP signaling at the nanometer scale. Cell 2020, 182, 1519–1530.e1517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Lu, T.-W.; Stolerman, L.M.; Tenner, B.; Yang, J.R.; Zhang, J.-F.; Falcke, M.; Rangamani, P.; Taylor, S.S.; Mehta, S.; et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 2020, 182, 1531–1544.e1515. [Google Scholar] [CrossRef] [PubMed]
- Klein, I.A.; Boija, A.; Afeyan, L.K.; Hawken, S.W.; Fan, M.; Dall’Agnese, A.; Agnese, A.; Oksuz, O.; Henninger, J.E.; Shrinivas, K.; et al. Partitioning of cancer therapeutics in nuclear condensates. Science 2020, 368, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Gross, I.; Durner, J. In search of enzymes with a role in 3’,5’-cyclic guanosine monophosphate metabolism in plants. Front. Plant Sci. 2016, 7, 576. [Google Scholar] [CrossRef]
- Diffley, P.E.; Geisbrecht, A.; Newton, R.P.; Oliver, M.; Smith, C.J.; Vaughan, J.; van Cleef, J.; van Geyschem, J.; Walton, T.J.; Bayliss, M.; et al. Variation in isomeric products of a phosphodiesterase from the chloroplasts of Phaseolus vulgaris in response to cations. Plant Biosystems 2001, 135, 143–156. [Google Scholar] [CrossRef]
- Isner, J.-C.; Olteanu, V.-A.; Hetherington, A.J.; Coupel-Ledru, A.; Sun, P.; Pridgeon, A.J.; Jones, G.S.; Oates, M.; Williams, T.A.; Maathuis, F.J.M.; et al. Short- and long-term effects of UVA on Arabidopsis are mediated by a novel cGMP phosphodiesterase. Curr. Biol. 2019, 29, 2580–2585.e2584. [Google Scholar] [CrossRef]
- Berman, H.M.; Ten Eyck, L.F.; Goodsell, D.S.; Haste, N.M.; Kornev, A.; Taylor, S.S. The cAMP binding domain: An ancient signaling module. Proc. Natl. Acad. Sci. USA 2005, 102, 45. [Google Scholar] [CrossRef]
- Ho, Y.S.; Burden, L.M.; Hurley, J.H. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000, 19, 5288–5299. [Google Scholar] [CrossRef]
- Bridges, D.; Fraser, M.E.; Moorhead, G.B. Cyclic nucleotide binding proteins in the Arabidopsis thaliana and Oryza sativa genomes. BMC bioinform. BMC Bioinform. 2005, 6, 6. [Google Scholar] [CrossRef]
- Hoshi, T. Regulation of voltage dependence of the KAT1 channel by intracellular factors. J. Gen. Physiol. 1995, 105, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Lemtiri-Chlieh, F.; Berkowitz, G.A. Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J. Biol. Chem. 2004, 279, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.; Mercier, R.W.; Hua, B.-G.; Fromm, H.; Berkowitz, G.A. Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 2002, 128, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.; Ma, W.; Lemtiri-Chlieh, F.; Tsaltas, D.; Leng, Q.; von Bodman, S.; Berkowitz, G.A. Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 2007, 19, 1081–1095. [Google Scholar] [CrossRef] [PubMed]
- Balagué, C.; Lin, B.; Alcon, C.; Flottes, G.; Malmström, S.; Köhler, C.; Neuhaus, G.; Pelletier, G.; Gaymard, F.; Roby, D. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide–gated channel ion channel family. Plant Cell 2003, 15, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef]
- Yu, X.; Xu, G.; Li, B.; de Souza Vespoli, L.; Liu, H.; Moeder, W.; Chen, S.; de Oliveira, M.V.V.; Ariádina de Souza, S.; Shao, W.; et al. The receptor kinases BAK1/SERK4 regulate Ca2+ channel-mediated cellular homeostasis for cell death containment. Curr. Biol. 2019, 29, 3778–3790.e3778. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wang, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T.; et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef]
- Dietrich, P.; Moeder, W.; Yoshioka, K. Plant cyclic nucleotide-gated channels: New insights on their functions and regulation. Plant Physiol. 2020, 184, 27–38. [Google Scholar] [CrossRef]
- Wang, Y.-F.; Munemasa, S.; Nishimura, N.; Ren, H.-M.; Robert, N.; Han, M.; Puzõrjova, I.; Kollist, H.; Lee, S.; Mori, I.; et al. Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 2013, 163, 578–590. [Google Scholar] [CrossRef]
- Bowler, C.; Neuhaus, G.; Yamagata, H.; Chua, N.-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 1994, 77, 73–81. [Google Scholar] [CrossRef]
- Szmidt-Jaworska, A.; Jaworski, K.; Kopcewicz, J. Involvement of cyclic GMP in phytochrome-controlled flowering of Pharbitis nil. J. Plant Physiol. 2008, 165, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Volotovski, I.D.; Dubovskaya, L.V.; Molchan, O.V. Photorecptor phytochrome regulates the cyclic guanosine 3’,5’-monophosphate synthesis in Avena sativa L. cells. Bul. J. Plant Physiol. 2003, 29, 3–12. [Google Scholar]
- Hughes, J. Phytochrome cytoplasmic signaling. Annu. Rev. Plant Biol. 2013, 64, 377–402. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, L.; Meier, S.; Gehring, C. The Arabidopsis cyclic nucleotide interactome. Cell Commun. Signal. 2016, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, T.D.; Hillmann, K.B. Enzyme promiscuity, metabolite damage, and metabolite damage control systems of the tricarboxylic acid cycle. FEBS J. 2020, 287, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Aram, L.; Braun, T.; Braverman, C.; Kaplan, Y.; Ravid, L.; Levin-Zaidman, S.; Arama, E. A Krebs cycle component limits caspase activation rate through mitochondrial surface restriction of CRL activation. Dev. Cell 2016, 37, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Wang, X.; Kaufman, B.A.; Butow, R.A. Aconitase Couples Metabolic Regulation to Mitochondrial DNA Maintenance. Science 2005, 307, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef]
- Pharmawati, M.; Billington, T.; Gehring, C.A. Stomatal guard cell responses to kinetin and natriuretic peptides are cGMP dependent. Cell. Mol. Life Sci. 1998, 54, 272–276. [Google Scholar] [CrossRef]
- Maathuis, F.J.M. cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 2006, 45, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Janse van Rensburg, H.C.; Van den Ende, W.; Signorelli, S. Autophagy in plants: Both a puppet and a puppet master of sugars. Front. Plant Sci. 2019, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Marshall, R.S.; Li, F. Understanding and exploiting the roles of autophagy in plants through multi-omics approaches. Plant Sci. 2018, 274, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Irani, N.G.; Di Rubbo, S.; Mylle, E.; Van den Begin, J.; Schneider-Pizoń, J.; Hniliková, J.; Šíša, M.; Buyst, D.; Vilarrasa-Blasi, J.; Szatmári, A.M.; et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 2012, 8, 583–589. [Google Scholar] [CrossRef]
- Mbengue, M.; Bourdais, G.; Gervasi, F.; Beck, M.; Zhou, J.; Spallek, T.; Bartels, S.; Boller, T.; Ueda, T.; Kuhn, H.; et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. USA 2016, 113, 11034–11039. [Google Scholar] [CrossRef]
- Ortiz-Morea, F.A.; Savatin, D.V.; Dejonghe, W.; Kumar, R.; Luo, Y.; Adamowski, M.; Van den Begin, J.; Dressano, K.; Pereira de Oliveira, G.; Zhao, X.; et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc. Natl. Acad. Sci. USA 2016, 113, 11028–11033. [Google Scholar] [CrossRef]
- Geldner, N.; Hyman, D.L.; Wang, X.; Schumacher, K.; Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007, 21, 1598–1602. [Google Scholar] [CrossRef]
- Martins, S.; Dohmann, E.M.N.; Cayrel, A.; Johnson, A.; Fischer, W.; Pojer, F.; Satiat-Jeunemaître, B.; Jaillais, Y.; Chory, J.; Geldner, N.; et al. Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 2015, 6, 6151. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, D.; Wang, P.; Ma, X.; Lin, W.; Chen, S.; Mishev, K.; Lu, D.; Kumar, R.; Vanhoutte, I.; et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 2018, 115, E1906–E1915. [Google Scholar] [CrossRef]
- Grubb, L.E.; Derbyshire, P.; Dunning, K.; Zipfel, C.; Menke, F.L.H.; Monaghan, J. Large-scale identification of ubiquitination sites on membrane-associated proteins in Arabidopsis thaliana seedlings. bioRxiv 2020. [Google Scholar] [CrossRef]
- VerPlank, J.J.S.; Tyrkalska, S.D.; Fleming, A.; Rubinsztein, D.C.; Goldberg, A.L. cGMP via PKG activates 26S proteasomes and enhances degradation of proteins, including ones that cause neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 2020, 117, 14220–14230. [Google Scholar] [CrossRef] [PubMed]
- Nan, W.; Wang, X.; Yang, L.; Hu, Y.; Wei, Y.; Liang, X.; Mao, L.; Bi, Y. Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J. Exp. Bot. 2014, 65, 1571–1583. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turek, I.; Irving, H. Moonlighting Proteins Shine New Light on Molecular Signaling Niches. Int. J. Mol. Sci. 2021, 22, 1367. https://doi.org/10.3390/ijms22031367
Turek I, Irving H. Moonlighting Proteins Shine New Light on Molecular Signaling Niches. International Journal of Molecular Sciences. 2021; 22(3):1367. https://doi.org/10.3390/ijms22031367
Chicago/Turabian StyleTurek, Ilona, and Helen Irving. 2021. "Moonlighting Proteins Shine New Light on Molecular Signaling Niches" International Journal of Molecular Sciences 22, no. 3: 1367. https://doi.org/10.3390/ijms22031367
APA StyleTurek, I., & Irving, H. (2021). Moonlighting Proteins Shine New Light on Molecular Signaling Niches. International Journal of Molecular Sciences, 22(3), 1367. https://doi.org/10.3390/ijms22031367






