Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion
Abstract
1. Introduction
2. Results
2.1. Isolation of SUP35 Mutants That Induce [PSI+] without Their Overproduction
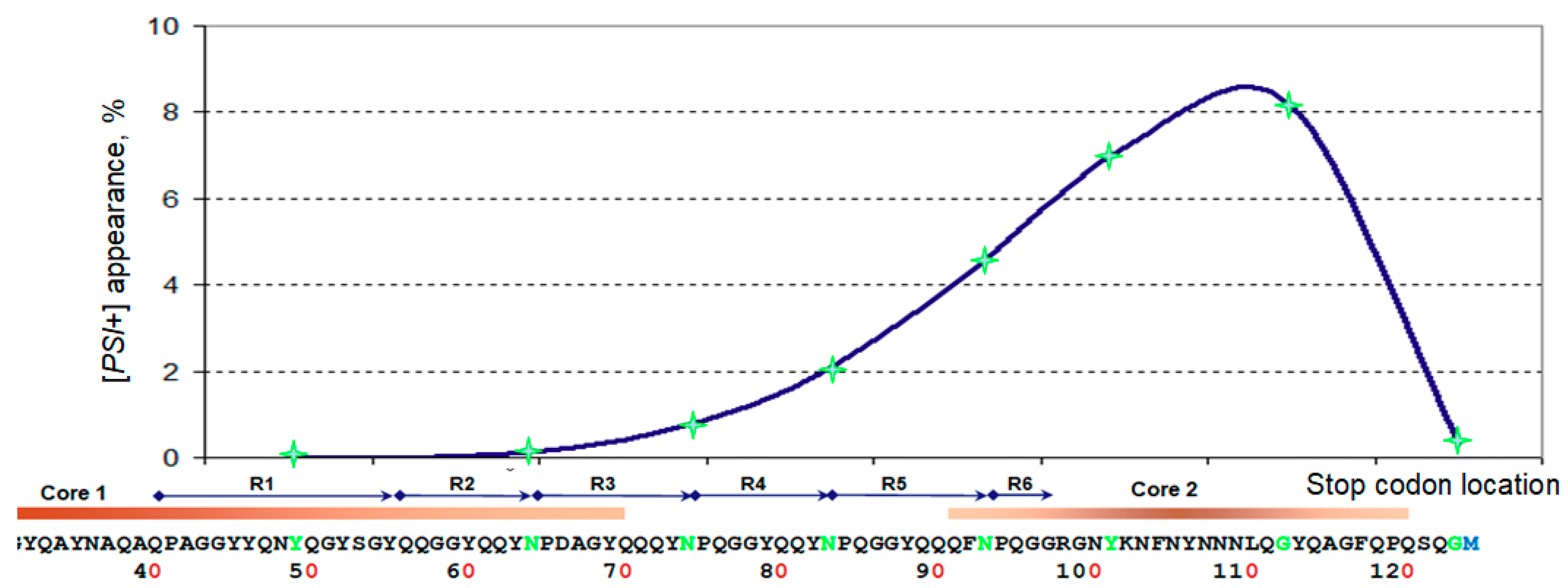
2.2. [PSI+] Induction by Sup35 N-Terminal Fragments
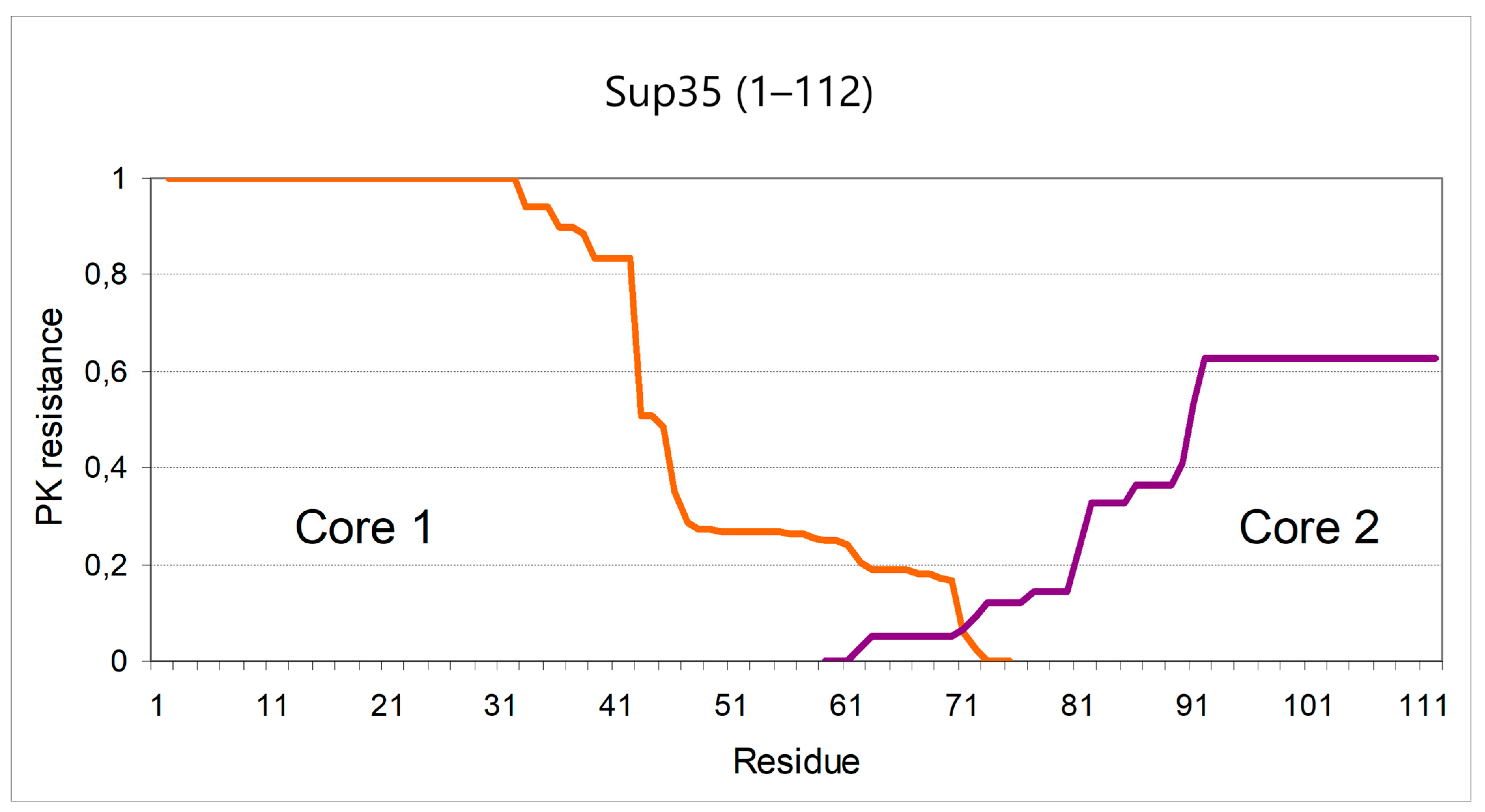
2.3. Structure of the Inducer Fragment
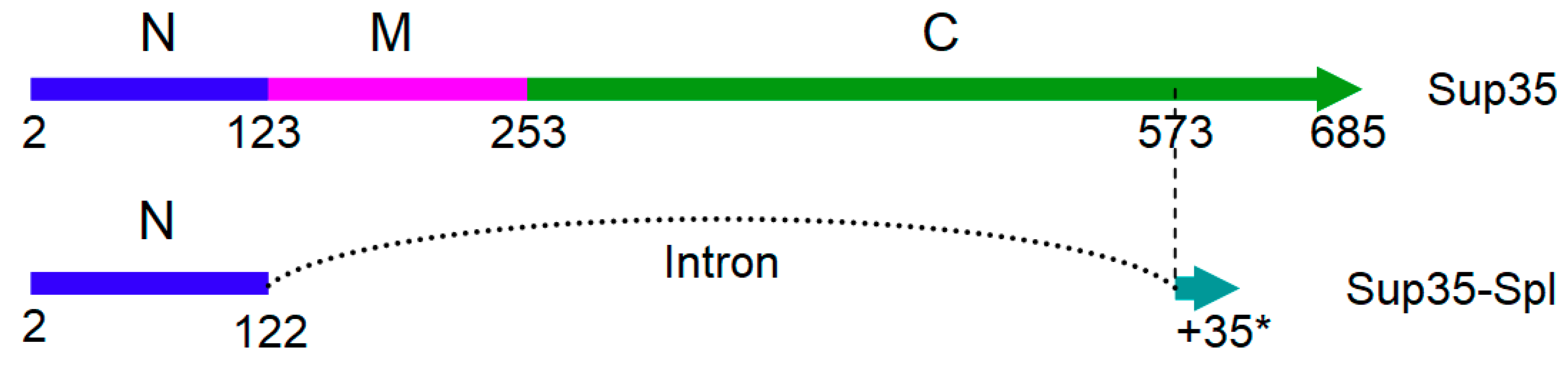
2.4. SUP35 Splicing
3. Discussion
3.1. The “Hot-Spot” of Sup35 Prionogenesis and Its Possible Structural Explanations
3.2. Sup35 Splicing and Its Effect on Prionogenesis
3.3. Support for the “Somatic Mutation” Hypothesis of Sporadic Amyloidoses
4. Materials and Methods
4.1. Yeast Strains and Media
4.2. Plasmids
4.3. SUP35 Mutagenesis and Obtaining Mutants with Increased [PSI+] Appearance
4.4. Evaluation of PSI+ Induction Rates
4.5. PK Digestion, Mass Spectrometry an Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PK | Proteinase K |
| MALDI-TOF | Matrix-assisted Laser Desorption/Ionization Time of Flight |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| ALS | Amyotrophic lateral sclerosis |
| SEM | Standard error of the mean |
References
- Chiti, F.; Dobson, C.M. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Arata, H.; Takashima, H.; Hirano, R.; Tomimitsu, H.; Machigashira, K.; Izumi, K.; Kikuno, M.; Ng, A.R.; Umehara, F.; Arisato, T.; et al. Early clinical signs and imaging findings in Gerstmann-Straussler-Scheinker syndrome (Pro102Leu). Neurology 2006, 66, 1672–1678. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.J. The prevalence and causes of dementia in people under the age of 65 years. J. Neurol. Neurosurg. Psychiatry 2003, 74, 1206–1209. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Brice, A. Parkinson’s disease: From monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 2009, 18, R48–R59. [Google Scholar] [CrossRef]
- Barykin, E.P.; Mitkevich, V.A.; Kozin, S.A.; Makarov, A.A. Amyloid β modification: A key to the sporadic alzheimer’s disease? Front. Genet. 2017, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, G.; Veltman, J.A. The role of de novo mutations in adult-onset neurodegenerative disorders. Acta Neuropathol. 2019, 137, 183–207. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Brundin, P. Parkinson’s disease and alpha synuclein: Is Parkinson’s disease a prion-like disorder? Mov. Disord. 2013, 28, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Masuda-Suzukake, M.; Falcon, B. Like prions: The propagation of aggregated tau and α-synuclein in neurodegeneration. Brain 2017, 140, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Son, M.; Bezsonov, E.E.; DeWilde, M.; Ducatez, M. Yeast prions compared to functional prions and amyloids. J. Mol. Biol. 2018, 430, 3707–3719. [Google Scholar] [CrossRef]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Frederick, K.K.; Debelouchina, G.T.; Kayatekin, C.; Dorminy, T.; Jacavone, A.C.; Griffin, R.G.; Lindquist, S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem. Biol. 2014, 21, 295–305. [Google Scholar] [CrossRef]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef]
- Dergalev, A.; Alexandrov, A.; Ivannikov, R.; Ter-Avanesyan, M.; Kushnirov, V. Yeast Sup35 prion structure: Two types, four parts, many variants. Int. J. Mol. Sci. 2019, 20, 2633. [Google Scholar] [CrossRef]
- Lancaster, A.K.; Bardill, J.P.; True, H.L.; Masel, J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] System. Genetics 2010, 184, 393–400. [Google Scholar] [CrossRef][Green Version]
- Tyedmers, J.; Madariaga, M.L.; Lindquist, S. Prion switching in response to environmental stress. PLoS Biol. 2008, 6, 2605–2613. [Google Scholar] [CrossRef]
- Salnikova, A.B.; Kryndushkin, D.S.; Smirnov, V.N.; Kushnirov, V.V.; Ter-Avanesyan, M.D. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J. Biol. Chem. 2005, 280, 8808–8812. [Google Scholar] [CrossRef]
- Wang, K.; Melki, R.; Kabani, M. A prolonged chronological lifespan is an unexpected benefit of the [PSI+] prion in yeast. PLoS ONE 2017, 12, e0184905. [Google Scholar] [CrossRef]
- Speldewinde, S.; Grant, C. The frequency of yeast [PSI+] prion formation is increased during chronological ageing. Microb. Cell 2017, 4, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Lindquist, S. Oligopeptide-repeat expansions modulate “protein-only” inheritance in yeast. Nature 1999, 400, 573–576. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Ter-Avanesyan, M.D.; Telckov, M.V.; Surguchov, A.P.; Smirnov, V.N.; Inge-Vechtomov, S.G. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene 1988, 66, 45–54. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Lin, J.-Y.; Lee, H.-C.; Wang, H.-L.; King, C.-Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc. Natl. Acad. Sci. USA. 2008, 105, 13345–13350. [Google Scholar] [CrossRef]
- Depace, A.H.; Santoso, A.; Hillner, P.; Weissman, J.S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998, 93, 1241–1252. [Google Scholar] [CrossRef]
- Sant’anna, R.; Fernández, M.R.; Batlle, C.; Navarro, S.; De Groot, N.S.; Serpell, L.; Ventura, S. Characterization of amyloid cores in prion domains. Sci. Rep. 2016, 6, 34274. [Google Scholar] [CrossRef]
- Hooks, K.B.; Delneri, D.; Griffiths-Jones, S. Intron evolution in Saccharomycetaceae. Genome Biol. Evol. 2014, 6, 2543–2556. [Google Scholar] [CrossRef]
- Qin, D.; Huang, L.; Wlodaver, A.; Andrade, J.; Staley, J.P. Sequencing of lariat termini in S. cerevisiae reveals 5′ splice sites, branch points, and novel splicing events. RNA 2016, 22, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN(+)]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.K.; Gavin-smyth, J.; Liebman, S.W. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 2009, 11, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Murata, K.; Hashimoto, W.; Kawai, S. Hsp104-dependent ability to assimilate mannitol and sorbitol conferred by a truncated Cyc8 with a C-terminal polyglutamine in Saccharomyces cerevisiae. PLoS ONE 2020, 15, e0242054. [Google Scholar] [CrossRef]
- Dagkesamanskaia, A.R.; Kushnirov, V.V.; Paushkin, S.V.; Ter-Avanesian, M.D. Fusion of glutathione S-transferase with the N-terminus of yeast Sup35p protein inhibits its prion-like properties. Genetika 1997, 33, 610–615. [Google Scholar] [PubMed]
- Kajava, A.V.; Baxa, U.; Wickner, R.B.; Steven, A.C. A model for Ure2p prion filaments and other amyloids: The parallel superpleated beta-structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7885–7890. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Hong, J.Y.; Kanneganti, V.; Park, S.-K.; Liebman, S.W. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet. 2015, 11, e1004814. [Google Scholar] [CrossRef]
- Matsuzono, K.; Ikeda, Y.; Liu, W.; Kurata, T.; Deguchi, S.; Deguchi, K.; Abe, K. A novel familial prion disease causing pan-autonomic-sensory neuropathy and cognitive impairment. Eur. J. Neurol. 2013, 20, e67–e69. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Adachi, Y.; Nakashima, K. Japanese familial amyotrophic lateral sclerosis family with a two-base deletion in the superoxide dismutase-1 gene. Neuropathology 2001, 21, 61–66. [Google Scholar] [CrossRef]
- Weskamp, K.; Tank, E.M.; Miguez, R.; McBride, J.P.; Gómez, N.B.; White, M.; Lin, Z.; Gonzalez, C.M.; Serio, A.; Sreedharan, J.; et al. Shortened TDP43 isoforms upregulated by neuronal hyperactivity drive TDP43 pathology in ALS. J. Clin. Investig. 2020, 130, 1139–1155. [Google Scholar] [CrossRef]
- Ghetti, B.; Piccardo, P.; Spillantini, M.G.; Ichimiya, Y.; Porro, M.; Perini, F.; Kitamoto, T.; Tateishi, J.; Seiler, C.; Frangione, B.; et al. Vascular variant of prion protein cerebral amyloidosis with tau-positive neurofibrillary tangles: The phenotype of the stop codon 145 mutation in PRNP. Proc. Natl. Acad. Sci. USA 1996, 93, 744–748. [Google Scholar] [CrossRef]
- Guerreiro, R.; Brás, J.; Wojtas, A.; Rademakers, R.; Hardy, J.; Graff-Radford, N. Nonsense mutation in PRNP associated with clinical Alzheimer’s disease. Neurobiol. Aging 2014, 35, 2656.e13–2656.e16. [Google Scholar] [CrossRef]
- Nakaya, T.; Maragkakis, M. Amyotrophic Lateral Sclerosis associated FUS mutation shortens mitochondria and induces neurotoxicity. Sci. Rep. 2018, 8, 15575. [Google Scholar] [CrossRef]
- Calvo, A.; Moglia, C.; Canosa, A.; Brunetti, M.; Barberis, M.; Traynor, B.J.; Carrara, G.; Valentini, C.; Restagno, G.; Chiò, A. De novo nonsense mutation of the FUS gene in an apparently familial amyotrophic lateral sclerosis case. Neurobiol. Aging 2014, 35, 1513.e7–1513.e11. [Google Scholar] [CrossRef]
- Corrado, L.; D’Alfonso, S.; Bergamaschi, L.; Testa, L.; Leone, M.; Nasuelli, N.; Momigliano-Richiardi, P.; Mazzini, L. SOD1 gene mutations in Italian patients with Sporadic Amyotrophic Lateral Sclerosis (ALS). Neuromuscul. Disord. 2006, 16, 800–804. [Google Scholar] [CrossRef]
- Gaudette, M.; Hirano, M.; Siddique, T. Current status of SOD1 mutations in familial amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 83–89. [Google Scholar] [CrossRef]
- Milholland, B.; Dong, X.; Zhang, L.; Hao, X.; Suh, Y.; Vijg, J. Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 2017, 8, 15183. [Google Scholar] [CrossRef]
- Spires-Jones, T.L.; Attems, J.; Thal, D.R. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017, 134, 187–205. [Google Scholar] [CrossRef]
- Ren, B.; Zhang, Y.; Zhang, M.; Liu, Y.; Zhang, D.; Gong, X.; Feng, Z.; Tang, J.; Chang, Y.; Zheng, J. Fundamentals of cross-seeding of amyloid proteins: An introduction. J. Mater. Chem. B 2019, 7, 7267–7282. [Google Scholar] [CrossRef]
- Häggqvist, B.; Näslund, J.; Sletten, K.; Westermark, G.T.; Mucchiano, G.; Tjernberg, L.O.; Nordstedt, C.; Engström, U.; Westermark, P. Medin: An integral fragment of aortic smooth muscle cell-produced lactadherin forms the most common human amyloid. Proc. Natl. Acad. Sci. USA 1999, 96, 8669–8674. [Google Scholar] [CrossRef] [PubMed]
- Migrino, R.Q.; Karamanova, N.; Truran, S.; Serrano, G.E.; Davies, H.A.; Madine, J.; Beach, T.G. Cerebrovascular medin is associated with Alzheimer’s disease and vascular dementia. Alzheimer’s Dement. 2020, 12, e12072. [Google Scholar] [CrossRef] [PubMed]
- Demaegd, K.; Schymkowitz, J.; Rousseau, F. Transcellular spreading of Tau in Tauopathies. Chembiochem 2018, 19, 2424–2432. [Google Scholar] [CrossRef] [PubMed]
- Groveman, B.R.; Foliaki, S.T.; Orru, C.D.; Zanusso, G.; Carroll, J.A.; Race, B.; Haigh, C.L. Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids. Acta Neuropathol. Commun. 2019, 7, 90. [Google Scholar] [CrossRef]
- Salman, M.M.; Marsh, G.; Kusters, I.; Delincé, M.; Di Caprio, G.; Upadhyayula, S.; de Nola, G.; Hunt, R.; Ohashi, K.G.; Gray, T.; et al. Design and validation of a human brain endothelial microvessel-on-a-chip open microfluidic model enabling advanced optical imaging. Front. Bioeng. Biotechnol. 2020, 8, 573775. [Google Scholar] [CrossRef] [PubMed]
- Centeno, E.G.Z.; Cimarosti, H.; Bithell, A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 2018, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Bailleul, P.A.; Newnam, G.P.; Steenbergen, J.N.; Chernoff, Y.O. Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics 1999, 153, 81–94. [Google Scholar] [PubMed]
- Parham, S.N.; Resende, C.G.; Tuite, M.F. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 2001, 20, 2111–2119. [Google Scholar] [CrossRef]
- Geissmann, Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 2013, 8, e54072. [Google Scholar] [CrossRef]

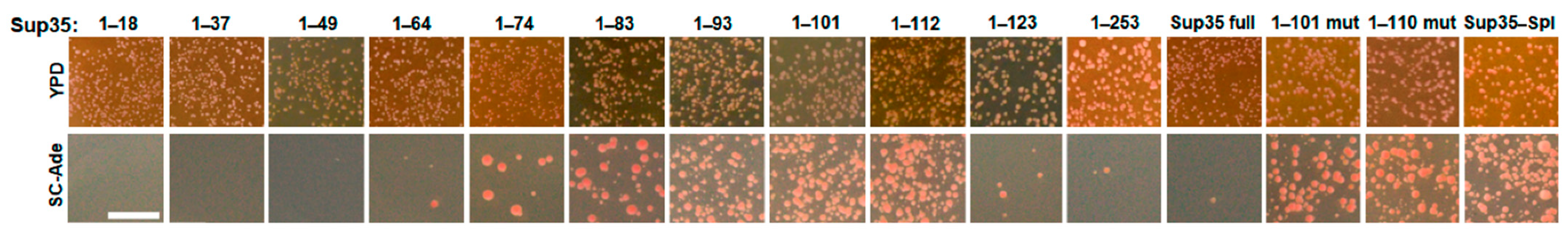

| Sup35 Fragment | [PSI+] Appearance, 10−5 |
|---|---|
| 1–18 | <1 * |
| 1–37 | <1 * |
| 1–49 | <1 * |
| 1–64 | 18.76 ± 12.23 |
| 1–74 | 393.0 ± 222.6 |
| 1–83 | 1572 ± 266.0 |
| 1–93 | 4044 ± 313.5 |
| 1–101 | 7201 ± 1227 |
| 1–112 | 8171 ± 1613 |
| 1–123 | 308.5 ± 269.3 |
| 1–252 | 5.13 ± 1.99 |
| Full Sup35 | 1.28 ± 0.08 |
| 1–101 mutant | 2222 ± 857.6 |
| 1–110 mutant | 1530 ± 372.2 |
| Spliced (1–122+) | 3692 ± 1504 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dergalev, A.A.; Urakov, V.N.; Agaphonov, M.O.; Alexandrov, A.I.; Kushnirov, V.V. Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion. Int. J. Mol. Sci. 2021, 22, 1542. https://doi.org/10.3390/ijms22041542
Dergalev AA, Urakov VN, Agaphonov MO, Alexandrov AI, Kushnirov VV. Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion. International Journal of Molecular Sciences. 2021; 22(4):1542. https://doi.org/10.3390/ijms22041542
Chicago/Turabian StyleDergalev, Alexander A., Valery N. Urakov, Michael O. Agaphonov, Alexander I. Alexandrov, and Vitaly V. Kushnirov. 2021. "Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion" International Journal of Molecular Sciences 22, no. 4: 1542. https://doi.org/10.3390/ijms22041542
APA StyleDergalev, A. A., Urakov, V. N., Agaphonov, M. O., Alexandrov, A. I., & Kushnirov, V. V. (2021). Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion. International Journal of Molecular Sciences, 22(4), 1542. https://doi.org/10.3390/ijms22041542







