Emerging Role of T-cell Receptor Constant β Chain-1 (TRBC1) Expression in the Flow Cytometric Diagnosis of T-cell Malignancies
Abstract
1. Introduction
2. Overview of T-Cell Receptor Constant β Chain Rearrangement
3. Development and Characterization of the JOVI-1 (anti-TRBC1) Antibody
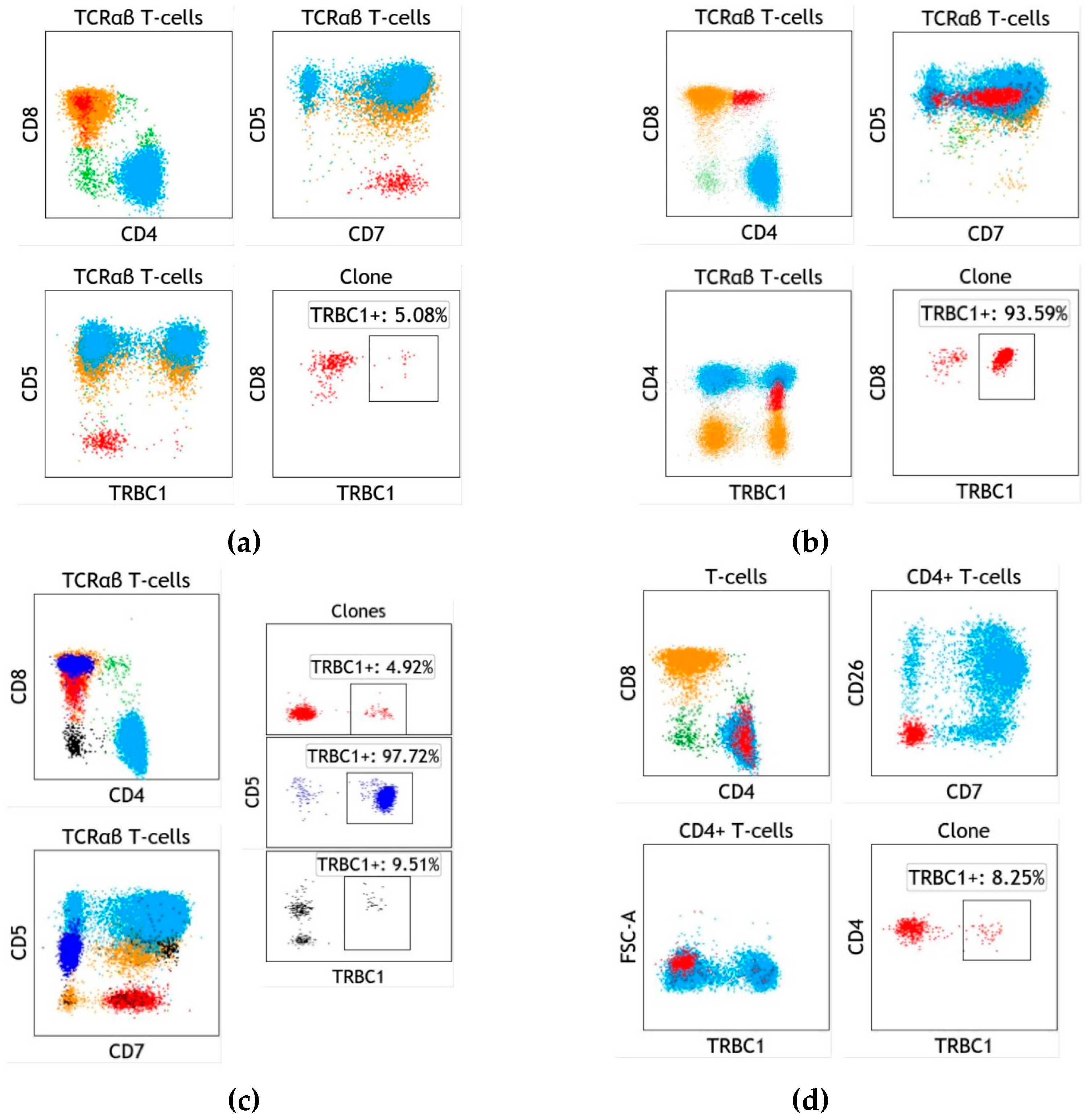
4. Assay Design and Implementation of TRBC1 for the Detection of Clonal T cells
- (1)
- Inability to detect clonal gamma/delta T-cells;
- (2)
- Limited ability to detect clonal T cells that are not immunophenotypically distinct with the combination of antigens studied;
- (3)
- Common detection of small T-cell clones of uncertain significance in patients without T-cell neoplasia (see below).
5. T-cell Clones of Uncertain Significance (T-CUS) Detected by TRBC1 Staining
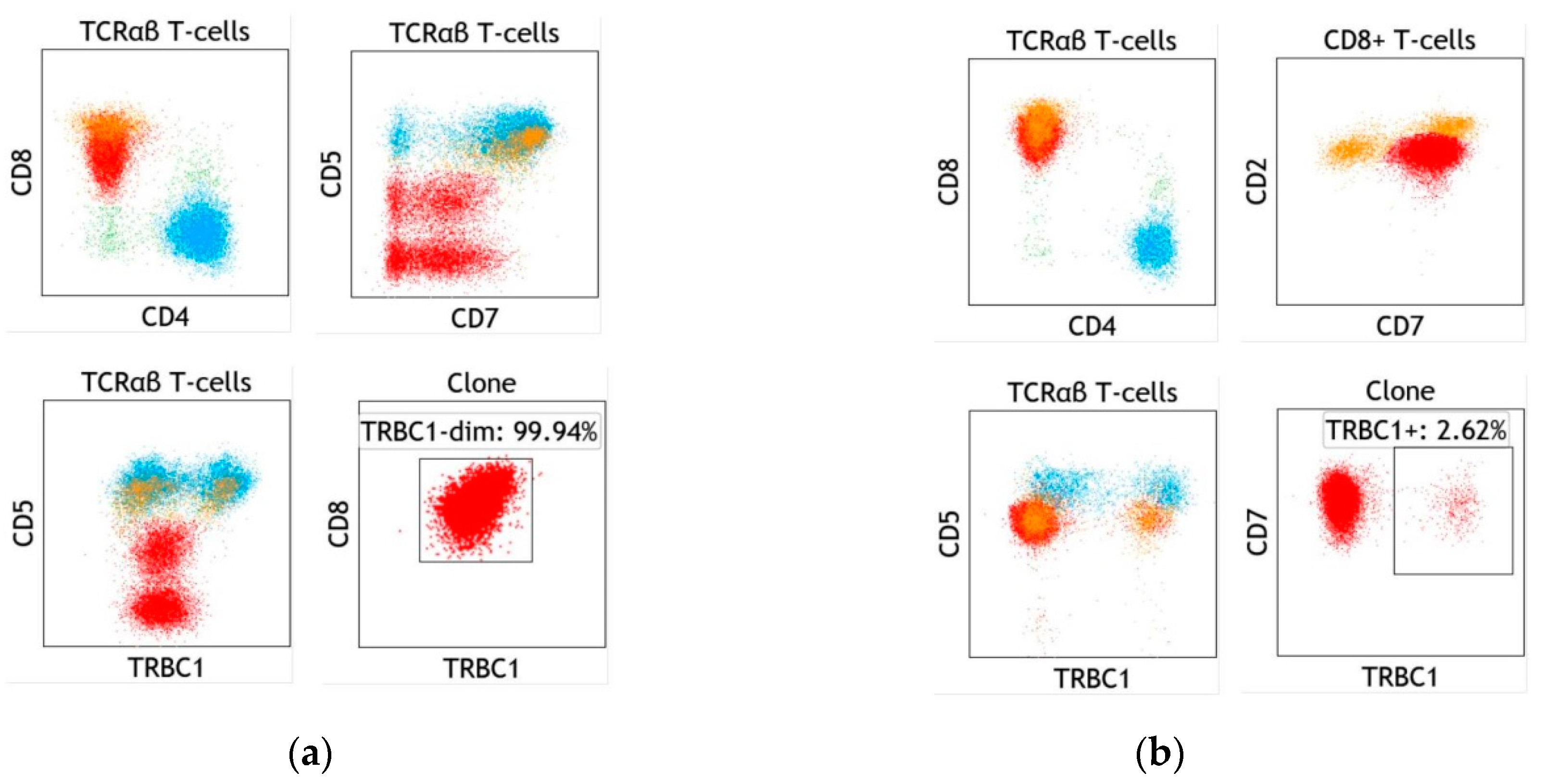
6. Specific Case of T-cell Large Granular Lymphocytic Leukemia
- (1)
- STAT3 and STAT5b mutations have been detected in approximately 50% and 2% of T-LGLL patients, respectively, indicating low sensitivity;
- (2)
- (3)
- Molecular study for STAT3 or STAT5b mutations is not broadly available and typically has a long turnaround time.
7. Specific Case of Blood Involvement by Cutaneous T-cell Lymphoma
- (1)
- Immunophenotypic analysis with assessment of clonality by TRBC1 has a far superior test performance for the detection of Sezary cells within the B0–B1 blood rating range, compared to the EORTC method;
- (2)
- The TRBC1 strategy can safely replace the EORTC method without significant impact on B2 blood rating;
- (3)
- Clonality assessment by TRBC1 effectively overcomes the limitations of comprehensive immunophenotypic analysis to identify Sezary cells lacking tumor-specific immunophenotypic abnormalities confidently.
8. Applications of TRBC1 Assessment for Tissue Analysis and Low-Cellularity Specimens
9. Future Developments
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bjorkman, P.J. MHC restriction in three dimensions: A view of T cell receptor/ligand interactions. Cell 1997, 89, 167–170. [Google Scholar] [CrossRef]
- Tonegawa, S. Somatic generation of antibody diversity. Nature 1983, 302, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, M.; White, H.; Gaulard, P.; Garcia-Sanz, R.; Gameiro, P.; Oeschger, S.; Jasani, B.; Ott, M.; Delsol, G.; Orfao, A.; et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia 2007, 21, 215–221. [Google Scholar] [CrossRef]
- Langerak, A.W.; Groenen, P.J.; Bruggemann, M.; Beldjord, K.; Bellan, C.; Bonello, L.; Boone, E.; Carter, G.I.; Catherwood, M.; Davi, F.; et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia 2012, 26, 2159–2171. [Google Scholar] [CrossRef]
- Wang, H.W.; Raffeld, M. Molecular assessment of clonality in lymphoid neoplasms. Semin. Hematol. 2019, 56, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.A.; Duncavage, E.J.; Mosbruger, T.L.; Szankasi, P.M.; Kelley, T.W. A comparison of deep sequencing of TCRG rearrangements vs traditional capillary electrophoresis for assessment of clonality in T-Cell lymphoproliferative disorders. Am. J. Clin. Pathol. 2014, 141, 348–359. [Google Scholar] [CrossRef]
- Morice, W.G.; Katzmann, J.A.; Pittelkow, M.R.; el-Azhary, R.A.; Gibson, L.E.; Hanson, C.A. A comparison of morphologic features, flow cytometry, TCR-Vbeta analysis, and TCR-PCR in qualitative and quantitative assessment of peripheral blood involvement by Sezary syndrome. Am. J. Clin. Pathol. 2006, 125, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Morice, W.G.; Kimlinger, T.; Katzmann, J.A.; Lust, J.A.; Heimgartner, P.J.; Halling, K.C.; Hanson, C.A. Flow cytometric assessment of TCR-Vbeta expression in the evaluation of peripheral blood involvement by T-cell lymphoproliferative disorders: A comparison with conventional T-cell immunophenotyping and molecular genetic techniques. Am. J. Clin. Pathol. 2004, 121, 373–383. [Google Scholar] [CrossRef]
- Morice, W.G.; Kurtin, P.J.; Leibson, P.J.; Tefferi, A.; Hanson, C.A. Demonstration of aberrant T-cell and natural killer-cell antigen expression in all cases of granular lymphocytic leukaemia. Br. J. Haematol. 2003, 120, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Olteanu, H.; Karandikar, N.J.; Eshoa, C.; Kroft, S.H. Laboratory findings in CD4(+) large granular lymphocytoses. Int. J. Lab. Hematol. 2010, 32, e9–e16. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Anderson, M.M.; Othus, M.; Wood, B.L. Clinical Experience With Modified, Single-Tube T-Cell Receptor Vbeta Flow Cytometry Analysis for T-Cell Clonality. Am. J. Clin. Pathol. 2016, 145, 467–485. [Google Scholar] [CrossRef]
- Maciocia, P.M.; Wawrzyniecka, P.A.; Philip, B.; Ricciardelli, I.; Akarca, A.U.; Onuoha, S.C.; Legut, M.; Cole, D.K.; Sewell, A.K.; Gritti, G.; et al. Targeting the T cell receptor beta-chain constant region for immunotherapy of T cell malignancies. Nat. Med. 2017, 23, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Viney, J.L.; Prosser, H.M.; Hewitt, C.R.; Lamb, J.R.; Owen, M.J. Generation of monoclonal antibodies against a human T cell receptor beta chain expressed in transgenic mice. Hybridoma 1992, 11, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, F.A.; Oettgen, H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010, 125, S33–S40. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.M.; Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 1988, 334, 395–402. [Google Scholar] [CrossRef]
- Krangel, M.S. Mechanics of T cell receptor gene rearrangement. Curr. Opin Immunol. 2009, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.J.; Langerak, A.W.; Bruggemann, M.; Evans, P.A.; Hummel, M.; Lavender, F.L.; Delabesse, E.; Davi, F.; Schuuring, E.; Garcia-Sanz, R.; et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003, 17, 2257–2317. [Google Scholar] [CrossRef]
- Cushman-Vokoun, A.M.; Connealy, S.; Greiner, T.C. Assay design affects the interpretation of T-cell receptor gamma gene rearrangements: Comparison of the performance of a one-tube assay with the BIOMED-2-based TCRG gene clonality assay. J. Mol. Diagn. 2010, 12, 787–796. [Google Scholar] [CrossRef]
- Mahe, E.; Pugh, T.; Kamel-Reid, S. T cell clonality assessment: Past, present and future. J. Clin. Pathol. 2018, 71, 195–200. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. The Complement System and Innate Immunity. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Toor, A.A.; Toor, A.A.; Rahmani, M.; Manjili, M.H. On the organization of human T-cell receptor loci: Log-periodic distribution of T-cell receptor gene segments. J. R Soc. Interface 2016, 13, 20150911. [Google Scholar] [CrossRef]
- Kuhns, M.S.; Davis, M.M.; Garcia, K.C. Deconstructing the form and function of the TCR/CD3 complex. Immunity 2006, 24, 133–139. [Google Scholar] [CrossRef]
- Chen, I.J.; Chen, H.L.; Demetriou, M. Lateral compartmentalization of T cell receptor versus CD45 by galectin-N-glycan binding and microfilaments coordinate basal and activation signaling. J. Biol. Chem 2007, 282, 35361–35372. [Google Scholar] [CrossRef]
- Gil, D.; Schamel, W.W.; Montoya, M.; Sanchez-Madrid, F.; Alarcon, B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 2002, 109, 901–912. [Google Scholar] [CrossRef]
- O’Shea, C.C.; Thornell, A.P.; Rosewell, I.R.; Hayes, B.; Owen, M.J. Exit of the pre-TCR from the ER/cis-Golgi is necessary for signaling differentiation, proliferation, and allelic exclusion in immature thymocytes. Immunity 1997, 7, 591–599. [Google Scholar] [CrossRef]
- Risueno, R.M.; Schamel, W.W.; Alarcon, B. T cell receptor engagement triggers its CD3epsilon and CD3zeta subunits to adopt a compact, locked conformation. PLoS ONE 2008, 3, e1747. [Google Scholar] [CrossRef]
- San Jose, E.; Alarcon, B. Receptor engagement transiently diverts the T cell receptor heterodimer from a constitutive degradation pathway. J. Biol. Chem 1999, 274, 33740–33746. [Google Scholar] [CrossRef] [PubMed]
- Toyonaga, B.; Yoshikai, Y.; Vadasz, V.; Chin, B.; Mak, T.W. Organization and sequences of the diversity, joining, and constant region genes of the human T-cell receptor beta chain. Proc. Natl. Acad. Sci. USA 1985, 82, 8624–8628. [Google Scholar] [CrossRef]
- Ohgami, R.S.; Arber, D.A.; Zehnder, J.L.; Natkunam, Y.; Warnke, R.A. Indolent T-lymphoblastic proliferation (iT-LBP): A review of clinical and pathologic features and distinction from malignant T-lymphoblastic lymphoma. Adv. Anat. Pathol. 2013, 20, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Juco, J.; Mann, K.P.; Holden, J.T. Flow cytometry in the differential diagnosis of lymphocyte-rich thymoma from precursor T-cell acute lymphoblastic leukemia/lymphoblastic lymphoma. Am. J. Clin. Pathol. 2004, 121, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Olteanu, H.; Jevremovic, D.; He, R.; Viswanatha, D.; Corley, H.; Horna, P. T-cell clones of uncertain significance are highly prevalent and show close resemblance to T-cell large granular lymphocytic leukemia. Implications for laboratory diagnostics. Mod. Pathol. 2020, 33, 2046–2057. [Google Scholar] [CrossRef] [PubMed]
- Novikov, N.D.; Griffin, G.K.; Dudley, G.; Drew, M.; Rojas-Rudilla, V.; Lindeman, N.I.; Dorfman, D.M. Utility of a Simple and Robust Flow Cytometry Assay for Rapid Clonality Testing in Mature Peripheral T-Cell Lymphomas. Am. J. Clin. Pathol. 2019, 151, 494–503. [Google Scholar] [CrossRef]
- Shi, M.; Jevremovic, D.; Otteson, G.E.; Timm, M.M.; Olteanu, H.; Horna, P. Single Antibody Detection of T-Cell Receptor alphabeta Clonality by Flow Cytometry Rapidly Identifies Mature T-Cell Neoplasms and Monotypic Small CD8-Positive Subsets of Uncertain Significance. Cytom. B Clin. Cytom. 2020, 98, 99–107. [Google Scholar] [CrossRef]
- van Dongen, J.J.; Lhermitte, L.; Bottcher, S.; Almeida, J.; van der Velden, V.H.; Flores-Montero, J.; Rawstron, A.; Asnafi, V.; Lecrevisse, Q.; Lucio, P.; et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 2012, 26, 1908–1975. [Google Scholar] [CrossRef]
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. [Google Scholar] [CrossRef]
- Arakawa, A.; Vollmer, S.; Tietze, J.; Galinski, A.; Heppt, M.V.; Burdek, M.; Berking, C.; Prinz, J.C. Clonality of CD4(+) Blood T Cells Predicts Longer Survival With CTLA4 or PD-1 Checkpoint Inhibition in Advanced Melanoma. Front. Immunol. 2019, 10, 1336. [Google Scholar] [CrossRef]
- Buchholz, V.R.; Neuenhahn, M.; Busch, D.H. CD8+ T cell differentiation in the aging immune system: Until the last clone standing. Curr. Opin. Immunol. 2011, 23, 549–554. [Google Scholar] [CrossRef]
- Blackman, M.A.; Woodland, D.L. The narrowing of the CD8 T cell repertoire in old age. Curr. Opin. Immunol. 2011, 23, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Shariff, N.; Cobbold, M.; Bruton, R.; Ainsworth, J.A.; Sinclair, A.J.; Nayak, L.; Moss, P.A. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol. 2002, 169, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Wedderburn, L.R.; Patel, A.; Varsani, H.; Woo, P. The developing human immune system: T-cell receptor repertoire of children and young adults shows a wide discrepancy in the frequency of persistent oligoclonal T-cell expansions. Immunology 2001, 102, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Maini, M.K.; Gudgeon, N.; Wedderburn, L.R.; Rickinson, A.B.; Beverley, P.C. Clonal expansions in acute EBV infection are detectable in the CD8 and not the CD4 subset and persist with a variable CD45 phenotype. J. Immunol. 2000, 165, 5729–5737. [Google Scholar] [CrossRef] [PubMed]
- Beverley, P.C.; Maini, M.K. Differences in the regulation of CD4 and CD8 T-cell clones during immune responses. Philos. Trans. R Soc. Lond B Biol. Sci. 2000, 355, 401–406. [Google Scholar] [CrossRef]
- Wack, A.; Cossarizza, A.; Heltai, S.; Barbieri, D.; D’Addato, S.; Fransceschi, C.; Dellabona, P.; Casorati, G. Age-related modifications of the human alphabeta T cell repertoire due to different clonal expansions in the CD4+ and CD8+ subsets. Int. Immunol. 1998, 10, 1281–1288. [Google Scholar] [CrossRef]
- Horna, P.; Olteanu, H.; Jevremovic, D.; Otteson, G.E.; Corley, H.; Ding, W.; Parikh, S.A.; Shah, M.V.; Morice, W.G.; Shi, M. Single-Antibody Evaluation of T-Cell Receptor beta Constant Chain Monotypia by Flow Cytometry Facilitates the Diagnosis of T-Cell Large Granular Lymphocytic Leukemia. Am. J. Clin. Pathol. 2021, in press. [Google Scholar] [CrossRef]
- Garrido, P.; Ruiz-Cabello, F.; Barcena, P.; Sandberg, Y.; Canton, J.; Lima, M.; Balanzategui, A.; Gonzalez, M.; Lopez-Nevot, M.A.; Langerak, A.W.; et al. Monoclonal TCR-Vbeta13.1+/CD4+/NKa+/CD8-/+dim T-LGL lymphocytosis: Evidence for an antigen-driven chronic T-cell stimulation origin. Blood 2007, 109, 4890–4898. [Google Scholar] [CrossRef]
- Rodriguez-Caballero, A.; Garcia-Montero, A.C.; Barcena, P.; Almeida, J.; Ruiz-Cabello, F.; Tabernero, M.D.; Garrido, P.; Munoz-Criado, S.; Sandberg, Y.; Langerak, A.W.; et al. Expanded cells in monoclonal TCR-alphabeta+/CD4+/NKa+/CD8-/+dim T-LGL lymphocytosis recognize hCMV antigens. Blood 2008, 112, 4609–4616. [Google Scholar] [CrossRef][Green Version]
- Horna, P.; Shi, M.; Jevremovic, D.; Craig, F.E.; Comfere, N.I.; Olteanu, H. Utility of TRBC1 Expression in the Diagnosis of Peripheral Blood Involvement by Cutaneous T-Cell Lymphoma. J. Invest. Dermatol. 2020, in press. [Google Scholar] [CrossRef]
- Lamy, T.; Moignet, A.; Loughran, T.P., Jr. LGL leukemia: From pathogenesis to treatment. Blood 2017, 129, 1082–1094. [Google Scholar] [CrossRef]
- Sanikommu, S.R.; Clemente, M.J.; Chomczynski, P.; Afable, M.G., 2nd; Jerez, A.; Thota, S.; Patel, B.; Hirsch, C.; Nazha, A.; Desamito, J.; et al. Clinical features and treatment outcomes in large granular lymphocytic leukemia (LGLL). Leuk Lymphoma 2018, 59, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.; Foucar, K.; Morice, W.G.; Matutes, E. T-cell large granular lymphocytic leukaemia. In WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Lee Harris, N., Jaffe, E.S., Pileri, S.A., Stein, H., Jurgen, T., Arber, D.A., Hasserjian, R.P., Le Beau, M.M., et al., Eds.; IARC: Lyon, France, 2017. [Google Scholar]
- Semenzato, G.; Zambello, R.; Starkebaum, G.; Oshimi, K.; Loughran, T.P., Jr. The lymphoproliferative disease of granular lymphocytes: Updated criteria for diagnosis. Blood 1997, 89, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Morice, W.G.; Kurtin, P.J.; Tefferi, A.; Hanson, C.A. Distinct bone marrow findings in T-cell granular lymphocytic leukemia revealed by paraffin section immunoperoxidase stains for CD8, TIA-1, and granzyme B. Blood 2002, 99, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, Y.; Almeida, J.; Gonzalez, M.; Lima, M.; Barcena, P.; Szczepanski, T.; van Gastel-Mol, E.J.; Wind, H.; Balanzategui, A.; van Dongen, J.J.; et al. TCRgammadelta+ large granular lymphocyte leukemias reflect the spectrum of normal antigen-selected TCRgammadelta+ T-cells. Leukemia 2006, 20, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Neff, J.L.; Rangan, A.; Jevremovic, D.; Nguyen, P.L.; Chiu, A.; Go, R.S.; Chen, D.; Morice, W.G.; Shi, M. Mixed-phenotype large granular lymphocytic leukemia: A rare subtype in the large granular lymphocytic leukemia spectrum. Hum. Pathol. 2018, 81, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Koskela, H.L.; Eldfors, S.; Ellonen, P.; van Adrichem, A.J.; Kuusanmaki, H.; Andersson, E.I.; Lagstrom, S.; Clemente, M.J.; Olson, T.; Jalkanen, S.E.; et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 2012, 366, 1905–1913. [Google Scholar] [CrossRef] [PubMed]
- Jerez, A.; Clemente, M.J.; Makishima, H.; Koskela, H.; Leblanc, F.; Peng Ng, K.; Olson, T.; Przychodzen, B.; Afable, M.; Gomez-Segui, I.; et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood 2012, 120, 3048–3057. [Google Scholar] [CrossRef] [PubMed]
- Savola, P.; Bruck, O.; Olson, T.; Kelkka, T.; Kauppi, M.J.; Kovanen, P.E.; Kytola, S.; Sokka-Isler, T.; Loughran, T.P.; Leirisalo-Repo, M.; et al. Somatic STAT3 mutations in Felty syndrome: An implication for a common pathogenesis with large granular lymphocyte leukemia. Haematologica 2018, 103, 304–312. [Google Scholar] [CrossRef]
- Jerez, A.; Clemente, M.J.; Makishima, H.; Rajala, H.; Gomez-Segui, I.; Olson, T.; McGraw, K.; Przychodzen, B.; Kulasekararaj, A.; Afable, M.; et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood 2013, 122, 2453–2459. [Google Scholar] [CrossRef]
- Kawakami, T.; Sekiguchi, N.; Kobayashi, J.; Imi, T.; Matsuda, K.; Yamane, T.; Nishina, S.; Senoo, Y.; Sakai, H.; Ito, T.; et al. Frequent STAT3 mutations in CD8(+) T cells from patients with pure red cell aplasia. Blood Adv. 2018, 2, 2704–2712. [Google Scholar] [CrossRef] [PubMed]
- Kucuk, C.; Jiang, B.; Hu, X.; Zhang, W.; Chan, J.K.; Xiao, W.; Lack, N.; Alkan, C.; Williams, J.C.; Avery, K.N.; et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat. Commun. 2015, 6, 6025. [Google Scholar] [CrossRef]
- Odejide, O.; Weigert, O.; Lane, A.A.; Toscano, D.; Lunning, M.A.; Kopp, N.; Kim, S.; van Bodegom, D.; Bolla, S.; Schatz, J.H.; et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood 2014, 123, 1293–1296. [Google Scholar] [CrossRef]
- Whittaker, S.J.; Cerroni, L.; Willemze, R.; Siebert, R. Sezary Syndrome. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; IARC: Lyon, France, 2017; pp. 390–391. [Google Scholar]
- Cerroni, L.; Sander, C.A.; Smoller, B.R.; Willemze, R.; Siebert, R. Mycosis Fungoides. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; IARC: Lyon, France, 2017; pp. 385–389. [Google Scholar]
- Scarisbrick, J.J.; Hodak, E.; Bagot, M.; Stranzenbach, R.; Stadler, R.; Ortiz-Romero, P.L.; Papadavid, E.; Evison, F.; Knobler, R.; Quaglino, P.; et al. Blood classification and blood response criteria in mycosis fungoides and Sezary syndrome using flow cytometry: Recommendations from the EORTC cutaneous lymphoma task force. Eur. J. Cancer 2018, 93, 47–56. [Google Scholar] [CrossRef]
- Olsen, E.; Vonderheid, E.; Pimpinelli, N.; Willemze, R.; Kim, Y.; Knobler, R.; Zackheim, H.; Duvic, M.; Estrach, T.; Lamberg, S.; et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: A proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007, 110, 1713–1722. [Google Scholar] [CrossRef]
- Agar, N.S.; Wedgeworth, E.; Crichton, S.; Mitchell, T.J.; Cox, M.; Ferreira, S.; Robson, A.; Calonje, E.; Stefanato, C.M.; Wain, E.M.; et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: Validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J. Clin. Oncol. 2010, 28, 4730–4739. [Google Scholar] [CrossRef]
- Benton, E.C.; Crichton, S.; Talpur, R.; Agar, N.S.; Fields, P.A.; Wedgeworth, E.; Mitchell, T.J.; Cox, M.; Ferreira, S.; Liu, P.; et al. A cutaneous lymphoma international prognostic index (CLIPi) for mycosis fungoides and Sezary syndrome. Eur. J. Cancer 2013, 49, 2859–2868. [Google Scholar] [CrossRef]
- Kelemen, K.; Guitart, J.; Kuzel, T.M.; Goolsby, C.L.; Peterson, L.C. The usefulness of CD26 in flow cytometric analysis of peripheral blood in Sezary syndrome. Am. J. Clin. Pathol. 2008, 129, 146–156. [Google Scholar] [CrossRef]
- Vaughan, J.; Harrington, A.M.; Hari, P.N.; Kroft, S.H.; Olteanu, H. Immunophenotypic stability of Sezary cells by flow cytometry: Usefulness of flow cytometry in assessing response to and guiding alemtuzumab therapy. Am. J. Clin. Pathol. 2012, 137, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Horna, P.; Deaver, D.M.; Qin, D.; Moscinski, L.C.; Sotomayor, E.M.; Glass, L.F.; Sokol, L. Quantitative flow cytometric identification of aberrant T cell clusters in erythrodermic cutaneous T cell lymphoma. Implications for staging and prognosis. J. Clin. Pathol. 2014, 67, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Haftcheshmeh, S.M.; Tajbakhsh, A.; Kazemi, M.; Esmaeili, S.A.; Mardani, F.; Fazeli, M.; Sahebkar, A. The clinical importance of CD4(+) CD7(-) in human diseases. J. Cell Physiol. 2019, 234, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Pulitzer, M.P.; Horna, P.; Almeida, J. Sezary syndrome and mycosis fungoides: An overview, including the role of immunophenotyping. Cytom. B Clin. Cytom. 2020, in press. [Google Scholar] [CrossRef]
- Horna, P.; Wang, S.A.; Wolniak, K.L.; Psarra, K.; Almeida, J.; Illingworth, A.J.; Johansson, U.; Craig, F.E.; Torres, R. Flow cytometric evaluation of peripheral blood for suspected Sezary syndrome or mycosis fungoides: International guidelines for assay characteristics. Cytom. B Clin. Cytom. 2020, in press. [Google Scholar] [CrossRef]
- Craig, F.E. It is time to adopt a multicolor immunophenotyping approach to evaluate blood for Sezary syndrome and mycosis fungoides. Cytom. B Clin. Cytom. 2020, in press. [Google Scholar] [CrossRef]
- Feng, B.; Jorgensen, J.L.; Jones, D.; Chen, S.S.; Hu, Y.; Medeiros, L.J.; Wang, S.A. Flow cytometric detection of peripheral blood involvement by mycosis fungoides and Sezary syndrome using T-cell receptor Vbeta chain antibodies and its application in blood staging. Mod. Pathol. 2010, 23, 284–295. [Google Scholar] [CrossRef]
- Lyapichev, K.A.; Bah, I.; Huen, A.; Duvic, M.; Routbort, M.J.; Wang, W.; Jorgensen, J.L.; Medeiros, L.J.; Vega, F.; Craig, F.E.; et al. Determination of immunophenotypic aberrancies provides better assessment of peripheral blood involvement by mycosis fungoides/Sezary syndrome than quantification of CD26- or CD7- CD4+ T-cells. Cytom. B Clin. Cytom 2020, in press. [Google Scholar] [CrossRef]
- Jamal, S.; Picker, L.J.; Aquino, D.B.; McKenna, R.W.; Dawson, D.B.; Kroft, S.H. Immunophenotypic analysis of peripheral T-cell neoplasms. A multiparameter flow cytometric approach. Am. J. Clin. Pathol. 2001, 116, 512–526. [Google Scholar] [CrossRef]
- Reichard, K.K.; Kroft, S.H. Flow Cytometry in the Assessment of Hematologic Disroders. In Neoplastic HematoPathol.ogy; Orazi, A., Foucar, K., Knowles, D.M., Eds.; Lippincott, Williams and Wilkins: Baltimore, MD, USA, 2013; pp. 119–145. [Google Scholar]
- Chen, W.; Kesler, M.V.; Karandikar, N.J.; McKenna, R.W.; Kroft, S.H. Flow cytometric features of angioimmunoblastic T-cell lymphoma. Cytom. B Clin. Cytom. 2006, 70, 142–148. [Google Scholar] [CrossRef]
- Jevremovic, D.; Olteanu, H. Flow Cytometry Applications in the Diagnosis of T/NK-Cell Lymphoproliferative Disorders. Cytom. B Clin. Cytom. 2019, 96, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.; Otteson, G.E.; Corley, H.; Shi, M.; Horna, P.; Jevremovic, D.; Olteanu, H. Flow Cytometric Evaluation of TRBC1 Expression in Tissue Specimens and Body Fluids is a Novel, Sensitive and Specific Method for Assessment of T-Cell Clonality and Diagnosis of T-Cell Neoplasms. Cytom. B Clin. Cytom. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M.; Baldan, V.; Ghongane, P.; Nicholson, A.; Bughda, R.; Akbar, Z.; Wawrzyniecka, P.; Maciocia, P.; Cordoba, S.; Thomas, S.; et al. Abstract 2183: Targeting TRBC1 and 2 for the treatment of T cell lymphomas. Cancer Res. 2020, 80, 2183. [Google Scholar] [CrossRef]



| Scenario | Utility of TRBC1 Staining |
|---|---|
| CD3+/TCRαβ+ T-cell neoplasias | Rapid demonstration of clonality on immunophenotypically distinct and expanded T-cell subsets, supporting a diagnosis of neoplasia and eliminating the need for a separate T-cell clonality assay. |
| Benign CD3+/TCRαβ+ T-cell subsets with immunophenotypic features concerning for neoplasia | Demonstration of TCR Cβ polytypia on benign subsets with atypical immunophenotypic features, rapidly and confidently ruling out neoplasia and preventing unnecessary additional work-up and/or misdiagnoses. |
| T-cell large granular lymphocytic leukemia (T-LGLL), and clonal T-cell large granular lymphocytic populations of uncertain significance (T-CUS) | Accurate identification and quantitation of clonal cytotoxic T-cell subsets, contributing to the diagnosis of T-LGLL and its distinction from T-CUS based on clone size. |
| Cutaneous T-cell lymphoma (CTCL) | Identification and quantitation of clonal CD4+ T-cell subsets, allowing for a confident diagnosis of CTCL involvement, and accurate assessment of the extent of blood involvement for staging purposes. |
| Fine needle aspirates and small biopsies | Rapid identification of immunophenotypically distinct T-cell clones, providing valuable information for the interpretation of limited specimens, and the need for additional work-up or an excisional biopsy. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horna, P.; Shi, M.; Olteanu, H.; Johansson, U. Emerging Role of T-cell Receptor Constant β Chain-1 (TRBC1) Expression in the Flow Cytometric Diagnosis of T-cell Malignancies. Int. J. Mol. Sci. 2021, 22, 1817. https://doi.org/10.3390/ijms22041817
Horna P, Shi M, Olteanu H, Johansson U. Emerging Role of T-cell Receptor Constant β Chain-1 (TRBC1) Expression in the Flow Cytometric Diagnosis of T-cell Malignancies. International Journal of Molecular Sciences. 2021; 22(4):1817. https://doi.org/10.3390/ijms22041817
Chicago/Turabian StyleHorna, Pedro, Min Shi, Horatiu Olteanu, and Ulrika Johansson. 2021. "Emerging Role of T-cell Receptor Constant β Chain-1 (TRBC1) Expression in the Flow Cytometric Diagnosis of T-cell Malignancies" International Journal of Molecular Sciences 22, no. 4: 1817. https://doi.org/10.3390/ijms22041817
APA StyleHorna, P., Shi, M., Olteanu, H., & Johansson, U. (2021). Emerging Role of T-cell Receptor Constant β Chain-1 (TRBC1) Expression in the Flow Cytometric Diagnosis of T-cell Malignancies. International Journal of Molecular Sciences, 22(4), 1817. https://doi.org/10.3390/ijms22041817






