The BIRC Family Genes Expression in Patients with Triple Negative Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Level of Expression of the BIRC Family Genes in Breast Cancer Tissue of Patients with TNBC Compared to Normal Tissue Surrounding the Tumor. Comparison of the Obtained Results with the Bioinformatic Analysis of Data Obtained from TCGA
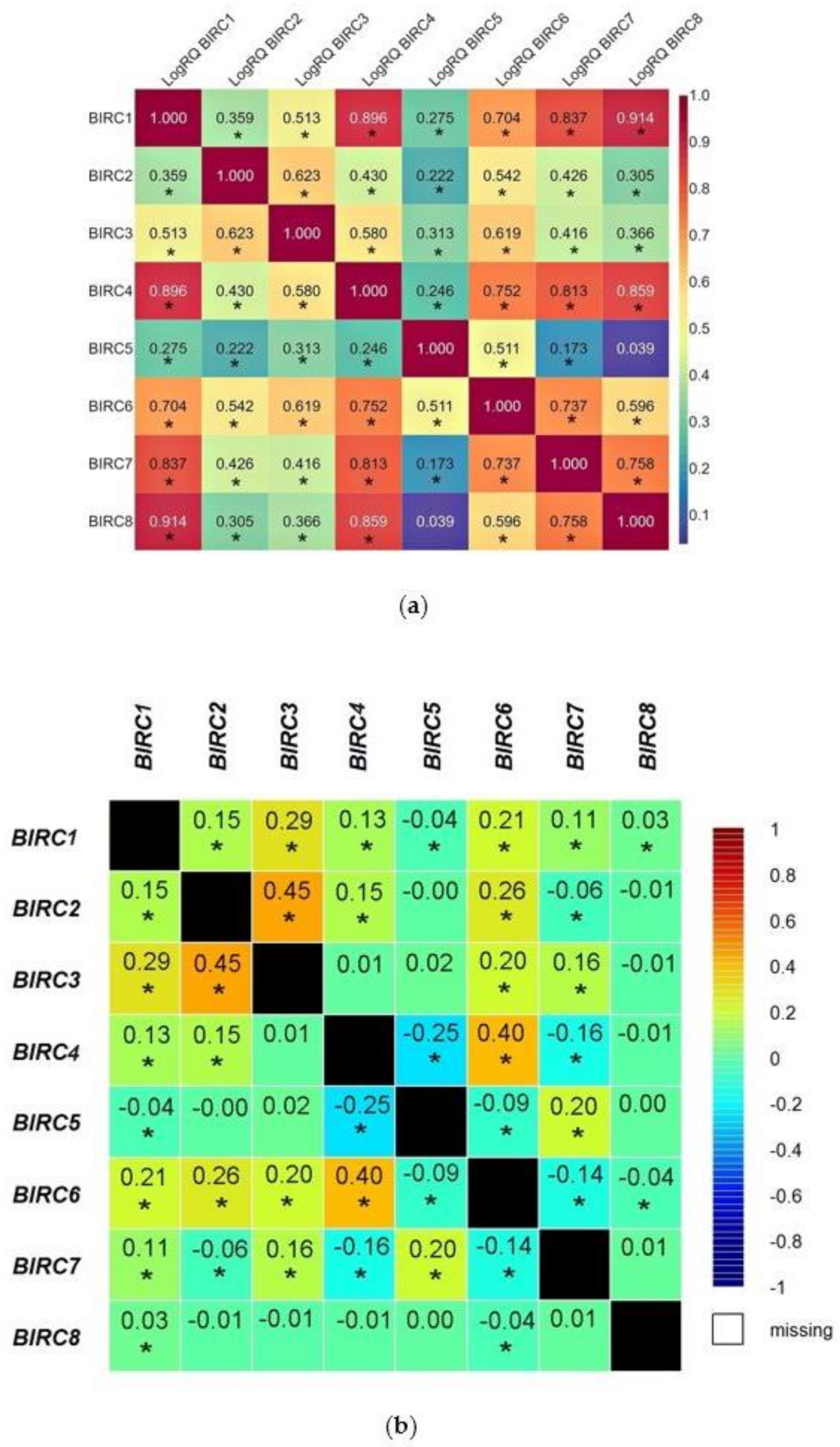
2.2. The Relationships between the Expression Levels of the Examined Genes in TNBC. Comparison of the Obtained Results with the Bioinformatic Analysis of Data Obtained from TCGA
2.3. The Analysis of the Dependence between Gene Expression and Clinical Data. Comparison of the Obtained Results with the Bioinformatic Analysis of Data Obtained from TCGA
2.3.1. Age
2.3.2. Lymphovascular Invasion
2.3.3. Cancer Cell Invasion of the Fat Tissue
2.3.4. Tumor Size
2.3.5. The Scarff-Bloom and Richardson (SBR) Grading System
2.4. Effect of the Expression Values of the BIRC Family Genes on Breast Cancer Patients Overall Survival
3. Discussion
4. Materials and Methods
4.1. Characteristics of the Study Group
4.2. Preparation of the Material for RNA Isolation
4.3. Tissue Homogenization
4.4. RNA Isolation and cDNA Reverse Transcription
4.5. Gene Expression Analysis
4.6. Methods of Statistical Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; Dandona, L.; et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [CrossRef] [PubMed]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef]
- Shao, Z.; Ma, X.; Zhang, Y.; Sun, Y.; Lv, W.; He, K.; Xia, R.; Wang, P.; Gao, X. CPNE1 predicts poor prognosis and promotes tumorigenesis and radioresistance via the AKT singling pathway in triple-negative breast cancer. Mol. Carcinog. 2020, 59, 533–544. [Google Scholar] [CrossRef]
- Carey, L.; Winer, E.; Viale, G.; Cameron, D.; Gianni, L. Triple-negative breast cancer: Disease entity or title of convenience? Nat. Rev. Clin. Oncol. 2010, 7, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Haffty, B.G.; Yang, Q.; Reiss, M.; Kearney, T.; Higgins, S.A.; Weidhaas, J.; Harris, L.; Hait, W.; Toppmeyer, D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J. Clin. Oncol. 2006, 24, 5652–5657. [Google Scholar] [CrossRef]
- Mehanna, J.; Haddad, F.G.H.; Eid, R.; Lambertini, M.; Kourie, H.R. Triple-negative breast cancer: Current perspective on the evolving therapeutic landscape. Int. J. Womens Health 2019, 11, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, F.; Yang, T.; Sheng, Y.; Zhong, T.; Chen, Y. Differential drug resistance acquisition to doxorubicin and paclitaxel in breast cancer cells. Cancer Cell Int. 2014, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Lehman, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, B.; Rafiee-Pour, H.A.; Khosravi, M.R.; Kefayat, A.; Baradaran, A.; Amjadi, E.; Goli, P. Prolactin receptor expression as a novel prognostic biomarker for triple negative breast cancer patients. Ann. Diagn. Pathol. 2020, 46, 151507. [Google Scholar] [CrossRef]
- Khosravi-Shahi, P.; Cabezón-Gutiérrez, L.; Custodio-Cabello, S. Metastatic triple negative breast cancer: Optimizing treatment options, new and emerging targeted therapies. Asia Pac. J. Clin. Oncol. 2018, 14, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Pietenpol, J.A. Clinical implications of molecular heterogeneity in triple negative breast cancer. Breast 2015, 24, 36–40. [Google Scholar] [CrossRef]
- Low, C.G.; Luk, I.S.; Lin, D.; Fazli, L.; Yang, K.; Xu, Y.; Gleave, M.; Gout, P.W.; Wang, Y. BIRC6 protein, an inhibitor of apoptosis: Role in survival of human prostate cancer cells. PLoS ONE 2013, 8, e55837. [Google Scholar] [CrossRef] [PubMed]
- LaCasse, E.C.; Baird, S.; Korneluk, R.G.; MacKenzie, A.E. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 1998, 17, 3247–3259. [Google Scholar] [CrossRef]
- Saleem, M.; Qadir, M.I.; Perveen, N.; Ahmad, B.; Saleem, U.; Irshad, T.; Ahmad, B. Inhibitors of Apoptotic Proteins: New Targets for Anticancer Therapy. Chem. Biol. Drug Des. 2013, 82, 243–251. [Google Scholar] [CrossRef]
- Liang, J.; Zhao, W.; Tong, P.; Li, P.; Zhao, Y.; Li, H.; Liang, J. Comprehensive molecular characterization of inhibitors of apoptosis proteins (IAPs) for therapeutic targeting in cancer. BMC Med. Genom. 2020, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Meier, P. To fight or die—inhibitor of apoptosis proteins at the crossroad of innate immunity and death. Curr. Opin. Cell Biol. 2010, 22, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Gyrd-Hansen, M.; Meier, P. IAPs: From caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat. Rev. Cancer 2010, 10, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Takahashi, R.; Salvesen, G.S.; Reed, J.C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 1997, 388, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Estornes, Y.; Bertrand, M. IAPs, regulators of innate immunity and inflammation. Semin. Cell Dev. Biol. 2015, 39, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Dueber, E.C.; Schoeffler, A.J.; Lingel, A.; Elliott, J.M.; Fedorova, A.V.; Giannetti, A.M.; Zobel, K.; Maurer, B.; Varfolomeev, E.; Wu, P.; et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 2011, 334, 376–380. [Google Scholar] [CrossRef]
- Middleton, A.J.; Budhidarmo, R.; Day, C.L. Use of E2~ Ubiquitin Conjugates for the Characterization of Ubiquitin Transfer by RING E3 Ligases Such as the Inhibitor of Apoptosis Proteins. Methods Enzymol. 2014, 545, 243–263. [Google Scholar] [CrossRef] [PubMed]
- Silke, J.; Vucic, D. IAP family of cell death and signaling regulators. Methods Enzymol. 2014, 545, 35–65. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.P.; Smith, L.; Smith, J.B. cIAP1 cooperatively inhibits procaspase-3 activation by the caspase-9 apoptosome. J. Biol. Chem. 2010, 285, 30061–30068. [Google Scholar] [CrossRef]
- McComb, S.; Cheung, H.H.; Korneluk, R.G.; Wang, S.; Krishnan, L.; Sad, S. cIAP1 and cIAP2 limit macrophage necrop-tosis by inhibiting Rip1 and Rip3 activation. Cell Death Differ. 2012, 19, 1791–1801. [Google Scholar] [CrossRef]
- Tenev, T.; Bianchi, K.; Darding, M.; Broemer, M.; Langlais, C.; Wallberg, F.; Zachariou, A.; Lopez, J.; MacFarlane, M.; Cain, K.; et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell 2011, 43, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, R.B.; Gyrd-Hansen, M. Inhibitor of apoptosis (IAP) proteins in regulation of inflammation and innate im-munity. Discov. Med. 2011, 11, 221–231. [Google Scholar]
- Weber, A.; Kirejczyk, Z.; Besch, R.; Potthoff, S.; Leverkus, M.; Hacker, G. Proapoptoticsignalling through Toll-like receptor-3 involves TRIF-dependent activation of caspase-8 and is under the control of inhibitor of apoptosis proteins in melanoma cells. Cell Death Differ. 2010, 17, 942–951. [Google Scholar] [CrossRef]
- Fukuda, K.; Sakakura, C.; Miyagawa, K.; Kuriu, Y.; Kin, S.; Nakase, Y.; Hagiwara, A.; Mitsufuji, S.; Okazaki, Y.; Hayashizaki, Y.; et al. Differential gene expression profiles of radioresistantoesophageal cancer cell lines established by continuous fractionated irradiation. Br. J. Cancer 2004, 91, 1543–1550. [Google Scholar] [CrossRef]
- Hingorani, P.; Dickman, P.; Garcia-Filion, P.; White-Collins, A.; Kolb, E.A.; Azorsa, D.O. BIRC5 expression is a poor prognostic marker in Ewing sarcoma. Pediatr. Blood Cancer 2013, 60, 35–40. [Google Scholar] [CrossRef]
- Tamm, I.; Kornblau, S.M.; Segall, H.; Krajewski, S.; Welsh, K.; Kitada, S.; Scudiero, D.A.; Tudor, G.; Qui, Y.H.; Monks, A.; et al. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin. Cancer Res. 2000, 6, 1796–1803. [Google Scholar]
- Aird, K.M.; Ghanayem, R.B.; Peplinski, S.; Lyerly, H.K.; Devi, G.R. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol. Cancer Ther. 2010, 9, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Small, S.; Keerthivasan, G.; Huang, Z.; Gurbuxani, S.; Crispino, J.D. Overexpression of survivin initiates hematologic malignancies in vivo. Leukemia 2010, 24, 1920–1926. [Google Scholar] [CrossRef]
- Falkenhorst, J.; Grunewald, S.; Mühlenberg, T.; Marino-Enriquez, A.; Reis, A.; Corless, C.; Heinrich, M.; Treckmann, J.; Erik Podleska, L.; Schuler, M.; et al. Inhibitor of Apoptosis Proteins (IAPs) are commonly dysregulated in GIST and can be pharmacologically targeted to enhance the pro-apoptotic activity of imatinib. Oncotarget 2016, 7, 41390–41403. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Iwamoto, S.; Gon, G.; Nohara, T.; Iwamoto, M.; Tanigawa, N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin. Cancer Res. 2000, 6, 127–134. [Google Scholar] [PubMed]
- Hinnis, A.R.; Luckett, J.C.; Walker, R.A. Survivin is an independent predictor of short-term survival in poor prognostic breast cancer patients. Br. J. Cancer 2007, 96, 639–645. [Google Scholar] [CrossRef]
- Ryan, B.M.; Konecny, G.E.; Kahlert, S.; Wang, H.J.; Untch, M.; Meng, G.; Pegram, M.D.; Podratz, K.C.; Crown, J.; Slamon, D.J.; et al. Survivin expression in breast cancer predicts clinical outcome and is associated with HER2, VEGF, urokinase plasminogen activator and PAI-1. Ann. Oncol. 2006, 17, 597–604. [Google Scholar] [CrossRef]
- Yamashita, S.; Masuda, Y.; Kurizaki, T.; Haga, Y.; Murayama, T.; Ikei, S.; Kamei, M.; Takeno, S.; Kawahara, K. Survivin expression predicts early recurrence in early-stage breast cancer. Anticancer Res. 2007, 27, 2803–2808. [Google Scholar] [PubMed]
- Masuda, P.N.; Sweep, F.C.; Wiegerinck, E.T.; Tjan-Heijnen, V.C.; Manders, P.; Beex, L.V.A.M.; de Kok, J. Survivin is an independent prognostic marker for risk stratification of breast cancer patients. Clin. Chem. 2004, 50, 1986–1993. [Google Scholar] [CrossRef][Green Version]
- Bartke, T.; Pohl, C.; Pyrowolakis, G.; Jentsch, S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell 2004, 14, 801–811. [Google Scholar] [CrossRef]
- Hao, Y.; Sekine, K.; Kawabata, A.; Nakamura, H.; Ishioka, T.; Ohata, H.; Katayama, R.; Hashimoto, C.; Zhang, X.; Noda, T.; et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat. Cell Biol. 2004, 6, 849–860. [Google Scholar] [CrossRef]
- Pohl, C.; Jentsch, S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell 2008, 132, 832–845. [Google Scholar] [CrossRef]
- Qiu, X.B.; Goldberg, A.L. The membrane-associated inhibitor of apoptosis protein, BRUCE/Apollon, antagonizes both the precursor and mature forms of Smac and caspase-9. J. Biol. Chem. 2005, 280, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.; Hao, Y.; Suzuki, Y.; Takahashi, R.; Tsuruo, T.; Naito, M. HtrA2 cleaves Apollon and induces cell death by IAP-binding motif in Apollon-deficient cells. Biochem. Biophys. Res. Commun. 2005, 330, 279–285. [Google Scholar] [CrossRef]
- Lamers, F.; Schild, L.; Koster, J.; Speleman, F.; Ora, I.; Westerhout, E.M.; van Sluis, P.; Versteeg, R.; Caron, H.N.; Molenaar, J.J. Identification of BIRC6 as a novel intervention target for neuroblastoma therapy. BMC Cancer 2012, 12, 285. [Google Scholar] [CrossRef]
- Tassi, E.; Zanon, M.; Vegetti, C.; Molla, A.; Bersani, I.; Perotti, V.; Pennati, M.; Zaffaroni, N.; Milella, M.; Ferrone, S.; et al. Role of Apollon in human melanoma resistance to antitumor agents that activate the intrinsic or the extrinsic apoptosis pathways. Clin. Cancer Res. 2012, 18, 3316–3327. [Google Scholar] [CrossRef]
- Dong, X.; Lin, D.; Low, C.; Vucic, E.A.; English, J.C.; Yee, J.; Murray, N.; Lam, W.L.; Ling, V.; Lam, S.; et al. Elevated expression of BIRC6 protein in non-small-cell lung cancers is associated with cancer recurrence and chemoresistance. J. Thorac. Oncol. 2013, 8, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Crnkovic-Mertens, I.; Muley, T.; Meister, M.; Hartenstein, B.; Semzow, J.; Butz, K.; Hoppe-Seyler, F. The anti-apoptotic livin gene is an important determinant for the apoptotic resistance of non-small cell lung cancer cells. Lung Cancer 2006, 54, 135–142. [Google Scholar] [CrossRef]
- Ashhab, Y.; Alian, A.; Polliack, A.; Panet, A.; Ben Yehuda, D. Two splicing variants of a new inhibitor of apoptosis gene with different biological properties and tissue distribution pattern. FEBS Lett. 2001, 495, 56–60. [Google Scholar] [CrossRef]
- Martinez-Ruiz, G.; Maldonado, V.; Ceballos-Cancino, G.; Grajeda, J.P.; Melendez-Zajgla, J. Role of Smac/DIABLO in cancer progression. J. Exp. Clin. Cancer Res. 2008, 27, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yin, X.; Xinrong, L.; Luo, X.; Xinliang, S.; Li, H.-Y.; Su, X.; Wang, X.-Y.; Chen, L.; Zheng, K.; et al. Livin promotes progression of breast cancer through induction of epithelial–mesenchymal transition and activation of AKT signaling. Cell. Signal. 2013, 25, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, L.; Feng, F.; Zheng, K.; Li, Y.; Wang, T.; Ren, G. Livin participates in resistance to trastuzumab therapy for breast cancer through ERK1/2 and AKT pathways and promotes EMT-like phenotype. RSC Adv. 2018, 50, 28588–28601. [Google Scholar] [CrossRef]
- Myung, D.S.; Park, Y.L.; Chung, C.Y.; Park, H.C.; Kim, J.S.; Cho, S.B.; Lee, W.S.; Lee, K.H.; Lee, J.H.; Joo, Y.E. Expression of Livin in colorectal cancer and its relationship to tumor cell behavior and prognosis. PLoS ONE 2013, 8, e73262. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cho, S.B.; Lee, W.S.; Park, Y.L.; Kim, N.; Oh, H.H.; Kim, M.; Oak, C.Y.; Chung, C.Y.; Park, H.C.; Kim, J.S.; et al. Livin is associated with the invasive and oncogenic phenotypes of human hepatocellular carcinoma cells. Hepatol. Res. 2015, 45, 448–457. [Google Scholar] [CrossRef]
- Hariu, H.; Hirohashi, Y.; Torigoe, T.; Asanuma, H.; Hariu, M.; Tamura, Y.; Aketa, K.; Nabeta, C.; Nakanishi, K.; Kamiguchi, K.; et al. Aberrant expression and potency as a cancer immunotherapy target of inhibitor of apoptosis protein family, Livin/ML-IAP in lung cancer. Clin. Cancer Res. 2005, 11, 1000–1009. [Google Scholar] [CrossRef]
- Kim, D.K.; Alvarado, C.S.; Abramowsky, C.R.; Gu, L.; Zhou, M.; Soe, M.M.; Sullivan, K.; George, B.; Schemankewitz, E.; Findley, H.W. Expression of inhibitor-of-apoptosis protein (IAP) livin by neuroblastoma cells: Correlation with prognostic factors and outcome. Pediatric Dev. Pathol. 2005, 8, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Altieri, B.; Sbiera, S.; Della Casa, S.; Weigand, I.; Wild, V.; Steinhauer, S.; Fadda, G.; Kocot, A.; Bekteshi, M.; Mambretti, E.M.; et al. Livin/BIRC7 expression as malignancy marker in adrenocortical tumors. Oncotarget 2017, 8, 9323–9338. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Cherniack, A.D.; Dewal, N.; Moffitt, R.A.; Danilova, L.; Murray, B.A.; Lerario, A.M.; Else, T.; Knijnenburg, T.A.; Ciriello, G.; et al. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer Cell 2016, 29, 723–736. [Google Scholar] [CrossRef]
- de Almagro, M.C.; Vucic, D. The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp. Oncol. 2012, 34, 200–211. [Google Scholar]
- Dubrez-Daloz, L.; Dupoux, A.; Cartier, J. IAPs—more than just inhibitors of apoptosis proteins. Cell Cycle 2008, 7, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Lau, R.; Pratt, M.A. The opposing roles of cellular inhibitor of apoptosis proteins in cancer. ISRN Oncol. 2012, 2012, 928120. [Google Scholar] [CrossRef]
- Wu, Q.; Li, B.; Li, Z.; Li, J.; Sun, S.; Sun, S. Cancer-associated adipocytes: Key players in breast cancer progression. J. Hematol. Oncol. 2019, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Paré, M.; Darini, C.Y.; Yao, X.; Chignon-Sicard, B.; Rekima, S.; Lachambre, S.; Virolle, V.; Aguilar-Mahecha, A.; Basik, M.; Dani, C.; et al. Breast cancer mammospheres secrete Adrenomedullin to induce lipolysis and browning of adjacent adipocytes. BMC Cancer 2020, 20, 784. [Google Scholar] [CrossRef]
- Dai, J.B.; Zhu, B.; Lin, W.J.; Gao, H.Y.; Dai, H.; Zheng, L.; Shi, W.H.; Chen, W.X. Identification of prognostic significance of BIRC5 in breast cancer using integrative bioinformatics analysis. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, X.; Shen, C.; Shi, Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. 2012, 31, 58. [Google Scholar] [CrossRef]
- Han, B.; Zhang, H.; Zhu, Y.; Han, X.; Wang, Z.; Gao, Z.; Yuan, Y.; Tian, R.; Zhanf, F.; Niu, R. Subtype-specific risk models for accurately predicting the prognosis of breast cancer using differentially expressed autophagy-related genes. AGING 2020, 12, 13318–13337. [Google Scholar] [CrossRef] [PubMed]
- Colak, D.; Nofal, A.; AlBakheet, A.; Nirmal, M.; Jeprel, H.; Eldali, A.; Taher, A.T.; Tulbah, A.; Ajarim, D.; Al Malik, O.; et al. Age-Specific Gene Expression Signatures for Breast Tumors and Cross-Species Conserved Potential Cancer Progression Markers in Young Women. PLoS ONE 2013, 8, e63204. [Google Scholar] [CrossRef]
- Dorman, S.N.; Baranova, K.; Knoll, J.H.; Urquhart, B.L.; Mariani, G.; Carcangiu, M.L.; Rogan, P.K. Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Mol. Oncol. 2016, 10, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Faversani, A.; Vaira, V.; Moro, G.P.; Tosi, D.; Lopergolo, A.; Schultz, D.C.; Rivadeneira, D.; Altieri, D.C.; Bosari, S. Survivin family proteins as novel molecular determinants of doxorubicin resistance in organotypic human breast tumors. Breast Cancer Res. 2014, 16, R55. [Google Scholar] [CrossRef]
- Rexhepaj, E.; Jirstrom, K.; O’Connor, D.P.; O’Brien, S.L.; Landberg, G.; Duffy, M.J.; Brennan, D.J.; Gallagher, W.M. Validation of cytoplasmic-to-nuclear ratio of survivin as an indicator of improved prognosis in breast cancer. BMC Cancer 2010, 10, 639. [Google Scholar] [CrossRef]
- Brennan, D.J.; Rexhepaj, E.; O’Brien, S.L.; McSherry, E.; O’Connor, D.P.; Fagan, A.; Culhane, A.C.; Higgins, D.G.; Jirstrom, K.; Millikan, R.C.; et al. Altered cytoplasmic-to-nuclear ratio of survivin is a prognostic indicator in breast cancer. Clin. Cancer Res. 2008, 14, 2681–2689. [Google Scholar] [CrossRef]
- Kennedy, S.M.; O’Driscoll, L.; Purcell, R.; Fitz-Simons, N.; McDermott, E.W.; Hill, A.D.; O’Higgins, N.J.; Parkinson, M.; Linehan, R.; Clynes, M. Prognostic importance of survivin in breast cancer. Br. J. Cancer 2003, 88, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Fu, F.; Lv, J.; Wang, M.; Li, Y.; Zhang, J.; Wang, C. Identification of potential key genes for HER-2 positive breast cancer based on bioinformatics analysis. Medicine 2020, 99, e18445. [Google Scholar] [CrossRef] [PubMed]
- Hamy, A.S.; Bieche, I.; Lehmann-Che, J.; Scott, V.; Bertheau, P.; Guinebretière, J.M.; Matthieu, M.C.; Sigal-Zafrani, B.; Tembo, O.; Marty, M.; et al. BIRC5 (survivin): A pejorative prognostic marker in stage II/III breast cancer with no response to neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2016, 159, 499–511. [Google Scholar] [CrossRef]
- Immunohistochemistry Standard Operating Protocol. 2011. Available online: https://edrn.nci.nih.gov/resources/standard-operating-procedures/assays/immunohistochemistry/immunoperoxidase-staining/sop-ihc.pdf/view (accessed on 4 December 2020).
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. The value of histological grade for 22.616 cases of breast cancer. The basis for a prognostic index. Cancer 1991, 8, 2142–2149. [Google Scholar]
- Lakhani, S.R.; Ellis, I.O.; Schnitt, S.; Tan, P.H.; van de Vijver, M. WHO Classification of Tumours of the Breast, 4th ed.; IARC: Lyon, France, 2012. [Google Scholar]
- Wittekind, C.; Meyer, H.J.; UICC (Hrsg). TNM: Classification of Malignant Tumors, 7th ed.; Wiley-VCH: Weinheim, Germany, 2010; Part VII; pp. 347–376. [Google Scholar]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinum thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 62, 156–159. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Jezequel, P.; Campone, M.; Gouraud, W.; Guérin-Charbonnel, C.; Leux, C.; Ricolleau, G.; Campion, L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2012, 131, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Jezequel, P.; Frenel, J.S.; Campion, L.; Guérin-Charbonnel, C.; Gouraud, W.; Ricolleau, G.; Campone, M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database 2013, bas060. [Google Scholar] [CrossRef] [PubMed]










| Gene | N Comparisons | Mean [logRQ] | SD [logRQ] | Median [logRQ] |
|---|---|---|---|---|
| BIRC1 | 740 | −0.431581 | 1.218730 | −0.386159 |
| BIRC2 | 690 | 0.009995 | 0.800377 | 0.005800 |
| BIRC3 | 554 | 0.129047 | 0.884598 | 0.192146 |
| BIRC4 | 720 | −0.197498 | 1.030632 | −0.142367 |
| BIRC5 | 522 | 0.683783 | 0.937065 | 0.648409 |
| BIRC6 | 780 | −0.069403 | 0.675635 | −0.051101 |
| BIRC7 | 277 | 0.034917 | 1.212470 | 0.026533 |
| BIRC8 | 566 | −0.442143 | 1.437147 | −0.386170 |
| Gene | T1 | T2 | T3 | p for Multiple Comparison | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| BIRC1 [LogRQ] | 0.141 | 1.4372 | −0.439 | 1.1611 | −0.597 | 1.2071 | T1*T2 = 0.006674 T1*T3 = 0.000178 T2*T3 = 0.165 |
| BIRC2 [LogRQ] | −0.117 | 0.9221 | 0.115 | 0.78002 | −0.158 | 0.7652 | T1*T2 = 0.338 T1*T3 = 0.999 T2*T3 = 0.000211 |
| BIRC3 [LogRQ] | −0.083 | 0.7279 | 0.215 | 0.9147 | −0.012 | 0.8259 | T1*T2 = 0.025384 T1*T3 = 0.999 T2*T3 = 0.000439 |
| BIRC4 [LogRQ] | 0.171 | 1.2865 | −0.257 | 0.9891 | −0.199 | 0.9956 | T1*T2 = 0.041191 T1*T3 = 0.193 T2*T3 = 0.999 |
| BIRC5 [LogRQ] | 0.625 | 0.8068 | 0.725 | 0.9314 | 0.611 | 0.9938 | T1*T2 = 0.999 T1*T3 = 0.999 T2*T3 = 0.993 |
| BIRC6 [LogRQ] | 0.285 | 0.8075 | −0.099 | 0.6315 | −0.128 | 0.6810 | T1*T2 = 0.000648 T1*T3 = 0.000204 T2*T3 = 0.999 |
| BIRC7 [LogRQ] | 0.086 | 0.9425 | 0.026 | 1.1819 | 0.038 | 1.3324 | T1*T2 = 0.999 T1*T3 = 0.999 T2*T3 = 0.999 |
| BIRC8 [LogRQ] | 0.083 | 1.5263 | −0.4401 | 1.4244 | −0.619 | 1.3987 | T1*T2 = 0.049377 T1*T3 = 0.005931 T2*T3 = 0.478 |
| Gene | pN0 | pN1 | pN2 | pN3 | p for Multiple Comparison | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| BIRC1 [LogRQ] | −0.329 | 1.2563 | −0.8809 | 0.9536 | −0.041 | 1.2437 | −1.106 | 0.8559 | pN0*pN1 = 0.000049 pN0*pN2 = 0.060788 pN0*pN3 = 0.000077 pN1*pN2 = 0.000000 pN1*pN3 = 0.924 pN2*pN3 = 0.000000 |
| BIRC2 [LogRQ] | 0.153 | 0.8322 | −0.3005 | 0.8336 | 0.061 | 0.6076 | −0.3603 | 0.5536 | pN0*pN1 = 0.000000 pN0*pN2 = 0.999 pN0*pN3 = 0.000124 pN1*pN2 = 0.001374 pN1*pN3 = 0.999 pN2*pN3 = 0.005638 |
| BIRC3 [LogRQ] | 0.3202 | 0.9284 | −0.1604 | 0.7963 | 0.092 | 0.7659 | −0.364 | 0.6195 | pN0*pN1 = 0.000000 pN0*pN2 = 0.188 pN0*pN3 = 0.000000 pN1*pN2 = 0.020712 pN1*pN3 = 0.203258 pN2*pN3 = 0.000174 |
| BIRC4 [LogRQ] | −0.113 | 1.0853 | −0.558 | 0.8206 | 0.166 | 0.9291 | −0.878 | 0.7969 | pN0*pN1 = 0.000117 pN0*pN2 = 0.00258 pN0*pN3 = 0.000001 pN1*pN2 = 0.000000 pN1*pN3 = 0.120 pN2*pN3 = 0.000000 |
| BIRC5 [LogRQ] | 0.864 | 0.8848 | 0.35001 | 0.9853 | 0.6305 | 0.9558 | 0.375 | 0.8014 | pN0*pN1 = 0.000178 pN0*pN2 = 0.302 pN0*pN3 = 0.014555 pN1*pN2 = 0.598 pN1*pN3 = 0.999 pN2*pN3 = 0.770 |
| BIRC6 [LogRQ] | 0.027 | 0.6647 | −0.345 | 0.58303 | 0.1404 | 0.6459 | −0.643 | 0.5425 | pN0*pN1 = 0.000000 pN0*pN2 = 0.404 pN0*pN3 = 0.000000 pN1*pN2 = 0.000000 pN1*pN3 = 0.03835 pN2*pN3 = 0.000000 |
| BIRC7 [LogRQ] | −0.0006 | 1.20208 | −0.144 | 1.1144 | 0.5592 | 1.24708 | −0.673 | 0.93806 | pN0*pN1 = 0.999 pN0*pN2 = 0.045489 pN0*pN3 = 0.121 pN1*pN2 = 0.032137 pN1*pN3 = 0.450 pN2*pN3 = 0.001058 |
| BIRC8 [LogRQ] | −0.429 | 1.4845 | −0.646 | 1.2053 | −0.017 | 1.5038 | −1.218 | 1.0467 | pN0*pN1 = 0.999 pN0*pN2 = 0.081 pN0*pN3 = 0. 012351 pN1*pN2 = 0.016192 pN1*pN3 = 0.167 pN2*pN3 = 0.000121 |
| Gene | SBR1 | SBR2 | SBR3 | p for Multiple Comparison | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| BIRC1 [LogRQ] | −0.063 | 1.5324 | −0.955 | 0.9574 | −0.361 | 1.1927 | SBR1*SBR2 = 0.000035 SBR1*SBR3 = 0.655 SBR2*SBR3= 0.000002 |
| BIRC2 [LogRQ] | 0.553 | 0.5956 | −0.583 | 0.6861 | 0.0709 | 0.7717 | SBR1*SBR2 = 0.000000 SBR1*SBR3 = 0.000002 SBR2*SBR3 = 0.000000 |
| BIRC3 [LogRQ] | 0.642 | 0.8845 | −0.128 | 0.7207 | 0.1402 | 0.89809 | SBR1*SBR2 = 0.000000 SBR1*SBR3 = 0.000027 SBR2*SBR3 = 0.000460 |
| BIRC4 [LogRQ] | 0.397 | 1.1951 | −0.737 | 0.804 | −0.155 | 0.9969 | SBR1*SBR2 = 0.000000 SBR1*SBR3 = 0.001191 SBR2*SBR3 = 0.000000 |
| BIRC5 [LogRQ] | 0.815 | 0.8244 | 0.346 | 0.9573 | 0.745 | 0.9317 | SBR1*SBR2 = 0.019157 SBR1*SBR3 = 0.999 SBR2*SBR3 = 0.003377 |
| BIRC6 [LogRQ] | 0.2504 | 0.8205 | −0.359 | 0.5693 | −0.046 | 0.6519 | SBR1*SBR2 = 0.000000 SBR1*SBR3 = 0.020894 SBR2*SBR3 = 0.000004 |
| BIRC7 [LogRQ] | −0.279 | 0.9576 | −0.303 | 1.1185 | 0.147 | 1.2398 | SBR1*SBR2 = 0.999 SBR1*SBR3 = 0.366 SBR2*SBR3 = 0.100 |
| BIRC8 [LogRQ] | −0.061 | 1.5664 | −1.049 | 1.2303 | −0.357 | 1.4276 | SBR1*SBR2 = 0.000345 SBR1*SBR3 = 0.599 SBR2*SBR3 = 0.000115 |
| Characteristic | Patients with TNBC (n = 30) |
|---|---|
| Age at diagnosis ≤50 >50 | 8 (≈26.67%) 22 (≈73.33%) |
| Familial history of cancer Yes No | 0 (0%) 30 (100%) |
| Adjuvant chemotherapy Yes No | 0 (0%) 30 (100%) |
| Gender: Male Female | 0 (0%) 30 (100%) |
| Lymphovascular invasion Yes No | 10 (33.33%) 20 (66.67%) |
| Invasion of the fat tissue Yes No | 5 (≈16.67%) 25 (≈83.33%) |
| Tumor size T1 T2 T3 | 3 (10%) 19 (≈63.33%) 8 (≈26.67%) |
| Lymph nodes N0 N1 N2 N3 | 17 (≈56.67%) 6 (20%) 5 (≈16.67%) 2 (≈6.67%) |
| SBR grade SBR1 SBR2 SBR3 | 3 (10%) 5 (≈16.67%) 22 (≈73.33%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makuch-Kocka, A.; Kocki, J.; Brzozowska, A.; Bogucki, J.; Kołodziej, P.; Płachno, B.J.; Bogucka-Kocka, A. The BIRC Family Genes Expression in Patients with Triple Negative Breast Cancer. Int. J. Mol. Sci. 2021, 22, 1820. https://doi.org/10.3390/ijms22041820
Makuch-Kocka A, Kocki J, Brzozowska A, Bogucki J, Kołodziej P, Płachno BJ, Bogucka-Kocka A. The BIRC Family Genes Expression in Patients with Triple Negative Breast Cancer. International Journal of Molecular Sciences. 2021; 22(4):1820. https://doi.org/10.3390/ijms22041820
Chicago/Turabian StyleMakuch-Kocka, Anna, Janusz Kocki, Anna Brzozowska, Jacek Bogucki, Przemysław Kołodziej, Bartosz J. Płachno, and Anna Bogucka-Kocka. 2021. "The BIRC Family Genes Expression in Patients with Triple Negative Breast Cancer" International Journal of Molecular Sciences 22, no. 4: 1820. https://doi.org/10.3390/ijms22041820
APA StyleMakuch-Kocka, A., Kocki, J., Brzozowska, A., Bogucki, J., Kołodziej, P., Płachno, B. J., & Bogucka-Kocka, A. (2021). The BIRC Family Genes Expression in Patients with Triple Negative Breast Cancer. International Journal of Molecular Sciences, 22(4), 1820. https://doi.org/10.3390/ijms22041820







