Interspecific Variation in One-Carbon Metabolism within the Ovarian Follicle, Oocyte, and Preimplantation Embryo: Consequences for Epigenetic Programming of DNA Methylation
Abstract
:1. Introduction
2. Results
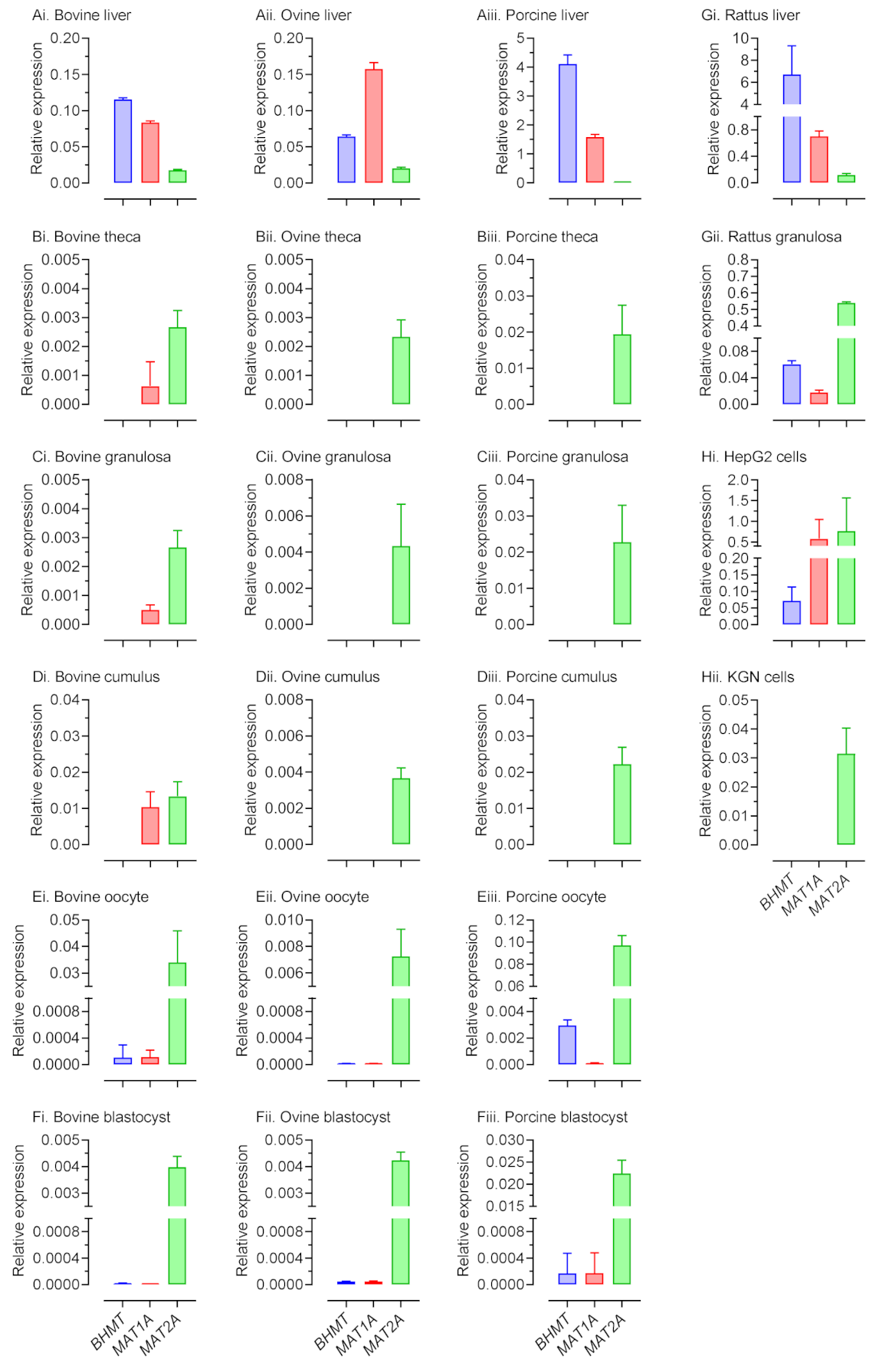
2.1. Expression of 1C metabolism Genes Differs between Species and Cell Type within Species
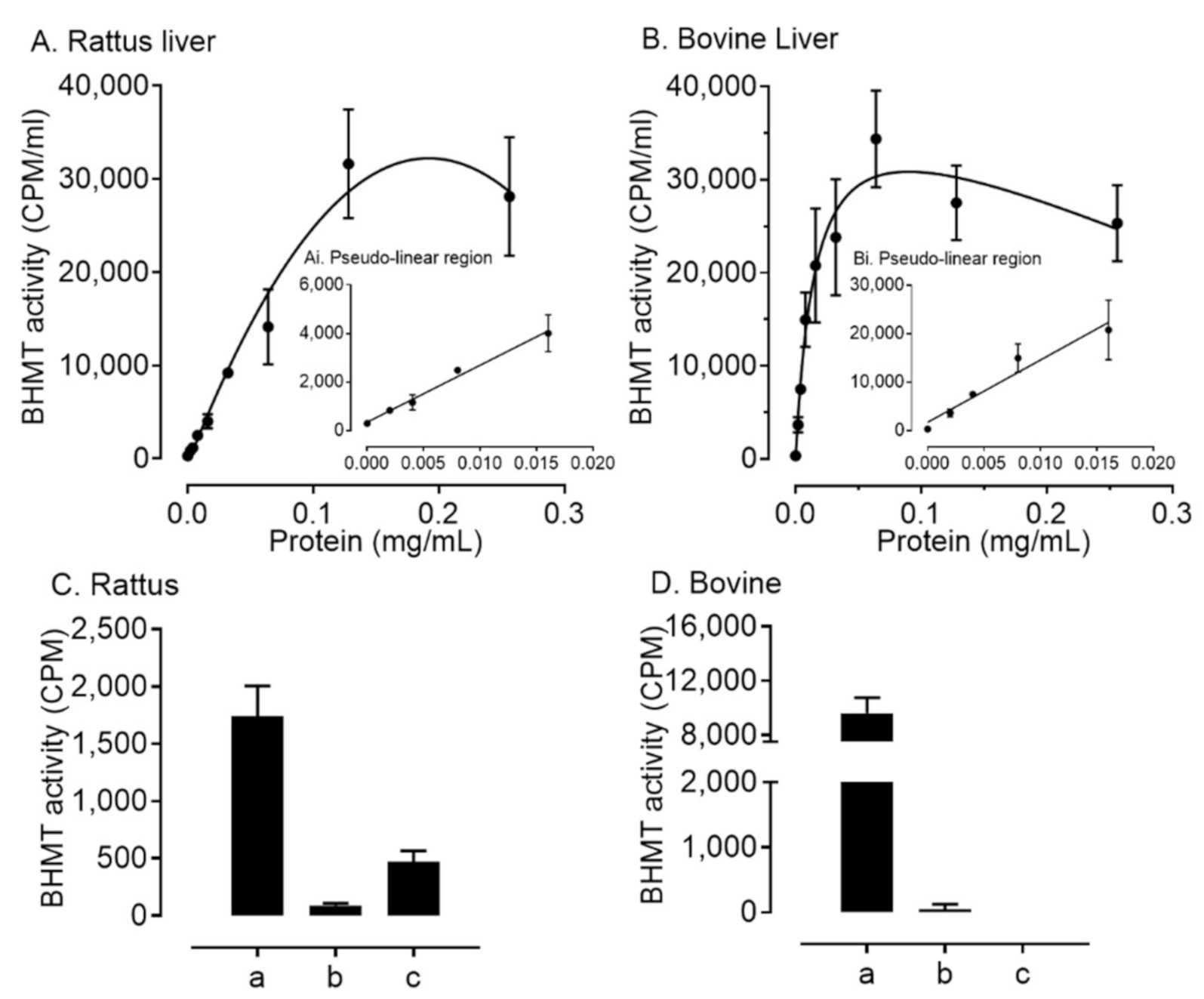
2.2. BHMT Expression and Activity within Ovarian Somatic Cells Differs between Species
2.3. MAT Expression and Activity is Sensitive to Physiological Levels of Methionine during Bovine Granulosa-Cell Culture
2.4. Bovine In Vitro Embryo Production (IVP) is Sensitive to Physiological Concentrations of Methionine
2.5. Methionine Concentration Affects Global DNA Methylation during Bovine IVP
2.6. Methionine Concentration Affects DNA Methylation of Specific Imprinted Genes
3. Discussion
3.1. Interspecific Variation in 1C Metabolism within the Ovarian Follicle and Preimplantation Embryo
3.2. Methionine Concentration during In Vitro Embryo Production Affects Global DNA Methylation
3.3. Methionine Concentration during In Vitro Embryo Production Affects Methylation of Imprinted Genes
3.4. Concluding Remarks
4. Materials and Methods
4.1. Tissue Collection and Primary Cell Isolation
4.2. Cell Culture
4.3. In Vitro Embryo Production (IVP), Immunostaining, and Sexing
4.4. Quantitative Real-Time PCR
4.5. Western Blots (BHMT)
4.6. Enzyme Activity Assays
4.7. LC-MS/MS
4.8. Reduced Representation Bisulfite Sequencing (RRBS)
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1C | one carbon |
| 5mC | 5-methylcytosine |
| ACTB | actin βeta |
| BHMT | betaine-homocysteine methyltransferase |
| BME | basal medium eagle |
| COC | cumulus–oocyte complex |
| CpG | cytosine-phosphate-guanine dinucleotide |
| FCS | fetal calf serum |
| GOI | gene of interest |
| Hcy | homocysteine |
| HepG2 | human liver cancer cell line |
| HPLC | high performance liquid chromatography |
| ICM | inner-cell mass |
| IVC | in vitro culture |
| IVF | in vitro fertilization |
| IVM | in vitro maturation |
| IVP | in vitro embryo production |
| KGN | human granulosa-like tumor cell line |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| LOS | large offspring syndrome |
| MAT1A | methionine adenosyltransferase 1A |
| MAT2A | methionine adenosyltransferase 2A |
| PBS | phosphate buffered saline |
| PVA | poly-vinyl alcohol |
| qPCR | quantitative real-time polymerase chain reaction |
| RRBS | reduced representation bisulfite sequencing |
| SAM | S-adenosylmethionine |
| SAH | S-adenosylhomocysteine |
| TE | trophectoderm |
References
- Xu, J.; Sinclair, K.D. One-carbon metabolism and epigenetic regulation of embryo development. Reprod. Fertil. Dev. 2015, 27, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Clare, C.E.; Brassington, A.H.; Kwong, W.Y.; Sinclair, K.D. One-Carbon Metabolism: Linking Nutritional Biochemistry to Epigenetic Programming of Long-Term Development. Annu. Rev. Anim. Biosci. 2019, 7, 263–287. [Google Scholar] [CrossRef] [PubMed]
- Steegers-Theunissen, R.P.; Twigt, J.; Pestinger, V.; Sinclair, K.D. The periconceptional period, reproduction and long-term health of offspring: The importance of one-carbon metabolism. Hum. Reprod. Update 2013, 19, 640–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, K.D.; Allegrucci, C.; Singh, R.; Gardner, D.S.; Sebastian, S.; Bispham, J.; Thurston, A.; Huntley, J.F.; Rees, W.D.; Maloney, C.A.; et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc. Natl. Acad. Sci. USA 2007, 104, 19351–19356. [Google Scholar] [CrossRef] [Green Version]
- Lambrot, R.; Xu, C.; Saint-Phar, S.; Chountalos, G.; Cohen, T.; Paquet, M.; Suderman, M.; Hallett, M.; Kimmins, S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat. Commun. 2013, 4, 2889. [Google Scholar] [CrossRef]
- Watkins, A.J.; Dias, I.; Tsuro, H.; Allen, D.; Emes, R.D.; Moreton, J.; Wilson, R.; Ingram, R.J.M.; Sinclair, K.D. Paternal diet programs offspring health through sperm- and seminal plasma-specific pathways in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 10064–10069. [Google Scholar] [CrossRef] [Green Version]
- Drake, A.J.; O’Shaughnessy, P.J.; Bhattacharya, S.; Monteiro, A.; Kerrigan, D.; Goetz, S.; Raab, A.; Rhind, S.M.; Sinclair, K.D.; Meharg, A.A.; et al. In utero exposure to cigarette chemicals induces sex-specific disruption of one-carbon metabolism and DNA methylation in the human fetal liver. BMC Med. 2015, 13, 18. [Google Scholar] [CrossRef] [Green Version]
- Lea, R.G.; Amezaga, M.R.; Loup, B.; Mandon-Pépin, B.; Stefansdottir, A.; Filis, P.; Kyle, C.; Zhang, Z.; Allen, C.; Purdie, L.; et al. The fetal ovary exhibits temporal sensitivity to a ‘real-life’ mixture of environmental chemicals. Sci. Rep. 2016, 6, 22279. [Google Scholar] [CrossRef] [Green Version]
- Haggarty, P.; McCallum, H.; McBain, H.; Andrews, K.; Duthie, S.; McNeill, G.; Templeton, A.; Haites, N.; Campbell, D.; Bhattacharya, S. Effect of B vitamins and genetics on success of in-vitro fertilisation: Prospective cohort study. Lancet 2006, 367, 1513–1519. [Google Scholar] [CrossRef]
- Rahimi, S.; Martel, J.; Karahan, G.; Angle, C.; Behan, N.A.; Chan, D.; MacFarlane, A.J.; Trasler, J.M. Moderate maternal folic acid supplementation ameliorates adverse embryonic and epigenetic outcomes associated with assisted reproduction in a mouse model. Hum. Reprod. 2019, 34, 851–862. [Google Scholar] [CrossRef]
- Hoek, J.; Steegers-Theunissen, R.P.M.; Willemsen, S.P.; Schoenmakers, S. Paternal Folate Status and Sperm Quality, Pregnancy Outcomes, and Epigenetics: A Systematic Review and Meta-Analysis. Mol. Nutr. Food Res. 2020, 64, e1900696. [Google Scholar] [CrossRef] [Green Version]
- Reed, M.C.; Nijhout, H.F.; Sparks, R.; Ulrich, C.M. A mathematical model of the methionine cycle. J. Theor. Biol. 2004, 226, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.C.; Gamble, M.V.; Hall, M.N.; Nijhout, H.F. Mathematical analysis of the regulation of competing methyltransferases. BMC Syst. Biol. 2015, 9, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwong, W.Y.; Adamiak, S.J.; Gwynn, A.; Singh, R.; Sinclair, K.D. Endogenous folates and single-carbon metabolism in the ovarian follicle, oocyte and pre-implantation embryo. Reproduction 2010, 139, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, K.D.; Singh, R. Modelling the developmental origins of health and disease in the early embryo. Theriogenology 2007, 67, 43–53. [Google Scholar] [CrossRef]
- Ikeda, S.; Namekawa, T.; Sugimoto, M.; Kume, S. Expression of methylation pathway enzymes in bovine oocytes and preimplantation embryos. J. Exp. Zool. A Ecol. Genet. Physiol. 2010, 313, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.B.; Kooistra, M.; Zhang, B.; Slow, S.; Fortier, A.L.; Garrow, T.A.; Lever, M.; Trasler, J.M.; Baltz, J.M. Betaine homocysteine methyltransferase is active in the mouse blastocyst and promotes inner cell mass development. J. Biol. Chem. 2012, 287, 33094–33103. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Denomme, M.M.; White, C.R.; Leung, K.Y.; Lee, M.B.; Greene, N.D.; Mann, M.R.; Trasler, J.M.; Baltz, J.M. Both the folate cycle and betaine-homocysteine methyltransferase contribute methyl groups for DNA methylation in mouse blastocysts. FASEB J. 2015, 29, 1069–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonilla, L.; Luchini, D.; Devillard, E.; Hansen, P.J. Methionine requirements for the preimplantation bovine embryo. J. Reprod. Dev. 2010, 56, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Shojaei Saadi, H.A.; Gagné, D.; Fournier, É.; Baldoceda Baldeon, L.M.; Sirard, M.A.; Robert, C. Responses of bovine early embryos to S-adenosyl methionine supplementation in culture. Epigenomics 2016, 8, 1039–1060. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Tomazou, E.M.; Brinkman, A.B.; Müller, F.; Simmer, F.; Gu, H.; Jäger, N.; Gnirke, A.; Stunnenberg, H.G.; Meissner, A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010, 28, 1106–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouse, S.D.; Nagarajan, R.O.; Costello, J.F. Genome-scale DNA methylation analysis. Epigenomics 2010, 2, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Xu, L.; Bickhart, D.M.; Abdel Hay, E.H.; Schroeder, S.G.; Connor, E.E.; Alexander, L.J.; Sonstegard, T.S.; Van Tassell, C.P.; Chen, H.; et al. Reduced representation bisulphite sequencing of ten bovine somatic tissues reveals DNA methylation patterns and their impacts on gene expression. BMC Genom. 2016, 17, 779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morbeck, D.E.; Paczkowski, M.; Fredrickson, J.R.; Krisher, R.L.; Hoff, H.S.; Baumann, N.A.; Moyer, T.; Matern, D. Composition of protein supplements used for human embryo culture. J. Assist. Reprod. Genet. 2014, 31, 1703–1711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarahomi, M.; Vaz, F.M.; van Straalen, J.P.; Schrauwen, F.A.P.; van Wely, M.; Hamer, G.; Repping, S.; Mastenbroek, S. The composition of human preimplantation embryo culture media and their stability during storage and culture. Hum. Reprod. 2019, 34, 1450–1461. [Google Scholar] [CrossRef]
- Steele, W.; Allegrucci, C.; Singh, R.; Lucas, E.; Priddle, H.; Denning, C.; Sinclair, K.; Young, L. Human embryonic stem cell methyl cycle enzyme expression: Modelling epigenetic programming in assisted reproduction? Reprod. Biomed. Online 2005, 10, 755–766. [Google Scholar] [CrossRef]
- Nishi, Y.; Yanase, T.; Mu, Y.; Oba, K.; Ichino, I.; Saito, M.; Nomura, M.; Mukasa, C.; Okabe, T.; Goto, K.; et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 2001, 142, 437–445. [Google Scholar] [CrossRef]
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 1979, 282, 615–616. [Google Scholar] [CrossRef] [PubMed]
- Wonnacott, K.E.; Kwong, W.Y.; Hughes, J.; Salter, A.M.; Lea, R.G.; Garnsworthy, P.C.; Sinclair, K.D. Dietary omega-3 and -6 polyunsaturated fatty acids affect the composition and development of sheep granulosa cells, oocytes and embryos. Reproduction 2010, 139, 57–69. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.; Kwong, W.Y.; Li, D.; Salter, A.M.; Lea, R.G.; Sinclair, K.D. Effects of omega-3 and -6 polyunsaturated fatty acids on ovine follicular cell steroidogenesis, embryo development and molecular markers of fatty acid metabolism. Reproduction 2011, 141, 105–118. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Li, J.; He, B.; Jia, Y.; Niu, Y.; Wang, C.; Zhao, R. Abnormally activated one-carbon metabolic pathway is associated with mtDNA hypermethylation and mitochondrial malfunction in the oocytes of polycystic gilt ovaries. Sci. Rep. 2016, 6, 19436. [Google Scholar] [CrossRef] [Green Version]
- Ramani, K.; Lu, S.C. Methionine adenosyltransferases in liver health and diseases. Liver Res. 2017, 1, 103–111. [Google Scholar] [CrossRef]
- Dalto, B.D.; Tsoi, S.; Audet, I.; Dyck, M.K.; Foxcroft, G.R.; Matte, J.J. Gene expression of porcine blastocysts from gilts fed organic or inorganic selenium and pyridoxine. Reproduction 2015, 149, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Young, L.E.; Fernandes, K.; McEvoy, T.G.; Butterwith, S.C.; Gutierrez, C.G.; Carolan, C.; Broadbent, P.J.; Robinson, J.J.; Wilmut, I.; Sinclair, K.D. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat. Genet. 2001, 27, 153–154. [Google Scholar] [CrossRef]
- Chen, Z.; Hagen, D.E.; Elsik, C.G.; Ji, T.; Morris, C.J.; Moon, L.E.; Rivera, R.M. Characterization of global loss of imprinting in fetal overgrowth syndrome induced by assisted reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 4618–4623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafontaine, S.; Labrecque, R.; Palomino, J.M.; Blondin, P.; Sirard, M.A. Specific imprinted genes demethylation in association with oocyte donor’s age and culture conditions in bovine embryos assessed at day 7 and 12 post insemination. Theriogenology 2020, 158, 321–330. [Google Scholar] [CrossRef]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Şenergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Farmer, W.T.; Farin, P.W.; Piedrahita, J.A.; Bischoff, S.R.; Farin, C.E. Expression of antisense of insulin-like growth factor-2 receptor RNA non-coding (AIRN) during early gestation in cattle. Anim. Reprod. Sci. 2013, 138, 64–73. [Google Scholar] [CrossRef]
- Shiraki, N.; Shiraki, Y.; Tsuyama, T.; Obata, F.; Miura, M.; Nagae, G.; Aburatani, H.; Kume, K.; Endo, F.; Kume, S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014, 19, 780–794. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, S.; Sugimoto, M.; Kume, S. Importance of methionine metabolism in morula-to-blastocyst transition in bovine preimplantation embryos. J. Reprod. Dev. 2012, 58, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, S.; Kawahara-Miki, R.; Iwata, H.; Sugimoto, M.; Kume, S. Role of methionine adenosyltransferase 2A in bovine preimplantation development and its associated genomic regions. Sci. Rep. 2017, 7, 3800. [Google Scholar] [CrossRef] [Green Version]
- Lane, M.; Gardner, D.K.; Hasler, M.J.; Hasler, J.F. Use of G1.2/G2.2 media for commercial bovine embryo culture: Equivalent development and pregnancy rates compared to co-culture. Theriogenology 2003, 60, 407–419. [Google Scholar] [CrossRef]
- Peñagaricano, F.; Souza, A.H.; Carvalho, P.D.; Driver, A.M.; Gambra, R.; Kropp, J.; Hackbart, K.S.; Luchini, D.; Shaver, R.D.; Wiltbank, M.C.; et al. Effect of maternal methionine supplementation on the transcriptome of bovine preimplantation embryos. PLoS ONE 2013, 8, e72302. [Google Scholar] [CrossRef] [Green Version]
- Arthur, J.S.; Elce, J.S.; Hegadorn, C.; Williams, K.; Greer, P.A. Disruption of the murine calpain small subunit gene, Capn4: Calpain is essential for embryonic development but not for cell growth and division. Mol. Cell. Biol. 2000, 20, 4474–4481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutt, P.; Croall, D.E.; Arthur, J.S.; Veyra, T.D.; Williams, K.; Elce, J.S.; Greer, P.A. m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 2006, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Takeo, S.; Goto, H.; Kuwayama, T.; Monji, Y.; Iwata, H. Effect of maternal age on the ratio of cleavage and mitochondrial DNA copy number in early developmental stage bovine embryos. J. Reprod. Dev. 2013, 59, 174–179. [Google Scholar] [CrossRef] [Green Version]
- Noli, L.; Khorsandi, S.E.; Pyle, A.; Giritharan, G.; Fogarty, N.; Capalbo, A.; Devito, L.; Jovanovic, V.M.; Khurana, P.; Rosa, H.; et al. Effects of thyroid hormone on mitochondria and metabolism of human preimplantation embryos. Stem Cells 2020, 38, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.E.; Hwang, K.C.; Sun, S.C.; Xu, Y.N.; Kim, N.H. Modulation of autophagy influences development and apoptosis in mouse embryos developing in vitro. Mol. Reprod. Dev. 2011, 78, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.Y.; Jones, A.; Yang, C.; Zhai, L.; Smith, A.D., 4th; Zhang, Z.; Chandrasekharan, M.B.; Sun, Z.W.; Renfrow, M.B.; Wang, Y.; et al. Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 2011, 286, 7190–7201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.J.; Conti, M.; Ramalho-Santos, M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 2013, 140, 3624–3634. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.M.; Wen, J.X.; Yuan, J.L.; Cang, M.; Liu, D.J. Knockdown of IGF-IR by siRNA injection during bovine preimplantation embryonic development. Cytotechnology 2012, 64, 165–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, K.; Mestdagh, P.; Lefever, S.; Van Poucke, M.; Van Zeveren, A.; Van Soom, A.; Vandesompele, J.; Peelman, L. Regulatory microRNA network identification in bovine blastocyst development. Stem Cells Dev. 2013, 22, 1907–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tribulo, P.; Leão, B.C.D.S.; Lehloenya, K.C.; Mingoti, G.Z.; Hansen, P.J. Consequences of endogenous and exogenous WNT signaling for development of the preimplantation bovine embryo. Biol. Reprod. 2017, 96, 1129–1141. [Google Scholar] [CrossRef]
- Negrón-Pérez, V.M.; Hansen, P.J. Role of yes-associated protein 1, angiomotin, and mitogen-activated kinase kinase 1/2 in development of the bovine blastocyst. Biol. Reprod. 2018, 98, 170–183. [Google Scholar] [CrossRef] [Green Version]
- Mok, H.J.; Shin, H.; Lee, J.W.; Lee, G.K.; Suh, C.S.; Kim, K.P.; Lim, H.J. Age-Associated Lipidome Changes in Metaphase II Mouse Oocytes. PLoS ONE 2016, 11, e0148577. [Google Scholar] [CrossRef]
- Hao, Y.; Zhang, Z.; Han, D.; Cao, Y.; Zhou, P.; Wei, Z.; Lv, M.; Chen, D. Gene expression profiling of human blastocysts from in vivo and ‘rescue IVM’ with or without melatonin treatment. Mol. Med. Rep. 2017, 16, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.E.; Ishida, M.; Demetriou, C.; Al-Olabi, L.; Leon, L.J.; Thomas, A.C.; Abu-Amero, S.; Frost, J.M.; Stafford, J.L.; Chaoqun, Y.; et al. The role and interaction of imprinted genes in human fetal growth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140074. [Google Scholar] [CrossRef] [Green Version]
- Argyraki, M.; Damdimopoulou, P.; Chatzimeletiou, K.; Grimbizis, G.F.; Tarlatzis, B.C.; Syrrou, M.; Lambropoulos, A. In-utero stress and mode of conception: Impact on regulation of imprinted genes, fetal development and future health. Hum. Reprod. Update 2019, 25, 777–801. [Google Scholar] [CrossRef]
- Wutz, A.; Smrzka, O.W.; Schweifer, N.; Schellander, K.; Wagner, E.F.; Barlow, D.P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 1997, 389, 745–749. [Google Scholar] [CrossRef]
- Thurston, A.; Taylor, J.; Gardner, J.; Sinclair, K.D.; Young, L.E. Monoallelic expression of nine imprinted genes in the sheep embryo occurs after the blastocyst stage. Reproduction 2008, 135, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Cruz, N.T.; Wilson, K.J.; Cooney, M.A.; Tecirlioglu, R.T.; Lagutina, I.; Galli, C.; Holland, M.K.; French, A.J. Putative imprinted gene expression in uniparental bovine embryo models. Reprod. Fertil. Dev. 2008, 20, 589–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Hagen, D.E.; Ji, T.; Elsik, C.G.; Rivera, R.M. Global misregulation of genes largely uncoupled to DNA methylome epimutations characterizes a congenital overgrowth syndrome. Sci. Rep. 2017, 7, 12667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregson, E.; Webb, R.; Sheldrick, E.L.; Campbell, B.K.; Mann, G.E.; Liddell, S.; Sinclair, K.D. Molecular determinants of a competent bovine corpus luteum: First- vs. final-wave dominant follicles. Reproduction 2016, 151, 563–575. [Google Scholar] [CrossRef] [Green Version]
- Zeebaree, B.K.; Kwong, W.Y.; Mann, G.E.; Gutierrez, C.G.; Sinclair, K.D. Physiological responses of cultured bovine granulosa cells to elevated temperatures under low and high oxygen in the presence of different concentrations of melatonin. Theriogenology 2018, 105, 107–114. [Google Scholar] [CrossRef]
- Goodhand, K.L.; Watt, R.G.; Staines, M.E.; Hutchinson, J.S.; Broadbent, P.J. In vivo oocyte recovery and in vitro embryo production from bovine donors aspirated at different frequencies or following FSH treatment. Theriogenology 1999, 51, 951–961. [Google Scholar] [CrossRef]
- Stringfellow, D.A.; Givens, M.D. Manual of the International Embryo Transfer Society (IETS), 4th ed.; IETS: Champaign, IL, USA, 2010. [Google Scholar]
- Nichols, J.; Silva, J.; Roode, M.; Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 2009, 136, 3215–3222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rattanasuk, S.; Parnpai, R.; Ketudat-Cairns, M. Multiplex polymerase chain reaction used for bovine embryo sex determination. J. Reprod. Dev. 2011, 57, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.C.; Huang, H.Y. Comparison of sulfur amino acid utilization for GSH synthesis between HepG2 cells and cultured rat hepatocytes. Biochem. Pharmacol. 1994, 47, 859–869. [Google Scholar] [CrossRef]
- Torres, L.; Avila, M.A.; Carretero, M.V.; Latasa, M.U.; Caballería, J.; López-Rodas, G.; Boukaba, A.; Lu, S.C.; Franco, L.; Mato, J.M. Liver-specific methionine adenosyltransferase MAT1A gene expression is associated with a specific pattern of promoter methylation and histone acetylation: Implications for MAT1A silencing during transformation. FASEB J. 2000, 14, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Kotb, M.; Kredich, N.M. S-Adenosylmethionine synthetase from human lymphocytes. Purification and characterization. J. Biol. Chem. 1985, 260, 3923–3930. [Google Scholar] [CrossRef]
- Xu, J.; Clare, C.E.; Brassington, A.H.; Sinclair, K.D.; Barrett, D.A. Comprehensive and quantitative profiling of B vitamins and related compounds in the mammalian liver. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2020, 1136, 121884. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Fiziev, P.; Yan, W.; Cokus, S.; Sun, X.; Zhang, M.Q.; Chen, P.Y.; Pellegrini, M. BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genom. 2013, 14, 774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Akalin, A.; Garrett-Bakelman, F.E.; Kormaksson, M.; Busuttil, J.; Zhang, L.; Khrebtukova, I.; Milne, T.A.; Huang, Y.; Biswas, D.; Hess, J.L.; et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012, 8, e1002781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akalin, A.; Franke, V.; Vlahoviček, K.; Mason, C.E.; Schübeler, D. Genomation: A toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 2015, 31, 1127–1129. [Google Scholar] [CrossRef]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef] [Green Version]
- Slieker, R.C.; Roost, M.S.; van Iperen, L.; Suchiman, H.E.; Tobi, E.W.; Carlotti, F.; de Koningm, E.J.; Slagboom, P.E.; Heijmans, B.T.; Chuva de Sousa Lopes, S.M. DNA Methylation Landscapes of Human Fetal Development. PLoS Genet. 2015, 11, e1005583. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Tutt, D.A.R.; Silvestri, G.; Serrano-Albal, M.; Simmons, R.J.; Kwong, W.Y.; Guven-Ates, G.; Canedo-Ribeiro, C.; Labrecque, R.; Blondin, P.; Handyside, A.H.; et al. Analysis of bovine blastocysts indicates ovarian stimulation does not induce chromosome errors, nor discordance between inner-cell mass and trophectoderm lineages. Theriogenology 2021, 161, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, K.D.; Young, L.E.; Wilmut, I.; McEvoy, T.G. In-utero overgrowth in ruminants following embryo culture: Lessons from mice and a warning to men. Hum. Reprod. 2000, 15 (Suppl. 5), 68–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Methionine (µM) | 10 | 50 | p-Value |
|---|---|---|---|
| Total oocytes matured | 1269 | 1289 | |
| A. Embryo development | 19 replicates | ||
| Cleaved of inseminated | 0.803 ± 0.0108 | 0.779 ± 0.0111 | - |
| Day 7 of: inseminated | 0.115 ± 0.0073 | 0.129 ± 0.0075 | - |
| :cleaved | 0.143 ± 0.0089 | 0.165 ± 0.0095 | - |
| Day 8 of: inseminated | 0.214 ± 0.0086 | 0.240 ± 0.0098 | 0.053 |
| :cleaved | 0.267 ± 0.0104 | 0.307 ± 0.0109 | 0.015 |
| Day 8 (IETS ‡ Stages 7–8) of | |||
| :total blastocysts | 0.570 ± 0.0167 | 0.627 ± 0.0157 | 0.017 |
| B. Day 8 blastomeres, n | 3 replicates | ||
| Total | 95.1 ± 1.80 | 101.7 ± 1.53 | 0.006 |
| Trophectoderm | 69.0 ± 1.60 | 72.9 ± 1.32 | 0.065 |
| Inner-cell mass | 28.0 ± 1.01 | 30.0 ± 0.85 | - |
| :Epiblast | 12.7 ± 0.66 | 14.3 ± 0.59 | 0.080 |
| :Hypoblast | 15.0 ± 0.74 | 15.2 ± 0.59 | - |
| C. Sex distribution † | 3 replicates | ||
| Males of Day 8 blastocysts | 0.577 ± 0.1159 | 0.611 ± 0.1157 | - |
| A. Cell lineage | B. Methionine | |||
|---|---|---|---|---|
| ICM | TE | 10 µM | 50 µM | |
| 10 v 50 µM | 10 v 50 µM | ICM v TE | ICM v TE | |
| CpG count | 9991 | 13,123 | 12,213 | 8088 |
| ↑ Methylation | 2449 (24.5) | 2361 (18.0) | 6671 (54.6) | 3365 (41.6) |
| ↓ Methylation | 7542 (75.5) | 10,762 (82.0) | 5542 (45.4) | 4723 (58.4) |
| Transcripts | 1576 | 1743 | 1773 | 1427 |
| Genes | 1573 | 1738 | 1768 | 1425 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clare, C.E.; Pestinger, V.; Kwong, W.Y.; Tutt, D.A.R.; Xu, J.; Byrne, H.M.; Barrett, D.A.; Emes, R.D.; Sinclair, K.D. Interspecific Variation in One-Carbon Metabolism within the Ovarian Follicle, Oocyte, and Preimplantation Embryo: Consequences for Epigenetic Programming of DNA Methylation. Int. J. Mol. Sci. 2021, 22, 1838. https://doi.org/10.3390/ijms22041838
Clare CE, Pestinger V, Kwong WY, Tutt DAR, Xu J, Byrne HM, Barrett DA, Emes RD, Sinclair KD. Interspecific Variation in One-Carbon Metabolism within the Ovarian Follicle, Oocyte, and Preimplantation Embryo: Consequences for Epigenetic Programming of DNA Methylation. International Journal of Molecular Sciences. 2021; 22(4):1838. https://doi.org/10.3390/ijms22041838
Chicago/Turabian StyleClare, Constance E., Valerie Pestinger, Wing Yee Kwong, Desmond A. R. Tutt, Juan Xu, Helen M. Byrne, David A. Barrett, Richard D. Emes, and Kevin D. Sinclair. 2021. "Interspecific Variation in One-Carbon Metabolism within the Ovarian Follicle, Oocyte, and Preimplantation Embryo: Consequences for Epigenetic Programming of DNA Methylation" International Journal of Molecular Sciences 22, no. 4: 1838. https://doi.org/10.3390/ijms22041838
APA StyleClare, C. E., Pestinger, V., Kwong, W. Y., Tutt, D. A. R., Xu, J., Byrne, H. M., Barrett, D. A., Emes, R. D., & Sinclair, K. D. (2021). Interspecific Variation in One-Carbon Metabolism within the Ovarian Follicle, Oocyte, and Preimplantation Embryo: Consequences for Epigenetic Programming of DNA Methylation. International Journal of Molecular Sciences, 22(4), 1838. https://doi.org/10.3390/ijms22041838






