The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Anchors the Peptidoglycan Layer by Spanning It with All Subunits
Abstract
:1. Introduction
2. Results
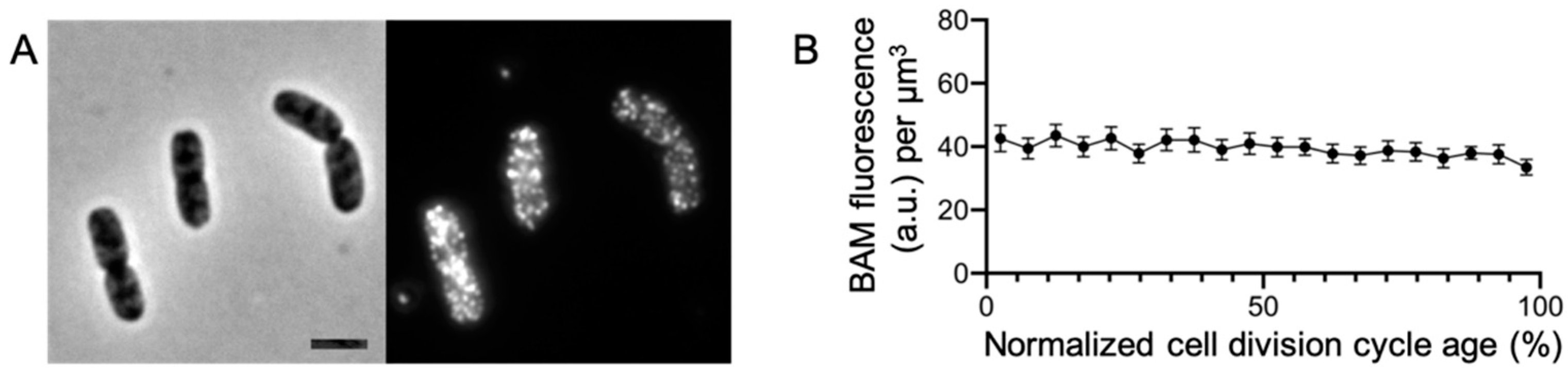
2.1. The BAM Complex Is Evenly Distributed in the Envelope
2.2. The Peptidoglycan Impairs BAM Immunolabelling
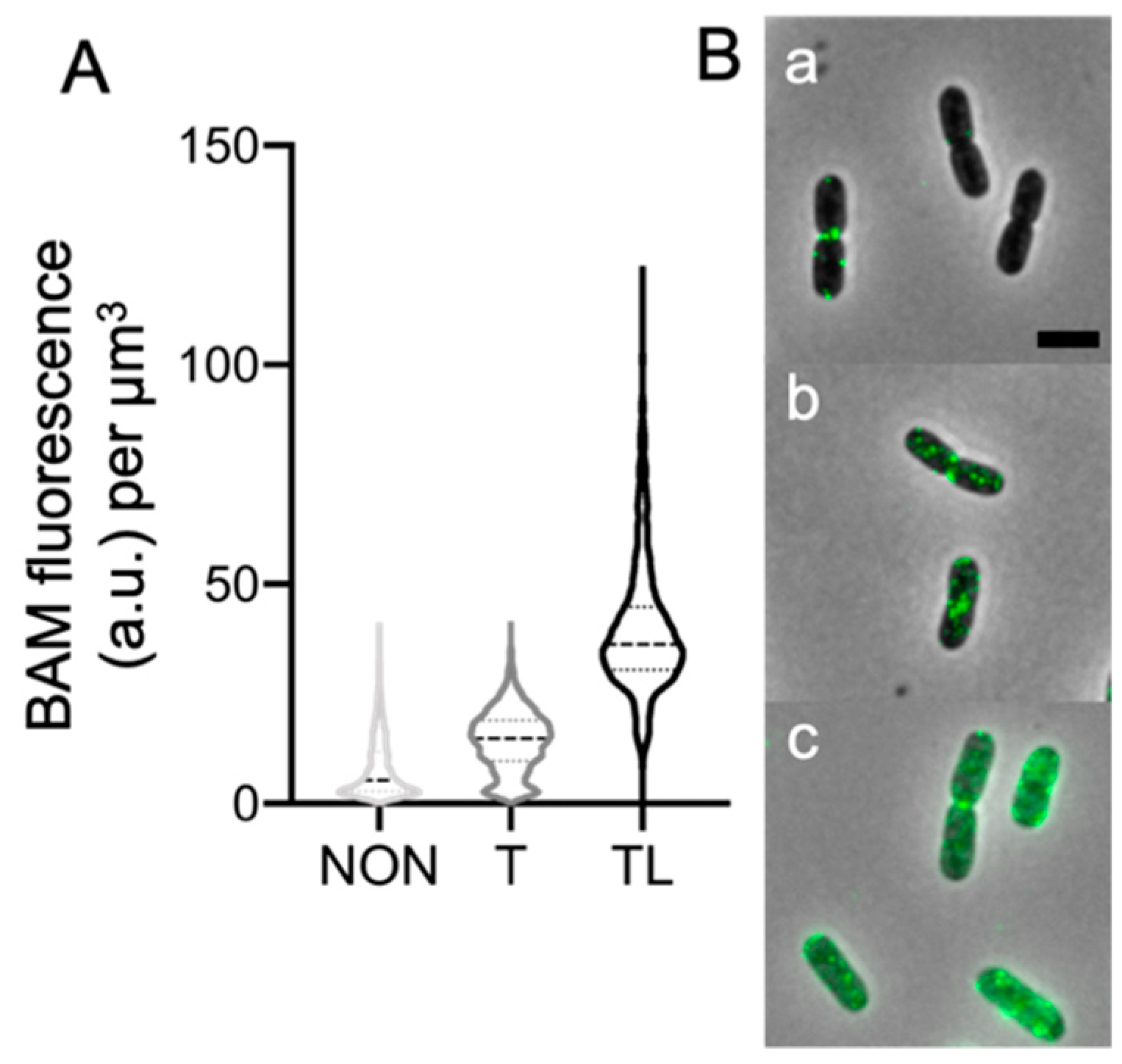
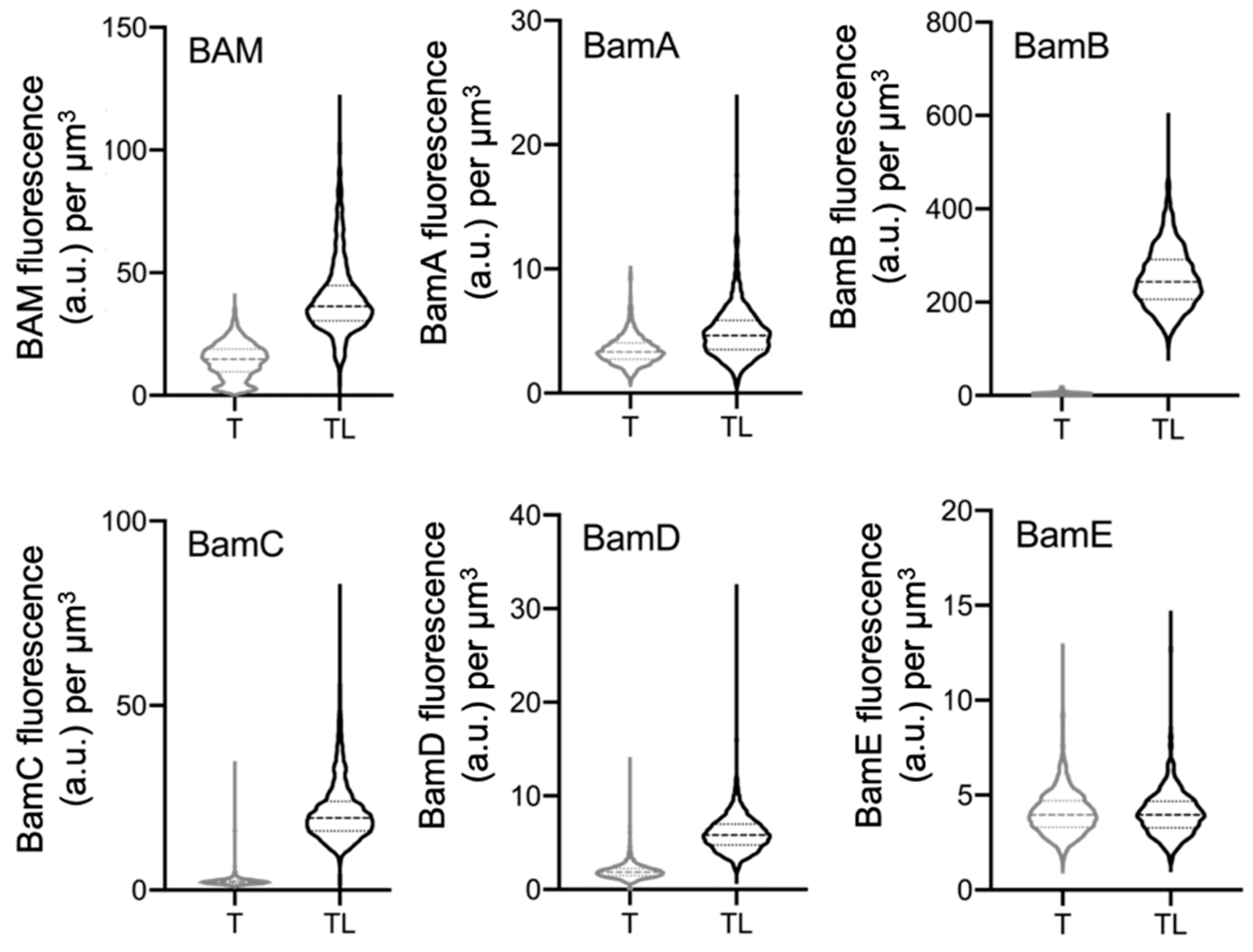
2.3. Peptidoglycan Digestion Is Needed to Access Each BAM Subunit
2.4. POTRA 1 Domain and BamE Require Peptidoglycan Digestion
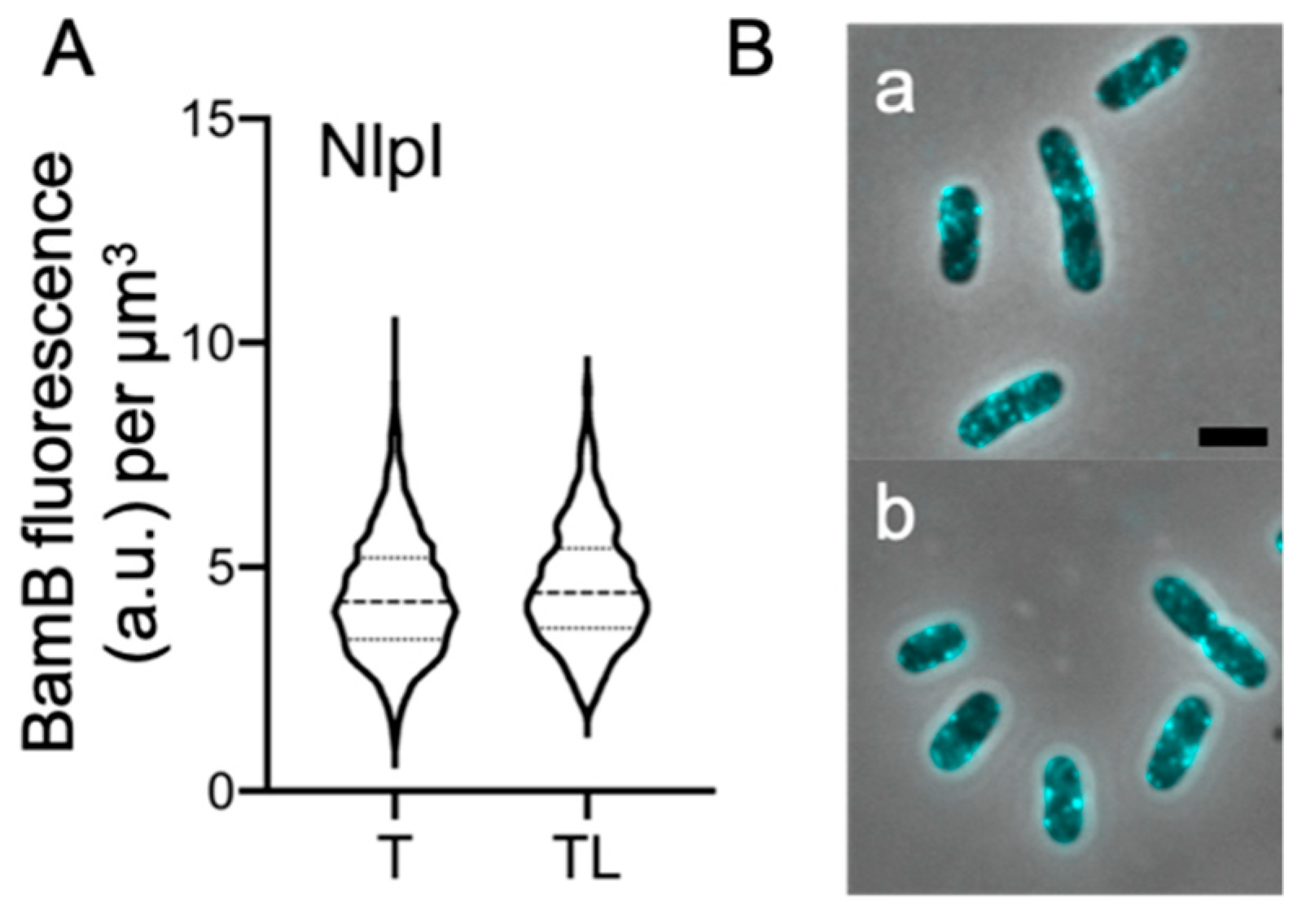
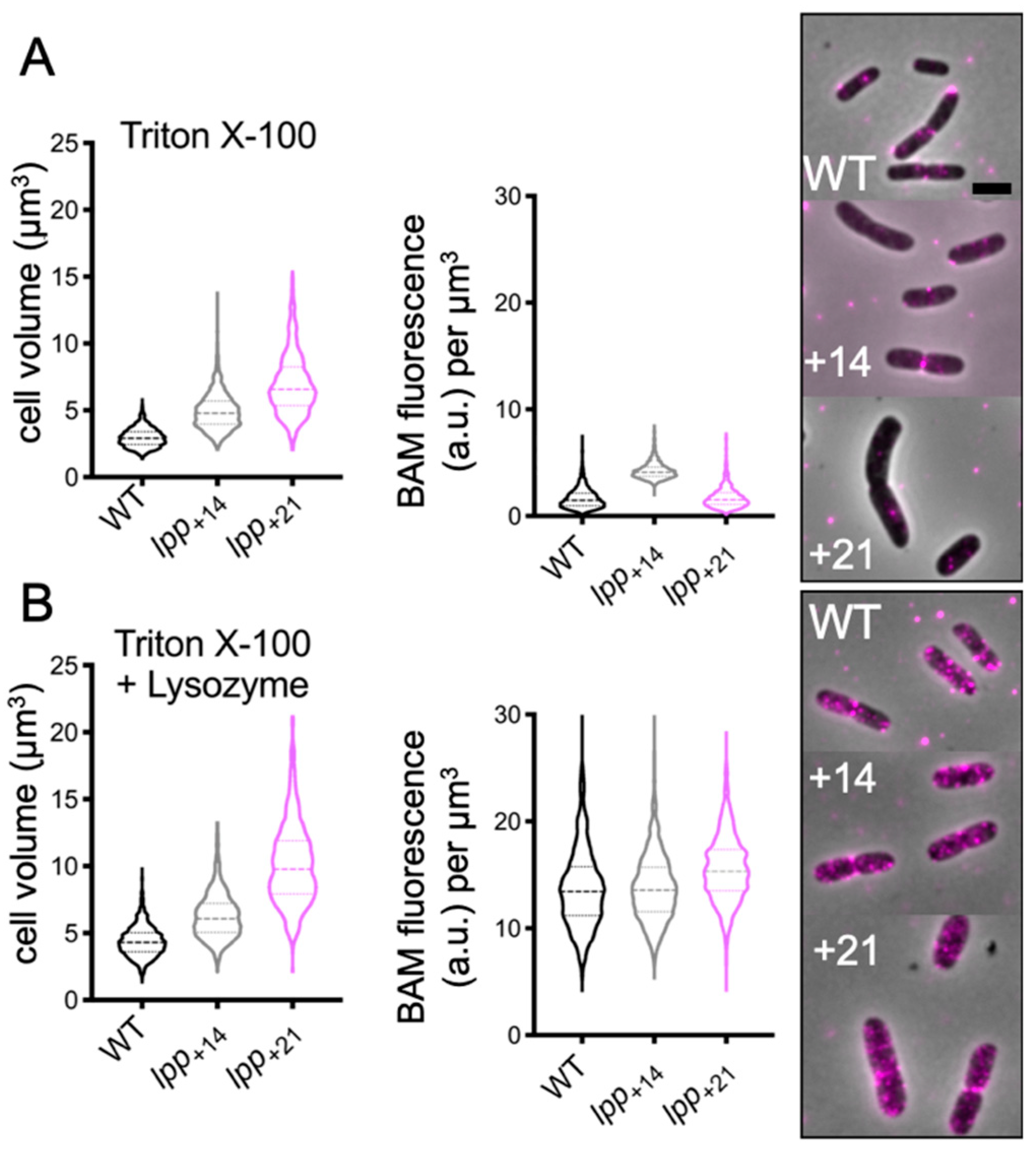
2.5. Peptidoglycan Digestion Is Not Required for All OM-Lipoproteins
2.6. Increasing the Distance between the Outer Membrane and Peptidoglycan Is Not Sufficient to Access the BAM Complex
3. Discussion and Conclusions
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Antibodies Production
4.3. Immunolabelling
4.4. Microscopy
4.5. Image Analysis
4.6. Construction of HA Tagged BAM Subunits
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sklar, J.G.; Wu, T.; Kahne, D.; Silhavy, T.J. Defining the roles of the periplasmic chaperones SurA, Skp and DegP in Escherichia coli. Genes Dev. 2007, 21, 2473–2484. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Malinverni, J.; Ruiz, N.; Kim, S.; Silhavy, T.J.; Kahne, D. Identification of a Multicomponent Complex Required for Outer Membrane Biogenesis in Escherichia coli. Cell 2005, 121, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinverni, J.C.; Werner, J.; Kim, S.; Sklar, J.G.; Kahne, D.; Misra, R.; Silhavy, T.J. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 2006, 61, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Charlson, E.S.; Werner, J.N.; Misra, R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 2006, 188, 7186–7194. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.H.; Szewczyk, J.; Pesavento, C.; Zietek, M.; Banzhaf, M.; Roszczenko, P.; Asmar, A.; Laloux, G.; Hov, A.K.; Leverrier, P.; et al. Detecting envelope stress by monitoring β-barrel assembly. Cell 2014, 159, 1652–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konovalova, A.; Mitchell, A.M.; Silhavy, T.J. A lipoprotein/b-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 2016, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hart, E.M.; Gupta, M.; Wühr, M.; Silhavy, T.J. The synthetic phenotype of ∆bamB ∆bamE double mutants results from a lethal jamming of the bam complex by the lipoprotein RcsF. MBio 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Létoquart, J.; Rodríguez-Alonso, R.; Nguyen, V.S.; Louis, G.; Calabrese, A.N.; Iorga, B.I.; Radford, S.E.; Cho, S.H.; Remaut, H.; Collet, J.F. Structural insight into the formation of lipoprotein-β-barrel complexes. Nat. Chem. Biol. 2020, 16, 1019–1025. [Google Scholar] [CrossRef]
- Vollmer, W.; Seligman, S.J. Architecture of peptidoglycan: More data and more models. Trends Microbiol. 2010, 18, 59–66. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [Green Version]
- Braun, V.; Rehn, K. Chemical Characterization, Spatial Distribution and Function of a Lipoprotein (Murein-Lipoprotein) of the E. coli Cell Wall: The Specific Effect of Trypsin on the Membrane Structure. Eur. J. Biochem. 1969, 10, 426–438. [Google Scholar] [CrossRef]
- Li, G.-W.; Burkhardt, D.; Gross, C.; Weissman, J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 2014, 157, 624–635. [Google Scholar] [CrossRef] [Green Version]
- Gerding, M.A.; Ogata, Y.; Pecora, N.D.; Niki, H.; De Boer, P.A.J. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 2007, 63, 1008–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asmar, A.T.; Ferreira, J.L.; Cohen, E.J.; Cho, S.H.; Beeby, M.; Hughes, K.T.; Collet, J.F. Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol. 2017, 15, e2004303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, E.J.; Ferreira, J.L.; Ladinsky, M.S.; Beeby, M.; Hughes, K.T. Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 2017, 356, 197–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, C.T.; Selkrig, J.; Perry, A.J.; Noinaj, N.; Buchanan, S.K.; Lithgow, T. Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J. Mol. Biol. 2012, 422, 545–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunasinghe, S.D.; Shiota, T.; Stubenrauch, C.J.; Schulze, K.E.; Webb, C.T.; Fulcher, A.J.; Dunstan, R.A.; Hay, I.D.; Naderer, T.; Whelan, D.R.; et al. The WD40 Protein BamB Mediates Coupling of BAM Complexes into Assembly Precincts in the Bacterial Outer Membrane. Cell Rep. 2018, 23, 2782–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vischer, N.O.E.; Verheul, J.; Postma, M.; van den Berg van Saparoea, B.; Galli, E.; Natale, P.; Gerdes, K.; Luirink, J.; Vollmer, W.; Vicente, M.; et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front. Microbiol. 2015, 6, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Buddelmeijer, N.; Aarsman, M.E.G.; Den Blaauwen, T. Immunolabeling of Proteins. Bio-Protocol 2013, 3, 1–5. [Google Scholar]
- Bakelar, J.; Buchanan, S.K.; Noinaj, N. The structure of the β-barrel assembly machinery complex. Science 2016, 351, 180–186. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Zheng, J.; Wang, Y.; Yang, X.; Liu, Y.; Sun, C.; Cao, B.; Zhou, H.; Ni, D.; Lou, J.; et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 192–196. [Google Scholar] [CrossRef]
- Matias, V.R.F.; Al-amoudi, A.; Dubochet, J.; Beveridge, T.J. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 6112–6118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banzhaf, M.; Yau, H.C.; Verheul, J.; Lodge, A.; Kritikos, G.; Mateus, A.; Cordier, B.; Hov, A.K.; Stein, F.; Wartel, M.; et al. Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J. 2020, 39, 1–20. [Google Scholar] [CrossRef]
- Demchick, P.; Koch, A.L. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 1996, 178, 768–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pink, D.; Moeller, J.; Quinn, B.; Jericho, M.; Beveridge, T. On the architecture of the gram-negative bacterial murein sacculus. J. Bacteriol. 2000, 182, 5925–5930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, A.L.; Woeste, S. Elasticity of the sacculus of Escherichia coli. J. Bacteriol. 1992, 174, 4811–4819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowles, T.J.; Jeeves, M.; Bobat, S.; Dancea, F.; McClelland, D.; Palmer, T.; Overduin, M.; Henderson, I.R. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 2008, 68, 1216–1227. [Google Scholar] [CrossRef]
- Bennion, D.; Charlson, E.S.; Coon, E.; Misra, R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 2010, 77, 1153–1171. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Malinverni, J.C.; Sliz, P.; Silhavy, T.J.; Harrison, S.C.; Kahne, D. Structure and Function of an Essential Component of the Outer Membrane Protein Assembly Machine. Science 2007, 317, 961–965. [Google Scholar] [CrossRef] [Green Version]
- Gatzeva-Topalova, P.Z.; Warner, L.R.; Pardi, A.; Sousa, M.C. Structure and Flexibility of the Complete Periplasmic Domain of BamA: The Protein Insertion Machine of the Outer Membrane. Structure 2010, 18, 1492–1501. [Google Scholar] [CrossRef] [Green Version]
- Gatzeva-Topalova, P.Z.; Walton, T.A.; Sousa, M.C. Crystal Structure of YaeT: Conformational Flexibility and Substrate Recognition. Structure 2008, 16, 1873–1881. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, R.; Jin, F.; Liu, Y.; Yu, J.; Fu, X.; Chang, Z. A supercomplex spanning the inner and outer membranes mediates the biogenesis of β-barrel outer membrane proteins in bacteria. J. Biol. Chem. 2016, 291, 16720–16729. [Google Scholar] [CrossRef] [Green Version]
- Alvira, S.; Watkins, D.; Troman, L.; Allen, W.; Lorriman, J.; Degliesposti, G.; Cohen, E.; Beeby, M.; Daum, B.; Gold, V.; et al. Inter-membrane association of the Sec and BAM translocons for bacterial outer-membrane biogenesis. eLife 2020, 60669. [Google Scholar] [CrossRef]
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.H.; Couloux, A.; Lee, S.W.; Yoon, S.H.; Cattolico, L.; et al. Genome Sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652. [Google Scholar] [CrossRef]
- Rossiter, A.E.; Leyton, D.L.; Tveen-Jensen, K.; Browning, D.F.; Sevastsyanovich, Y.; Knowles, T.J.; Nichols, K.B.; Cunningham, A.F.; Overduin, M.; Schembri, M.A.; et al. The essential β-barrel assembly machinery complex components bamd and bama are required for autotransporter biogenesis. J. Bacteriol. 2011, 193, 4250–4253. [Google Scholar] [CrossRef] [Green Version]
- Rigel, N.W.; Schwalm, J.; Ricci, D.P.; Silhavy, T.J. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J. Bacteriol. 2012, 194, 1002–1008. [Google Scholar] [CrossRef] [Green Version]
- Jensen, K.A.J.F. The Escherichia coli K-12 ‘Wild Types’ W3110 and MG1655 Have an rph Frameshift Mutation That Leads to Pyrimidine Starvation Due to LowpyrE Expression Levels. J. Bacteriol. 1993, 175, 3401–3407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppelman, C.M.; Aarsman, M.E.G.; Postmus, J.; Pas, E.; Muijsers, A.O.; Scheffers, D.J.; Nanninga, N.; Den Blaauwen, T. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 2004, 51, 645–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelstein, A.; Amodaj, N.; Hoover, K.; Vale, R.; Stuurman, N. Computer Control of Microscopes Using μ Manager. Curr. Protoc. Mol. Biol. 2010, 92, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, L.; Stern, A.S.; Berg, H.C. Growth of flagellar filaments of Escherichia coli is independent of filament length. J. Bacteriol. 2012, 194, 2437–2442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meiresonne, N.Y.; van der Ploeg, R.; Hink, M.A.; Den Blaauwen, T. Activity-Related Conformational Changes in D,D-Carboxypeptidases Revealed by In Vivo Periplasmic Förster Resonance Energy Transfer Assay in Escherichia coli. MBio 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain | Genotype | Reference 3 |
|---|---|---|
| BW25113 | lacI+rrnBT14 ΔlacZWJ16 hsdR514Δara-BADAH33 ΔrhaBADLD78 rph-1 Δ(araB–D)567 Δ(rhaD–B)568 ΔlacZ4787(::rrnB-3) hsdR514 rph-1 | [16] |
| BL21DE3 | F– ompT hsdSB (rB–, mB–) gal dcm (DE3) | [35] |
| ΔbamE | BW25113 ΔbamE::cam | [36] |
| bamA101 | DH300 bamA101 | [37] |
| MG1655 | F−, lambda−, rph-1 | [38] |
| Lpp+14 | DH300 Δlpp::lpp+14 | [14] |
| Lpp+21 | DH300 Δlpp::lpp+21 | [14] |
| Name | Primer Sequence |
|---|---|
| 100-EC- BamA*NcoI-FW | GCTGTAGGCGGTAACGCGATGGCGGTTGC |
| 101-EC- BamA*NcoI-RV | GCTGTAGGCGGTAACGCGATGGCGGTTGC |
| 106-EC-BamA-NcoI-FW | CGTATACCATGGCGATGAAAAAGTTGCTCATAGCGTCG |
| 107-EC-BamA-EcoRI-RV | CGTAATGAATTCCCAGGTTTTACCGATGTTAAACTGGAAC |
| 110-EC-HA-BamA-FW | GTGCCGGATGTGCCGGATTATGCGTTCGTAGTGAAAGATATTCATTTCGAAGGCCTTC |
| 111-EC-HA-BamA-RV | CACATCCGGCACATCATACGGATACCCTTCAGCACCGTATACGGTGGC |
| 127-EC-BamE-NcoI-FW | GCGCGCCATGGGCCGCTGTAAAACGCTGACTGC |
| 128-EC-BamE-HA-HindIII | GCGCGAAGCTTTTAAGCGTAATCTGGAACATCGTATGGGTAGTTA CCACTCAGCGCAGGTTTGTTATC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Consoli, E.; Collet, J.-F.; den Blaauwen, T. The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Anchors the Peptidoglycan Layer by Spanning It with All Subunits. Int. J. Mol. Sci. 2021, 22, 1853. https://doi.org/10.3390/ijms22041853
Consoli E, Collet J-F, den Blaauwen T. The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Anchors the Peptidoglycan Layer by Spanning It with All Subunits. International Journal of Molecular Sciences. 2021; 22(4):1853. https://doi.org/10.3390/ijms22041853
Chicago/Turabian StyleConsoli, Elisa, Jean-François Collet, and Tanneke den Blaauwen. 2021. "The Escherichia coli Outer Membrane β-Barrel Assembly Machinery (BAM) Anchors the Peptidoglycan Layer by Spanning It with All Subunits" International Journal of Molecular Sciences 22, no. 4: 1853. https://doi.org/10.3390/ijms22041853








