Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation
Abstract
1. Introduction
2. HKT Sequence and Structure
2.1. Evolution of the HKT Family
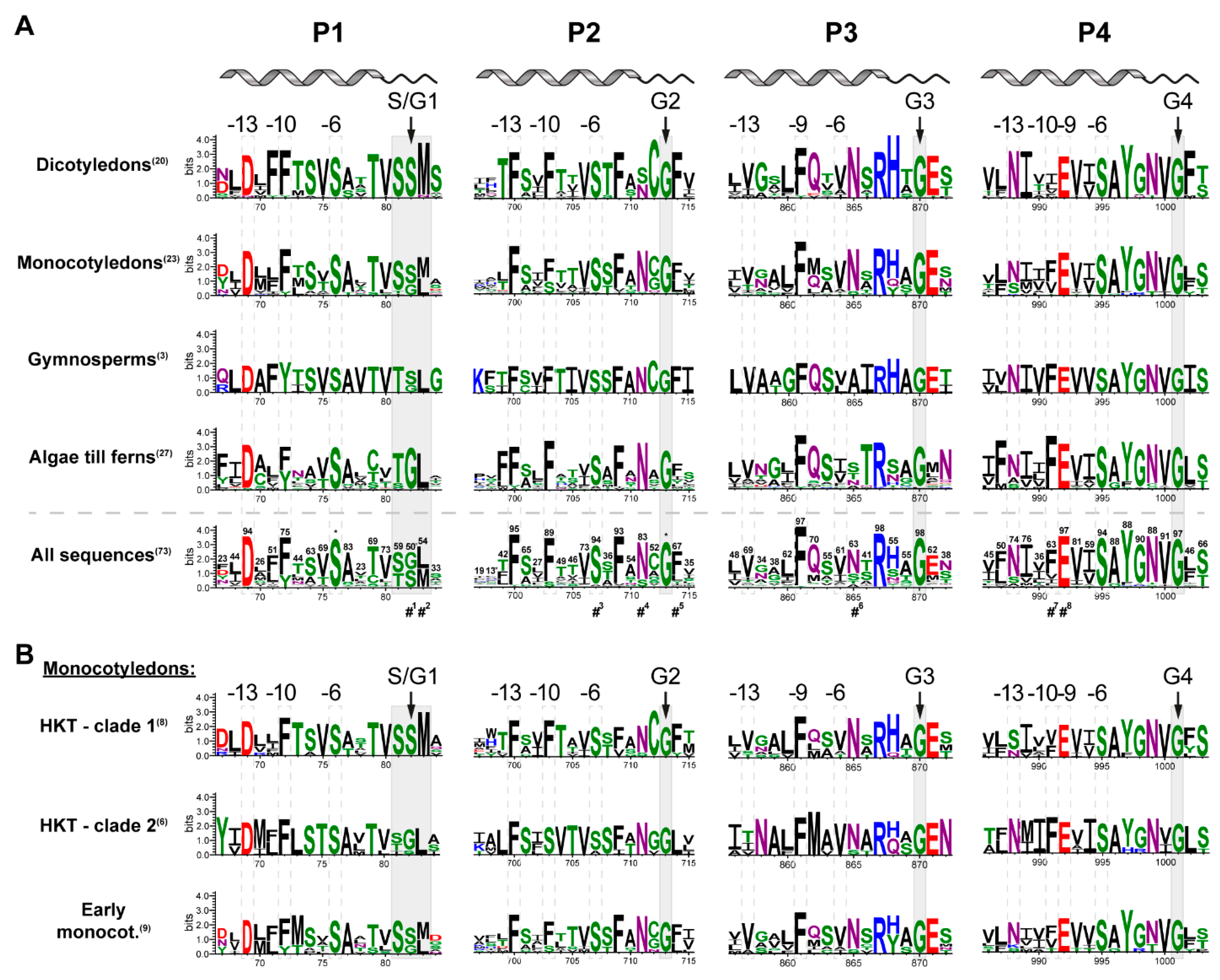
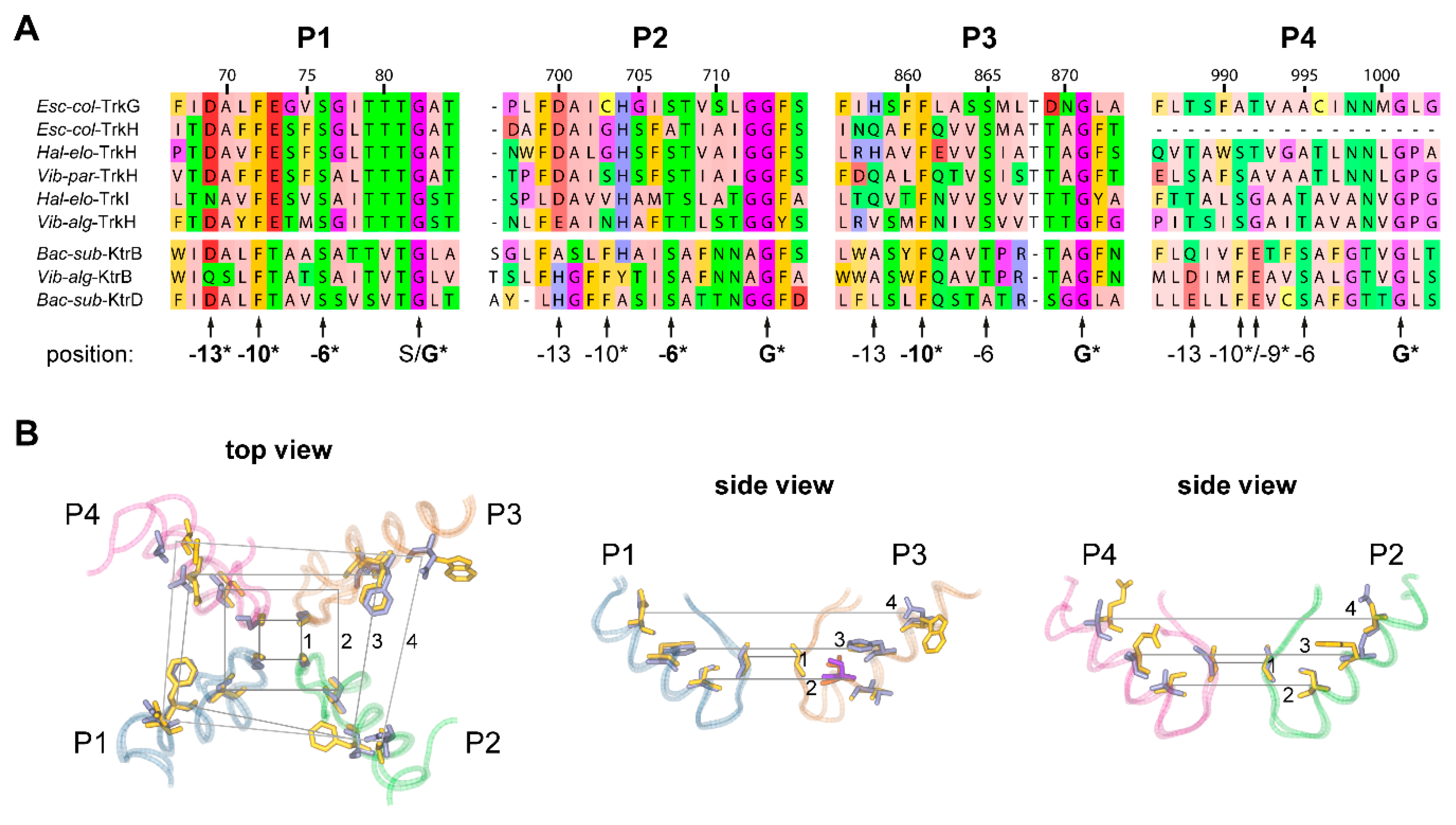
2.2. Pore Domains Bear a High Degree of Conservation
2.3. A Selectivity Filter Motif in the First Pore Region Changed
2.4. Monocotyledons Are Sequence- and Function-Wise Versatile
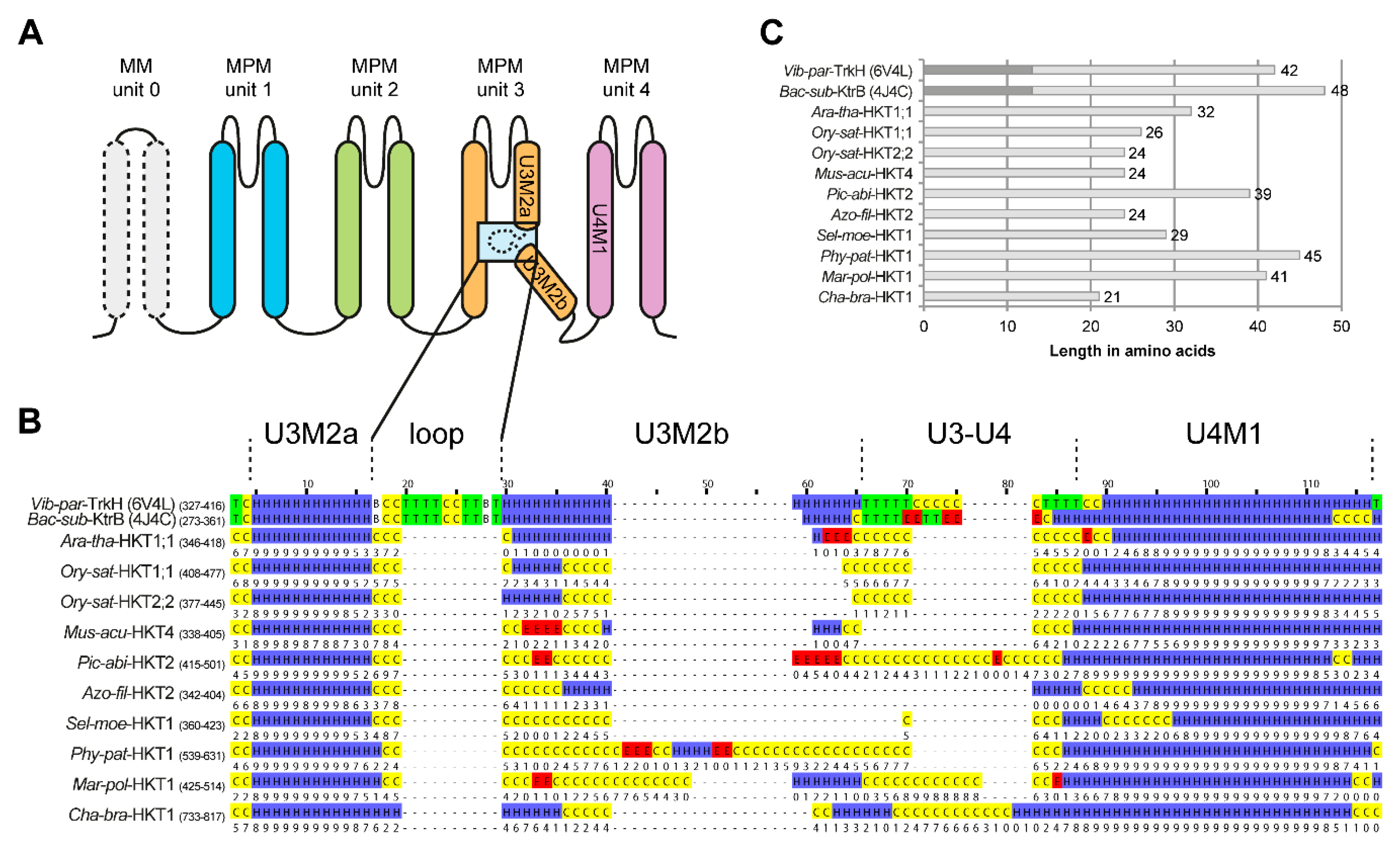
2.5. Structure of the Second Transmembrane Segment in Unit 3
3. HKT Gene Expression and Regulation
3.1. Gene Expression Regulation and Protein Localization
3.2. Regulation of AtHKT1;1 Gene in Arabidopsis
3.3. Regulation of OsHKT Gene Expression in Rice
3.4. Regulation of HKT Gene Expression in Other Plant Species
3.5. Non-Transcription Factor-Mediated HKT Gene Regulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HKT | High-affinity K+ (potassium) Transporters |
| Na+ | Sodium |
| K+ | Potassium |
References
- Schachtman, D.; Schroeder, J. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 1994, 370, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [CrossRef] [PubMed]
- Kronzucker, H.J.; Britto, D.T. Sodium transport in plants: A critical review. New Phytol. 2011, 189, 54–81. [Google Scholar] [CrossRef] [PubMed]
- Platten, J.D.; Cotsaftis, O.; Berthomieu, P.; Bohnert, H.; Davenport, R.J.; Fairbairn, D.J.; Horie, T.; Leigh, R.A.; Lin, H.-X.; Luan, S.; et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006, 11, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Haro, R.; Bañuelos, M.A.; Rodríguez-Navarro, A. High-affinity sodium uptake in land plants. Plant Cell Physiol. 2010, 51, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Porras, J.L.; Riaño-Pachón, D.M.; Benito, B.; Haro, R.; Sklodowski, K.; Rodríguez-Navarro, A.; Dreyer, I. Phylogenetic analysis of k(+) transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 2012, 3, 167. [Google Scholar] [CrossRef]
- Mäser, P.; Hosoo, Y.; Goshima, S.; Horie, T.; Eckelman, B.; Yamada, K.; Yoshida, K.; Bakker, E.P.; Shinmyo, A.; Oiki, S.; et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Natl. Acad. Sci. USA 2002, 99, 6428–6433. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef]
- Corratgé-Faillie, C.; Jabnoune, M.; Zimmermann, S.; Véry, A.-A.; Fizames, C.; Sentenac, H. Potassium and sodium transport in non-animal cells: The Trk/Ktr/HKT transporter family. Cell. Mol. Life Sci. 2010, 67, 2511–2532. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Park, H.C.; Ali, A.; Oh, D.-H.; Aman, R.; Kropornicka, A.; Hong, H.; Choi, W.; Chung, W.S.; Kim, W.-Y.; et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K(+) specificity in the presence of NaCl. Plant Physiol. 2012, 158, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Fairbairn, D.J.; Liu, W.; Schachtman, D.P.; Gomez-Gallego, S.; Day, S.R.; Teasdale, R.D. Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol. Biol. 2000, 43, 515–525. [Google Scholar] [CrossRef]
- Jabnoune, M.; Espeout, S.; Mieulet, D.; Fizames, C.; Verdeil, J.-L.; Conéjéro, G.; Rodríguez-Navarro, A.; Sentenac, H.; Guiderdoni, E.; Abdelly, C.; et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009, 150, 1955–1971. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Brodsky, D.E.; Costa, A.; Kaneko, T.; Schiavo, F.L.; Katsuhara, M.; Schroeder, J.I. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol. 2011, 156, 1493–1507. [Google Scholar] [CrossRef]
- Riedelsberger, J.; Vergara-Jaque, A.; Piñeros, M.; Dreyer, I.; Gonzalez, W. An extracellular cation coordination site influences ion conduction of OsHKT2;2. BMC Plant Biol. 2019, 19, 316. [Google Scholar] [CrossRef]
- Horie, T.; Yoshida, K.; Nakayama, H.; Yamada, K.; Oiki, S.; Shinmyo, A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001, 27, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Horie, T.; Xue, S.; Leung, H.-Y.; Katsuhara, M.; Brodsky, D.E.; Wu, Y.; Schroeder, J.I. Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol. 2010, 152, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.-Z.; Wang, W.; Wang, S.-M.; Li, L.-G.; Buchanan, B.B.; Lin, H.-X.; Gao, J.-P.; Luan, S. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proc. Natl. Acad. Sci. USA 2010, 107, 7089–7094. [Google Scholar] [CrossRef]
- Gassmann, W.; Rubio, F.; Schroeder, J.I. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 1996, 10, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Oomen, R.J.F.J.; Benito, B.; Sentenac, H.; Rodríguez-Navarro, A.; Talón, M.; Véry, A.-A.; Domingo, C. HKT2;2/1, a K⁺-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J. 2012, 71, 750–762. [Google Scholar] [CrossRef]
- Sassi, A.; Mieulet, D.; Khan, I.; Moreau, B.; Gaillard, I.; Sentenac, H.; Véry, A.-A. The rice monovalent cation transporter OsHKT2;4: Revisited ionic selectivity. Plant Physiol. 2012, 160, 498–510. [Google Scholar] [CrossRef]
- Wang, T.; Gassmann, W.; Rubio, F.; Schroeder, J.; Glass, A. Rapid Up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 1998, 118, 651–659. [Google Scholar] [CrossRef]
- Garciadeblás, B.; Senn, M.E.; Bañuelos, M.A.; Rodríguez-Navarro, A. Sodium transport and HKT transporters: The rice model. Plant J. 2003, 34, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.-Y.; Leung, H.-Y.; Hattori, K.; Konomi, M.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar]
- Almeida, P.; Katschnig, D.; de Boer, A.H. HKT transporters--state of the art. Int. J. Mol. Sci. 2013, 14, 20359–20385. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Saibo, N.J.M.; Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Berthomieu, P.; Conéjéro, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar] [CrossRef]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef]
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 2012, 7, e39865. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Horie, T.; Costa, A.; Kim, T.H.; Han, M.J.; Horie, R.; Leung, H.-Y.; Miyao, A.; Hirochika, H.; An, G.; Schroeder, J.I. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007, 26, 3003–3014. [Google Scholar] [CrossRef] [PubMed]
- Haro, R.; Bañuelos, M.A.; Senn, M.E.; Barrero-Gil, J.; Rodríguez-Navarro, A. HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol. 2005, 139, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Mian, A.; Oomen, R.J.F.J.; Isayenkov, S.; Sentenac, H.; Maathuis, F.J.M.; Véry, A.-A. Over-expression of an Na+-and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 2011, 68, 468–479. [Google Scholar] [CrossRef]
- Golldack, D.; Su, H.; Quigley, F.; Kamasani, U.R.; Muñoz Garay, C.; Balderas, E.; Popova, O.V.; Bennett, J.; Bohnert, H.J.; Pantoja, O. Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J. 2002, 31, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Benito, B.; Haro, R.; Amtmann, A.; Cuin, T.A.; Dreyer, I. The twins K+ and Na+ in plants. J. Plant Physiol. 2014, 171, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Raddatz, N.; Pardo, J.M.; Yun, D.-J. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol. Plant. 2020, 471003. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Ariyarathna, H.A.C.K.; Francki, M.G. Phylogenetic relationships and protein modelling revealed two distinct subfamilies of group II HKT genes between crop and model grasses. Genome 2016, 59, 509–517. [Google Scholar] [CrossRef]
- Diatloff, E.; Kumar, R.; Schachtman, D.P. Site directed mutagenesis reduces the Na+ affinity of HKT1, an Na+ energized high affinity K+ transporter. FEBS Lett. 1998, 432, 31–36. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Raddatz, N.; Aman, R.; Kim, S.; Park, H.C.; Jan, M.; Baek, D.; Khan, I.U.; Oh, D.-H.; Lee, S.Y.; et al. A Single Amino-Acid Substitution in the Sodium Transporter HKT1 Associated with Plant Salt Tolerance. Plant. Physiol. 2016, 171, 2112–2126. [Google Scholar] [CrossRef]
- Rubio, F.; Schwarz, M.; Gassmann, W.; Schroeder, J.I. Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J. Biol. Chem. 1999, 274, 6839–6847. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.M.F.; de Boer, G.-J.; de Boer, A.H. Assessment of natural variation in the first pore domain of the tomato HKT1;2 transporter and characterization of mutated versions of SlHKT1;2 expressed in Xenopus laevis oocytes and via complementation of the salt sensitive athkt1;1 mutant. Front. Plant Sci. 2014, 5, 1463. [Google Scholar] [CrossRef]
- Durell, S.R.; Guy, H.R. Structural Models of the KtrB, TrkH, and Trk1,2 Symporters Based on the Structure of the KcsA K+ Channel. Biophysical. J. 1999, 77, 789–807. [Google Scholar] [CrossRef]
- Cao, Y.; Jin, X.; Huang, H.; Derebe, M.G.; Levin, E.J.; Kabaleeswaran, V.; Pan, Y.; Punta, M.; Love, J.; Weng, J.; et al. Crystal structure of a potassium ion transporter, TrkH. Nature 2011, 471, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Pires, R.S.; Szollosi, A.; Morais-Cabral, J.H. The structure of the KtrAB potassium transporter. Nature 2013, 496, 323–328. [Google Scholar] [CrossRef]
- Tholema, N.; Vor der Brüggen, M.; Mäser, P.; Nakamura, T.; Schroeder, J.I.; Kobayashi, H.; Uozumi, N.; Bakker, E.P. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 2005, 280, 41146–41154. [Google Scholar] [CrossRef]
- Corratgé, C.; Zimmermann, S.; Lambilliotte, R.; Plassard, C.; Marmeisse, R.; Thibaud, J.-B.; Lacombe, B.; Sentenac, H. Molecular and functional characterization of a Na(+)-K(+) transporter from the Trk family in the ectomycorrhizal fungus Hebeloma cylindrosporum. J. Biol. Chem. 2007, 282, 26057–26066. [Google Scholar] [CrossRef]
- Cao, Y.; Pan, Y.; Huang, H.; Jin, X.; Levin, E.J.; Kloss, B.; Zhou, M. Gating of the TrkH ion channel by its associated RCK protein TrkA. Nature 2013, 496, 317–322. [Google Scholar] [CrossRef]
- Hänelt, I.; Wunnicke, D.; Müller-Trimbusch, M.; Vor der Brüggen, M.; Kraus, I.; Bakker, E.P.; Steinhoff, H.-J. Membrane region M2C2 in subunit KtrB of the K+ uptake system KtrAB from Vibrio alginolyticus forms a flexible gate controlling K+ flux: An electron paramagnetic resonance study. J. Biol. Chem. 2010, 285, 28210–28219. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Torrisi, M.; Kaleel, M.; Pollastri, G. Deeper Profiles and Cascaded Recurrent and Convolutional Neural Networks for state-of-the-art Protein Secondary Structure Prediction. Sci. Rep. 2019, 9, 12374-12. [Google Scholar] [CrossRef] [PubMed]
- Musci, M.; Maruccia, G.; Ferretti, M. A Review of Quasi-perfect Secondary Structure Prediction Servers. In Database and Expert Systems Applications, Proceedings of the DEXA 2019 International Workshops BIOKDD, IWCFS, MLKgraphs and TIR, Linz, Austria, 26–29 August 2019; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1062, pp. 21–26. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Sun, J.; Cao, H.; Cheng, J.; He, X.; Sohail, H.; Niu, M.; Huang, Y.; Bie, Z. Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber. Int. J. Mol. Sci. 2018, 19, 2648. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Balderas, E.; Vera-Estrella, R.; Golldack, D.; Quigley, F.; Zhao, C.; Pantoja, O.; Bohnert, H.J. Expression of the cation transporter McHKT1 in a halophyte. Plant Mol. Biol. 2003, 52, 967–980. [Google Scholar] [CrossRef]
- Pérez, N.J.; Pineda, B.; Sogo, B.G.; Atares, A.; Athman, A.; Byrt, C.S.; Olías, R.; Asins, M.J.; Gilliham, M.; Moreno, V.; et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017, 40, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.W.; Dunlevy, J.D.; Wu, Y.; Blackmore, D.H.; Walker, R.R.; Edwards, E.J.; Gilliham, M.; Walker, A.R. Functional differences in transport properties of natural HKT1;1 variants influence shoot Na+ exclusion in grapevine rootstocks. New Phytol. 2018, 217, 1113–1127. [Google Scholar] [CrossRef]
- Huang, L.; Kuang, L.; Wu, L.; Shen, Q.; Han, Y.; Jiang, L.; Wu, D.; Zhang, G. The HKT Transporter HvHKT1;5 Negatively Regulates Salt Tolerance. Plant Physiol. 2020, 182, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Mohamed, S.; Regnault, T.; Mieulet, D.; Guiderdoni, E.; Sentenac, H.; Véry, A.-A. Constitutive Contribution by the Rice OsHKT1;4 Na+ Transporter to Xylem Sap Desalinization and Low Na+ Accumulation in Young Leaves Under Low as High External Na+ Conditions. Front. Plant Sci. 2020, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Imran, S.; Horie, T.; Katsuhara, M. Expression and Ion Transport Activity of Rice OsHKT1;1 Variants. Plants 2020, 9, 16. [Google Scholar] [CrossRef]
- Rosas-Santiago, P.; Lagunas-Gómez, D.; Barkla, B.J.; Vera-Estrella, R.; Lalonde, S.; Jones, A.; Frommer, W.B.; Zimmermannova, O.; Sychrova, H.; Pantoja, O. Identification of rice cornichon as a possible cargo receptor for the Golgi-localized sodium transporter OsHKT1;3. J. Exp. Bot. 2015, 66, 2733–2748. [Google Scholar] [CrossRef] [PubMed]
- Do, P.T.; Hoang, Y.H.; Le, M.Q.; Tang, H.T.; Nguyen, D.H. OsHKT1;3 gene sequence polymorphisms and expression profile in rice (Oryza sativa L.). AJAR 2018, 13, 2659–2667. [Google Scholar]
- Suzuki, K.; Yamaji, N.; Costa, A.; Okuma, E.; Kobayashi, N.I.; Kashiwagi, T.; Katsuhara, M.; Wang, C.; Tanoi, K.; Murata, Y.; et al. OsHKT1;4-mediated Na + transport in stems contributes to Na + exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 2016, 16, 22. [Google Scholar] [CrossRef]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Kashino, M.F.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-Induced Tissue-Specific Cytosine Methylation Downregulates Expression of HKT Genes in Contrasting Wheat (Triticum aestivum L.) Genotypes. DNA Cell Biol. 2017, 36, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Xu, B.; Krishnan, M.; Lightfoot, D.J.; Athman, A.; Jacobs, A.K.; Watson-Haigh, N.S.; Plett, D.; Munns, R.; Tester, M.; et al. The Na(+) transporter, TaHKT1;5-D, limits shoot Na(+) accumulation in bread wheat. Plant J. 2014, 80, 516–526. [Google Scholar] [CrossRef]
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006, 142, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef]
- Hamamoto, S.; Horie, T.; Hauser, F.; Deinlein, U.; Schroeder, J.I.; Uozumi, N. HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotechnol. 2014, 32C, 113–120. [Google Scholar] [CrossRef]
- Tada, Y.; Endo, C.; Katsuhara, M.; Horie, T.; Shibasaka, M. High-Affinity K⁺ Transporters from a Halophyte, Sporobolus virginicus, Mediate Both K⁺ and Na⁺ Transport in Transgenic Arabidopsis, X. laevis Oocytes and …. Plant Cell Physiol. 2019, 60, 176–187. [Google Scholar] [CrossRef]
- Singh, A.; Bhushan, B.; Gaikwad, K.; Yadav, O.P.; Kumar, S.; Rai, R.D. Induced defence responses of contrasting bread wheat genotypes under differential salt stress imposition. Indian J. Biochem. Biophys. 2015, 52, 75–85. [Google Scholar]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Physiological, Biochemical, Epigenetic and Molecular Analyses of Wheat (Triticum aestivum) Genotypes with Contrasting Salt Tolerance. Front. Plant Sci. 2017, 8, 3389. [Google Scholar] [CrossRef]
- Cao, Y.; Liang, X.; Yin, P.; Zhang, M.; Jiang, C. A domestication-associated reduction in K+ -preferring HKT transporter activity underlies maize shoot K+ accumulation and salt tolerance. New Phytol. 2019, 222, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.-J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Jha, D.; Salt, D.E.; Tester, M.; Hill, K.; Kieber, J.J.; Schaller, G.E. Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 2010, 64, 753–763. [Google Scholar] [CrossRef]
- Shkolnik-Inbar, D.; Adler, G.; Bar-Zvi, D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013, 73, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-C.; Cheng, W.-H.; Hong, C.-Y.; Chang, Y.-S.; Chang, M.-C. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice 2018, 11, 50. [Google Scholar] [CrossRef]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.-J.; Hwang, I.; Liu, B.; Xu, Z.-Y. A DNA Methylation Reader-Chaperone Regulator-Transcription Factor Complex Activates OsHKT1;5 Expression during Salinity Stress. Plant Cell Online 2020, 32, 3535–3558. [Google Scholar] [CrossRef]
- Chen, N.; Tong, S.; Tang, H.; Zhang, Z.; Liu, B.; Lou, S.; Liu, J.; Liu, H.; Ma, T.; Jiang, Y. The PalERF109 transcription factor positively regulates salt tolerance via PalHKT1;2 in Populus alba var. pyramidalis. Tree Physiol. 2020, 40, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, K.; Rauf, M.; Ahmed, M.; Malik, Z.A.; Habib, I.; Ahmed, Z.; Mahmood, K.; Ali, R.; Masmoudi, K.; Lemtiri-Chlieh, F.; et al. Functional characterisation of an intron retaining K(+) transporter of barley reveals intron-mediated alternate splicing. Plant Biol. 2015, 17, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, I.; Sussmilch, F.C.; Fukushima, K.; Riadi, G.; Becker, D.; Schultz, J.; Hedrich, R. How to Grow a Tree: Plant Voltage-Dependent Cation Channels in the Spotlight of Evolution. Trends Plant Sci. 2021, 26, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]

| Gene Name | Class | Species | Cotyledons | Gene Expression Localization | Expression Under Stress | Subcellular Protein Localization | Reference |
|---|---|---|---|---|---|---|---|
| AtHKT1;1 | I | Arabidopsis thaliana | Di | Leaf and flower phloem, leaf xylem parenchyma; Root xylem parenchyma and phloem | Expression peaks at 30 mM K+ or Na+ in roots and shoots Lower or higher K+ and Na+ did not increase expression | PM | [23,26] |
| CmHKT1;1 | I | Cucurbita moschata | Di | NA | PM | [58] | |
| McHKT1;1 | I | Mesembryanthemum crystallinum | Di | Leaf, stems, flowers, and seedpods | High Na+: increased expression in leaves after 6 h, then decreased expression over 48 h | PM | [59] |
| SlHKT1;1 | I | Solanum lycopersicum | Di | Vascular bundles of the main and secondary veins of leaves | High Na+: decreased expression in leave and stems but increased expression in roots | NA | [60] |
| SlHKT1;2 | I | Solanum lycopersicum | Di | Vascular bundles of the main and secondary veins of leaf; Stellar cells of root | High Na+: decreased expression in leave and stems but increased expression in roots | NA | [60] |
| VisHKT1;1 | I | Vitis vinifera | Di | Root stele | NA | PM | [61] |
| HvHKT1;5 | I | Hordeum vulgare | Mono | Epidermis and parenchyma and pericycle cells adjacent to xylem vessels in the root stele | Low and high K+: increased root expression High Na+: increased expression in roots | PM | [62] |
| OsHTK1;1 | I | Oryza sativa | Mono | Leaf bulliform cells and vascular tissues; Root epidermis, exodermis, cortex, and stele (mainly phloem) | Higher expression in shoots than in roots High Na+: increasing expression in roots but decreasing expression in shoots [63] | PM | [12,63,64,65] |
| OsHTK1;3 | I | Oryza sativa | Mono | Leaf bulliform cells, vascular tissues, and mesophyll cells; Root cortex and stele vascular tissues | High Na+: increased expression in leaves and roots | Golgi membrane | [12,63,66,67] |
| OsHTK1;4 | I | Oryza sativa | Mono | Leaf sheaths | High Na+: increased expression in leaves and roots | PM | [28,63,68] |
| OsHTK1;5 | I | Oryza sativa | Mono | Parenchyma cells bordering xylem vessels in shoots and roots | High Na+: increased expression in leaves and roots | PM | [28,31,63,69] |
| TaHKT1;4 | I | Triticum aestivum | Mono | Roots | NA | NA | [70] |
| TaHKT1;5-D | I | Triticum aestivum | Mono | Roots | NA | PM | [71] |
| TmHKT1;4-A2 | I | Triticum monococcum | Mono | Leaf sheaths; Roots | NA | NA | [72] |
| TmHKT1;5-A | I | Triticum monococcum | Mono | Parenchyma and pericycle cells adjacent to xylem vessels in the root stele | NaCl treatment did not affect expression in roots | NA | [71,73] |
| HvHKT2;1 | II | Hordeum vulgare | Mono | Leaf blade and sheath; Root cortex cells | Low K+: increased expression in leaf sheaths, leaf blades, and roots High Na+: decreased expression in leaf sheaths and roots but increased expression in leaf blades | NA | [34] |
| OsHKT2;1 | II | Oryza sativa | Mono | Leaf bulliform cells, vascular tissues, and mesophyll cells; Root epidermis, endodermis, exodermis, cortex, and stele (mainly phloem) | Low K+: increased expression in roots High K+ or Na+: decreased expression in roots | PM | [12,19,33] |
| OsHKT2;2 | II | Oryza sativa | Mono | Roots | Low K+ and/or low Na+: increased expression High K+ and/or high Na+: decreased expression | NA | [19] |
| OsHKT2;3 | II | Oryza sativa | Mono | Leaf blade and sheath; Roots | NA | NA | [74] |
| OsHKT2;4 | II | Oryza sativa | Mono | Leaf blade and sheath, leaf epidermal cells, vasculature of spikelets, leaves, and stems; Root vasculature | NA | PM | [13,17,75] |
| SvHKT2;1 | II | Sporobolus virginicus | Mono | NA | NA | PM | [75] |
| SvHKT2;2 | II | Sporobolus virginicus | Mono | NA | NA | PM | [75] |
| TaHKT2;1 | II | Triticum aestivum | Mono | Vasculature tissue of leaf mesophyll; Root cortical cells | NA | NA | [1,76] |
| TaHKT2;3 | II | Triticum aestivum | Mono | Shoots; Roots | NA | NA | [76,77] |
| ZmHKT2 | II | Zea mays | Mono | Roots, with highest expression in the stele | NA | PM | [78] |
| Species | Transcription Factor | Regulator | HKT | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | ABI4 | Negative | AtHKT1;1 | [81] |
| ARR1 | Negative | AtHKT1;1 | [80] | |
| ARR12 | Negative | AtHKT1;1 | [80] | |
| AtbZIP24 | Negative | AtHKT1;1 | [79] | |
| Oryza sativa | Osbhlh035 | Positive | OsHKT1;3 and OsHKT1;5 | [82] |
| OsMYB106 (with OsBAG4 and OsSUVH7) | Positive | OsHKT1;5 | [83] | |
| OsMYBc | Positive | OsHKT1;1 | [64] | |
| Populus alba | PalERF109 | Positive | PalHKT1;2 | [84] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riedelsberger, J.; Miller, J.K.; Valdebenito-Maturana, B.; Piñeros, M.A.; González, W.; Dreyer, I. Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation. Int. J. Mol. Sci. 2021, 22, 1892. https://doi.org/10.3390/ijms22041892
Riedelsberger J, Miller JK, Valdebenito-Maturana B, Piñeros MA, González W, Dreyer I. Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation. International Journal of Molecular Sciences. 2021; 22(4):1892. https://doi.org/10.3390/ijms22041892
Chicago/Turabian StyleRiedelsberger, Janin, Julia K. Miller, Braulio Valdebenito-Maturana, Miguel A. Piñeros, Wendy González, and Ingo Dreyer. 2021. "Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation" International Journal of Molecular Sciences 22, no. 4: 1892. https://doi.org/10.3390/ijms22041892
APA StyleRiedelsberger, J., Miller, J. K., Valdebenito-Maturana, B., Piñeros, M. A., González, W., & Dreyer, I. (2021). Plant HKT Channels: An Updated View on Structure, Function and Gene Regulation. International Journal of Molecular Sciences, 22(4), 1892. https://doi.org/10.3390/ijms22041892







