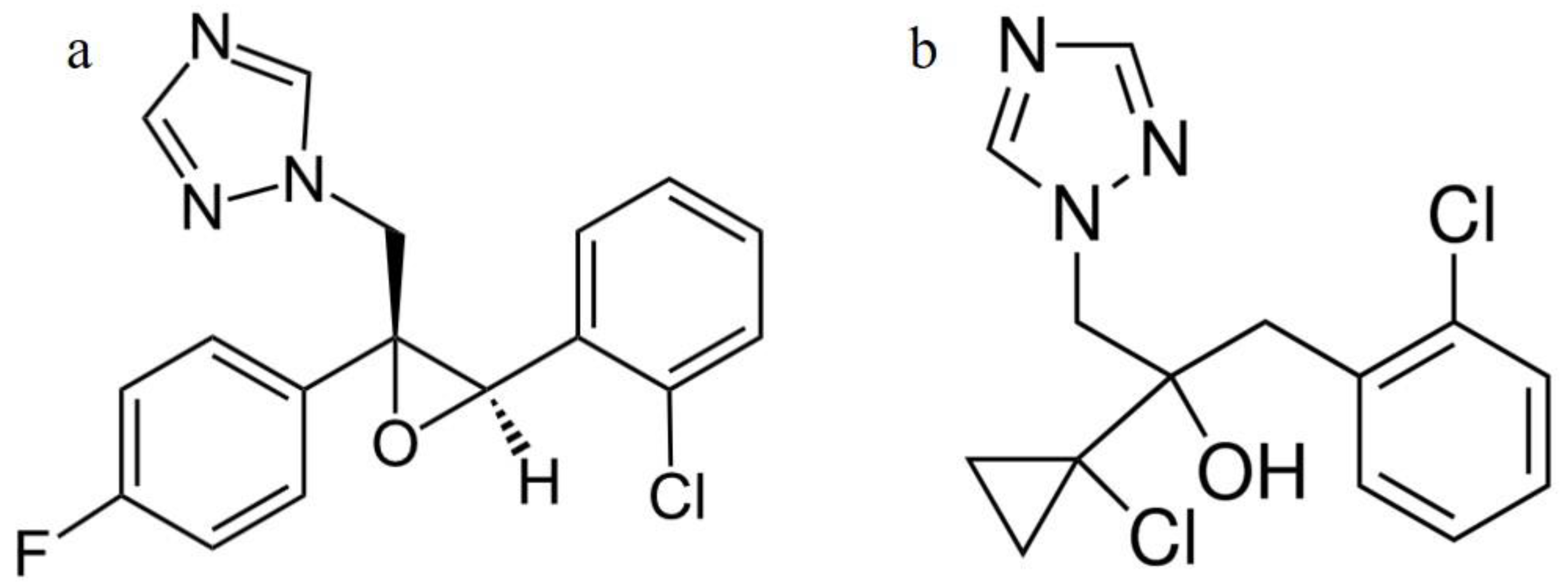
Interaction of Conazole Pesticides Epoxiconazole and Prothioconazole with Human and Bovine Serum Albumin Studied Using Spectroscopic Methods and Molecular Modeling
Abstract
:1. Introduction
2. Results and Discussion
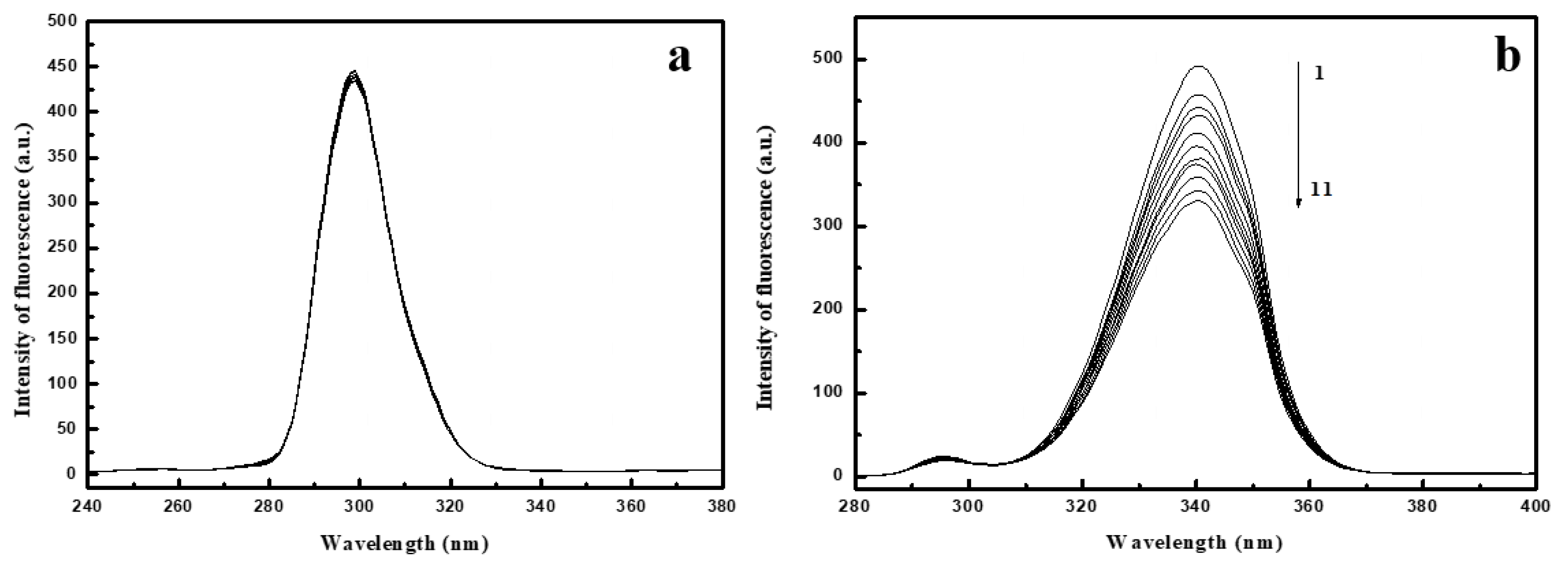
2.1. Fluorescence Spectra of Conazoles–SA System
2.2. Determination of Binding Parameters
2.3. Thermodynamic Parameters
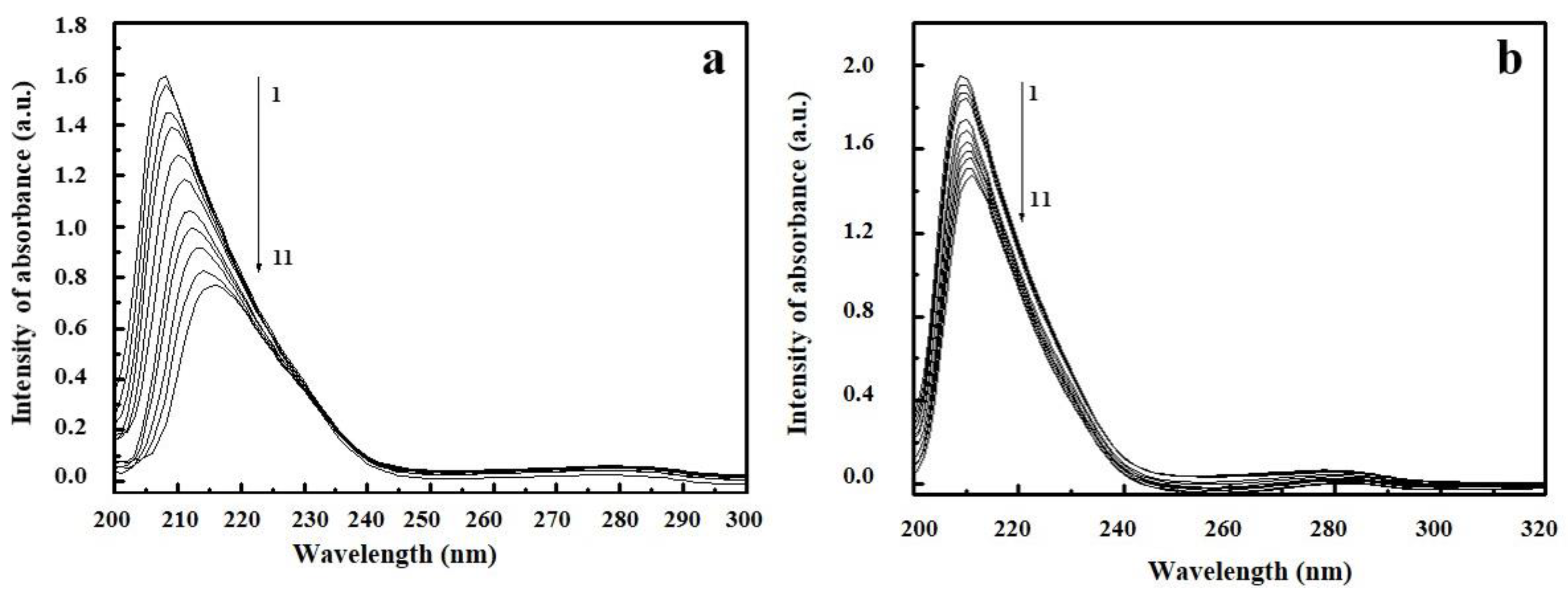
2.4. UV/Vis Spectroscopy
2.5. Synchronous Fluorescence Spectroscopy Studies
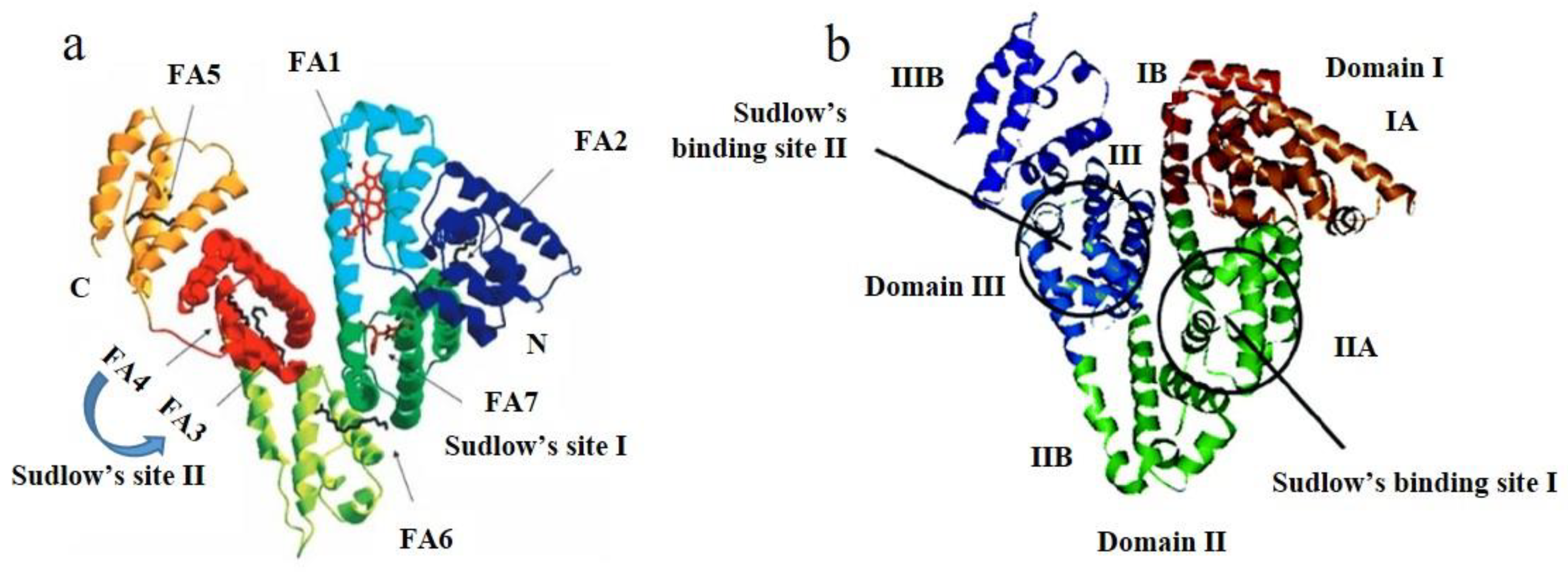
2.6. Site Marker Competitive Experiments
2.7. Three-Dimensional Fluorescence Spectroscopy
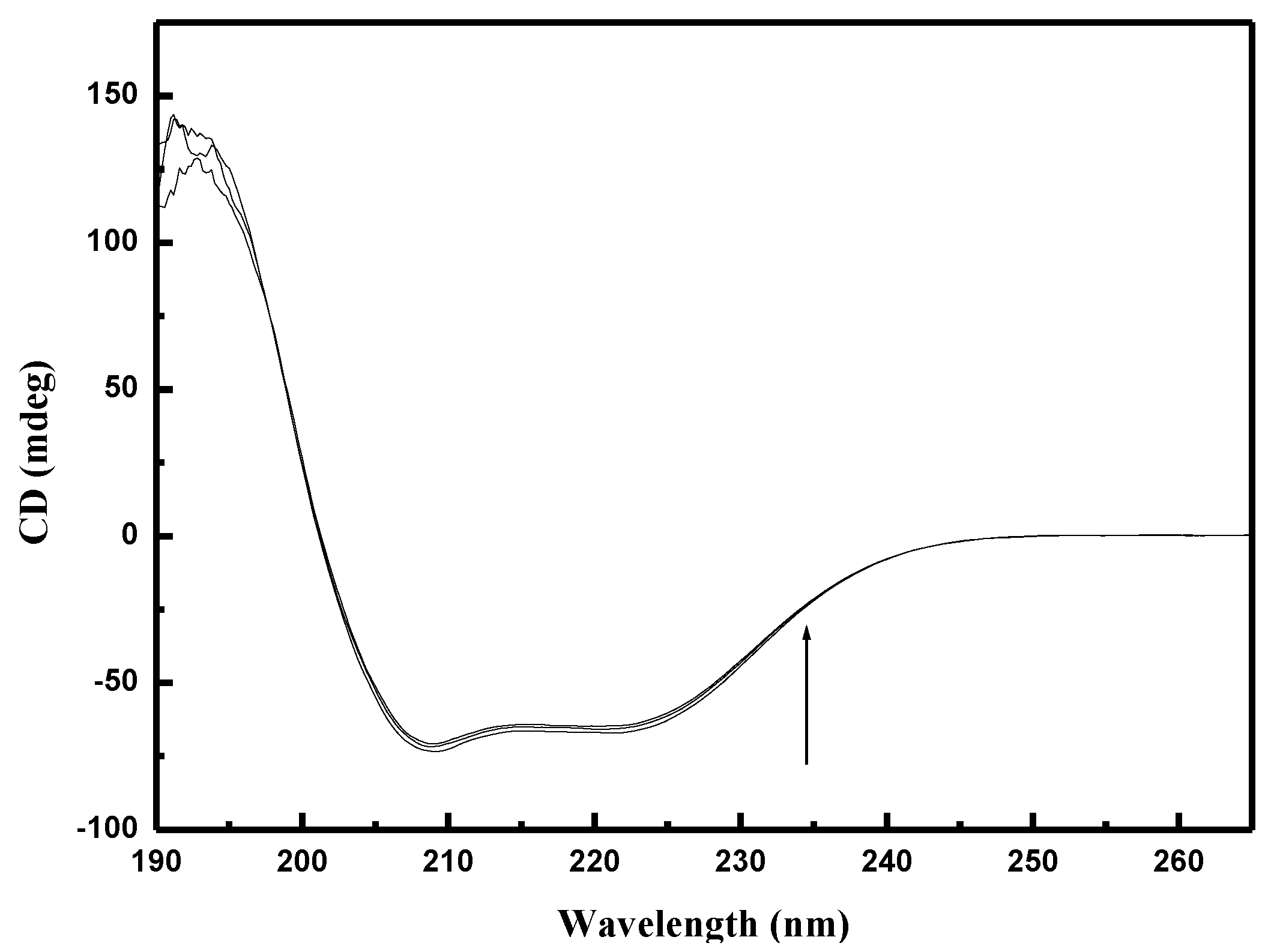
2.8. Circular Dichroism
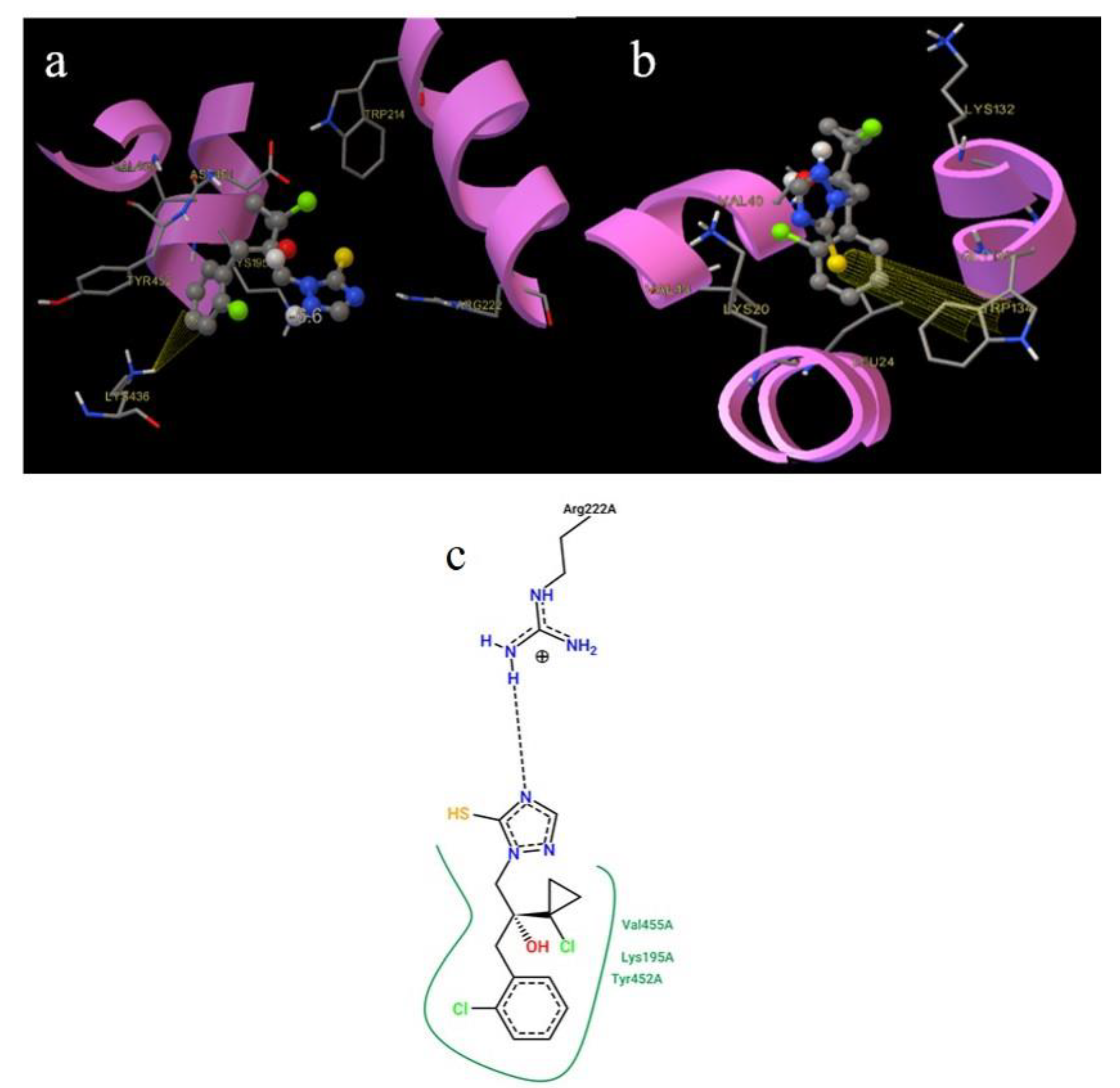
2.9. Molecular Docking
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Fluorescence Measurements
3.3. UV/Vis Measurements
3.4. Circular Dichroism (CD) Studies
3.5. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- U.S. Environmental Protection Agency. Available online: https://www3.epa.gov/pesticides/chem_search/reg_actions/registration/fs_PC-123909_01-Aug-06.pdf (accessed on 10 August 2006).
- Goetz, A.K.; Ren, H.; Schmid, J.E.; Blystone, C.R.; Thillainadarajah, I.; Best, D.S.; Nichols, H.P.; Strader, L.F.; Wolf, D.C.; Narotsky, M.G.; et al. Disruption of testosterone homeostasis as a mode of action for the reproductive toxicity of triazole fungicides in the male rat. Toxicol. Sci. 2006, 95:1, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Menegola, E.; Broccia, M.L.; Di Renzo, F.; Giavini, E. Postulated pathogenic pathway in triazole fungicide induced dysmorphogenic effects. Reprod. Toxicol. 2006, 22, 186–195. [Google Scholar] [CrossRef]
- Arthur, J.; Acierto, A.M.; Assaad, A. Epoxiconazole: Tolerance on Bananas and Coffee Exported to the US; US Environmental Protection Agency, Office of Pesticide Programs, Health Effect Division: Washington, DC, USA, 2005. [Google Scholar]
- Schneider, S.; Hofmann, T.; Stinchcombe, S.; Moreno, M.C.R.; Fegert, I.; Strauss, V.; Groters, S.; Fabian, E.; Thiaener, J.; Fussell, K.C.; et al. Species differences in developmental toxicity of epoxiconazole and its relevance to humans. Birth Defects Res. B Dev. Reprod. Toxicol. 2013, 98, 230–246. [Google Scholar] [CrossRef]
- Taxvig, C.; Hass, U.; Axelstad, M.; Dalgaard, M.; Boberg, J.; Andeasen, H.R.; Vinggaard, A.M. Endocrine-disrupting activities in vivo of the fungicides tebuconazole and epoxiconazole. Toxicol. Sci. 2007, 100, 464–473. [Google Scholar] [CrossRef] [Green Version]
- Silva, D.; Cortez, C.M.; Cunha-Bastos, J.; Louro, S.R. Methyl parathion interaction with human and bovine serum albumin. Toxicol. Lett. 2004, 147, 53–61. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, B.; He, Z.; Li, L.; Zhang, Q.; Kaziem, A.E.; Wang, M. Stereoselective bioactivity of the chiral triazole fungicide prothioconazole and its metabolite. Pestic. Biochem. Phys. 2019, 160, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Ghisari, M.; Long, M.; Tabbo, A.; Bonefeld-Jørgensen, E.C. Effects of currently used pesticides and their mixtures on the function of thyroid hormone and aryl hydrocarbon receptor in cell culture. Toxicol. Appl. Pharmacol. 2015, 284, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Schwarzbacherová, V.; Šiviková, K.; Drážovská, M.; Dianovský, J. Evaluation of DNA damage and cytotoxicity induced by triazole fungicide in cultured bovine lymphocytes. Caryologia 2015, 68, 233–238. [Google Scholar] [CrossRef]
- Carter, D.C.; Ho, J.X. Structure of serum albumin. Adv. Protein Chem. 1994, 45, 153–203. [Google Scholar] [PubMed]
- Tian, J.; Liu, J.; He, W.; Hu, Z.; Yao, X.; Chen, X. Probing the binding of scutellarin to human serum albumin by circular dichroism, fluorescence spectroscopy, FTIR, and molecular modelling method. Biomacromolecules 2004, 5, 1956–1961. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.E.; Christ, D.D. Plasma protein binding of drugs. Annu. Rep. Med. Chem. 1996, 31, 327–336. [Google Scholar]
- Bolel, P.; Mahapatra, N.; Halder, M. Optical spectroscopic exploration of binding of cochinealred A with two homologous serum albumins. J. Agric. Food Chem. 2012, 60, 3727–3734. [Google Scholar] [CrossRef] [PubMed]
- Mallick, A.; Haldar, B.; Chattopadhyay, N. Spectroscopic investigation on the interaction of ICT probe 3-acetyl-4-oxo-6,7-dihydro-12Hindolo-[2,3-a]quinolizine with serum albumins. J. Phys. Chem. B 2005, 109, 14683–14690. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Lü, W.J.; Su, L.; Shan, Z.J.; Chen, X.G. Binding of phthalate plasticizers to human serum albumin in vitro: A multispectroscopic approach and molecular modeling. J. Agric. Food Chem. 2012, 60, 1135–1145. [Google Scholar] [CrossRef]
- Sułkowska, A.; Maciążek-Jurczyk, M.; Bojko, B.; Rownicka, J.; Zubik-Skupień, I.; Temba, E.; Pentak, D.; Sułkowski, W.W. Competitive binding of phenylbutazone and colchicine to serum albumin in multidrug therapy: A spectroscopic study. J. Mol. Struct. 2008, 881, 97–106. [Google Scholar] [CrossRef]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Sułkowska, A. Interaction of drugs with bovine and human serum albumin. J. Mol. Struct. 2008, 614, 227–232. [Google Scholar] [CrossRef]
- Meloun, B.; Morávek, L.; Kostka, V. Complete amino acid sequence of human serum albumin. FEBS Lett. 1975, 58, 134–137. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Guo, J.; Xu, B.; Chen, Z. Binding of chlorpyrifos and cypermethrin to blood proteins. Pestic. Biochem. Phys. 2006, 85, 110–114. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Z.; Zhang, D.; Lv, J. Flow-injection analysis chemiluminescence detection combined with microdialysis sampling for studying protein binding of drug. Talanta 2001, 53, 835–841. [Google Scholar] [CrossRef]
- Fasano, M.; Curry, S.; Terreno, E.; Galliano, M.; Fanali, G.; Narciso, P.; Notari, S.; Ascenzi, P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life 2005, 57, 787–796. [Google Scholar] [CrossRef]
- Ascenzi, P.; Fasano, M. Allostery in a monomeric protein: The case of human serum albumin. Biophys. Chem. 2010, 148, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Fanali, G.; Masi, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human serum albumin: From bench to bedside. Mol. Aspects Med. 2012, 33, 209–290. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.D.J.B.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal structure analysis of warfarin binding to human serum albumin anatomy of drug site I. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef] [Green Version]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Zsila, F.; Bikari, Z.; Malik, D.; Hari, P.; Pechan, I.; Berces, A.; Hazai, E. Evaluation of drug–human serum albumin binding interactions with support vector machine aided online automated docking. Bioinformatics 2011, 27, 1806–1813. [Google Scholar] [CrossRef] [Green Version]
- Patra, S.; Santhosh, K.; Pabbathi, A.; Samanta, A. Diffision of organic dyes in bovine serum albumin solution studied by fluorescence correlation spectroscopy. RSC Adv. 2012, 2, 6079–6086. [Google Scholar] [CrossRef]
- Peters, T.J. The albumin molecule: Its structure and chemical properties. In All about Albumin: Biochemistry, Genetics and Medical Applications; Peters, T.J., Ed.; Academic Press: San Diego, CA, USA, 1996; pp. 9–75. [Google Scholar]
- Zhang, C.X.; Lippard, S.J. New metal complexes as potential therapeutics. Curr. Opin. Chem. Biol. 2003, 7, 481–489. [Google Scholar] [CrossRef]
- Meggers, E. Targeting proteins with metal complexes. Chem. Commun. 2009, 9, 1001–1010. [Google Scholar] [CrossRef] [Green Version]
- Alyar, H.; Alyar, S.; Űnal, A.; Őzbek, N.; Sahin, E.; Karacan, N. Synthesis, characterization and antimicrobial activity of mtoluenesulfonamide, N, N′-1, 2-ethanediylbis (mtsen) and [Cu (II)(phenanthroline) 2] mtsen complex. J. Mol. Struct. 2012, 1028, 116–125. [Google Scholar] [CrossRef]
- Krishnamoorthy, P.; Sathyadevi, P.; Cowley, A.H.; Butorac, R.R.; Dharmaraj, N. Evaluation of DNA binding, DNA cleavage, protein binding and in vitro cytotoxic activities of bivalent transition metal hydrazone complexes. Eur. J. Med. Chem. 2011, 46, 3376–3387. [Google Scholar] [CrossRef] [PubMed]
- Samari, F.; Hemmateenejad, B.; Shamsipur, M.; Rashidi, M.; Samouei, H. Affinity of two novel five-coordinated anticancer Pt (II) complexes to human and bovine serum albumins: A spectroscopic approach. Inorg. Chem. 2012, 51, 3454–3464. [Google Scholar] [CrossRef] [PubMed]
- Sathyadevi, P.; Krishnamoorthy, P.; Butorac, R.R.; Cowley, A.H.; Bhuvanesh, N.S.P.; Dharmarajn, N. Effect of substitution and planarity of the ligand on DNA/BSA interaction, free radical scavenging and cytotoxicity of diamagnetic Ni (II) complexes: A systematic investigation. Dalton Trans. 2011, 40, 9690–9702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, F.; Xie, C.Z.; Zhang, Y.W.; Qiao, Z.; Qiao, X.; Xu, J.Y.; Yan, S.P. Two new dicopper (II) complexes with oxamido-bridged ligand: Synthesis, crystal structures, DNA binding/cleavage and BSA binding activity. J. Inorg. Biochem. 2012, 115, 78–86. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Tang, B.P.; Zhang, H.M.; Zhou, Q.H.; Zhang, G.C. Studies on the interaction between imidacloprid and human serum albumin: Spectroscopic approach. J. Photochem. Photobiol. B 2009, 94, 183–190. [Google Scholar] [CrossRef]
- Yan, C.N.; Mei, P.; Guan, Z.J.; Liu, Y. Studies on thermodynamics features of the interaction between imidacloprid and bovine serum albumin. Chin. J. Chem. 2007, 25, 1085–1089. [Google Scholar] [CrossRef]
- Zhang, J.; Zhuang, S.; Tong, C.; Liu, W. Probing the molecular interaction of triazole fungicides with human serum albumin by multispectroscopic techniques and molecular modeling. J. Agric. Food Chem. 2013, 61, 7203–7211. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Quenching of fluorescence. In Principles of Fluorescence Spectroscopy, 3rd ed.; Lakowicz, J.R., Ed.; Plenum Press: New York, NY, USA, 1983; pp. 199–227. [Google Scholar]
- Lu, Y.; Dasog, M.; Leontowich, A.F.; Scott, R.W.; Paige, M.F. Fluorescently labeled gold nanoparticles with minimal fluorescence quenching. J. Phys. Chem. C 2010, 114, 17446–17454. [Google Scholar] [CrossRef]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981, 114, 199–227. [Google Scholar] [CrossRef]
- Bhattacharya, A.A.; Grüne, T.; Curry, S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Némethy, G.; Scheraga, H.A. The structure of water and hydrophobic bonding in proteins. iii. The thermodynamic properties of hydrophobic bonds in proteins1, 2. J. Phys. Chem. 1962, 66, 1773–1789. [Google Scholar]
- Congdon, R.W.; Muth, G.W.; Splittgerber, A.G. The binding interaction of Coomassie blue with proteins. Anal. Biochem. 1993, 213, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Kragh-Hansen, U. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981, 33, 17. [Google Scholar]
- Tao, W.S.; Li, W.; Jiang, Y.M. Protein Molecular Basic; The People’s Education Press: Beijing, China, 1981; p. 256. [Google Scholar]
- Cui, F.L.; Fan, J.; Li, J.P.; Hu, Z.D. Interactions between 1-benzoyl-4-p-chlorophenyl thiosemicarbazide and serum albumin: Investigation by fluorescence spectroscopy. Bioorg. Med. Chem. 2004, 12, 151–157. [Google Scholar] [CrossRef]
- Vekshin, N.L. Division of tyrosine and tryptophan fluorescent components by synchronous scanning method. Биофизика 1996, 41, 1179. [Google Scholar]
- Xu, T.; Guo, X.; Zhang, L.; Pan, F.; Lv, J.; Zhang, Y.; Jin, H. Multiple spectroscopic studies on the interaction between olaquindox, a feed additive, and bovine serum albumin. Food Chem. Toxicol. 2012, 50, 2540–2546. [Google Scholar] [CrossRef]
- Klajnert, B.; Bryszewska, M. Fluorescence studies on PAMAM dendrimers interactions with bovine serum albumin. Bioelectrochemistry 2002, 55, 33–35. [Google Scholar] [CrossRef]
- Pan, X.; Qin, P.; Liu, R.; Wang, J. Characterizing the interaction between tartrazine and two serum albumins by a hybrid spectroscopic approach. J. Agric. Food Chem. 2011, 59, 6650–6656. [Google Scholar] [CrossRef]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Spectroscopic techniques in the study of protein binding. A fluorescence technique for the evaluation of the albumin binding and displacement of warfarin and warfarin-alcohol. Clin. Exp. Pharmacol. Physiol. 1975, 2, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, M.G.; Poppi, R.J. Determination of doxorubicin in human plasma by excitation–emission matrix fluorescence and multi-way analysis. Anal. Chim. Acta 2003, 493, 69–81. [Google Scholar] [CrossRef]
- Kalaiarasi, G.; Rajkumar, S.R.J.; Dharani, S.; Malecko, J.G.; Prabhakaran, R. An investigation on 3-acetyl-7-methoxy-coumarin Schi_ bases and their Ru (II) metallates with potent antiproliferative activity and enhanced LDH and NO release. RSC Adv. 2018, 8, 1539–1561. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wang, Y.; Tang, L.; Liu, H.; Chen, W.; Zheng, Z.; Zou, G. Binding of puerarin to human serum albumin: A spectroscopic analysis and molecular docking. J. Fluoresc. 2008, 18, 433–442. [Google Scholar] [CrossRef]
- Lu, Z.X.; Cui, T.; Shi, Q.L. Applications of Circular Dichroism and Optical Rotatory Dispersion in Molecular Biology; Science Press: Beijing, China, 1987; pp. 79–82. [Google Scholar]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.T.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876. [Google Scholar] [CrossRef]
- Luis-Jeune, C.; Andrade-Navarro, M.A.; Perez-Iratxeta, C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 2012, 80, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Sun, X.; Ma, X. Interaction of tebuconazole with bovine serum albumin: Determination of the binding mechanism and binding site by spectroscopic methods. J. Environ. Sci. Health B 2020, 55, 1–8. [Google Scholar] [CrossRef]
- Alsalme, A.; Khan, R.A.; Alkathiri, A.M.; Ali, M.S.; Tabassum, S.; Jaafar, M.; Al-Lohedan, H.A. β-carboline silver compound binding studies with human serum albumin: A comprehensive multispectroscopic analysis and molecular modeling study. Bioinorg. Chem. Appl. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Feng, Q.; Cui, F.; Xing, W.; Zhang, G.; Yao, X. Interaction of 3′-azido-3′-deamoni daunorubicin with human serum albumin: Investigation by fluorescence spectroscopy and molecular modeling methods. Bioorg. Med. Chem. Lett. 2010, 20, 6899–6904. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.; Nusrat, S.; Alam, P.; Malik, S.; Chaturvedi, S.K.; Ajmal, M.R.; Abdelhameed, A.S. Investigating the site selective binding of busulfan to human serum albumin: Biophysical and molecular docking approaches. Int. J. Biol. Macromol. 2018, 107, 1414–1421. [Google Scholar] [CrossRef]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999, 12, 439–446. [Google Scholar] [CrossRef]
- Lee, P.; Wu, X. Modifications of human serum albumin and their binding effect. Curr. Pharm. Des. 2015, 21, 1862–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed] [Green Version]
- Stierand, K.; Maaß, P.; Rarey, M. Molecular Complexes at a Glance: Automated Generationa of two-dimensional Complex Diagrams. Bioinformatics 2006, 22, 1710–1716. [Google Scholar] [CrossRef] [PubMed]

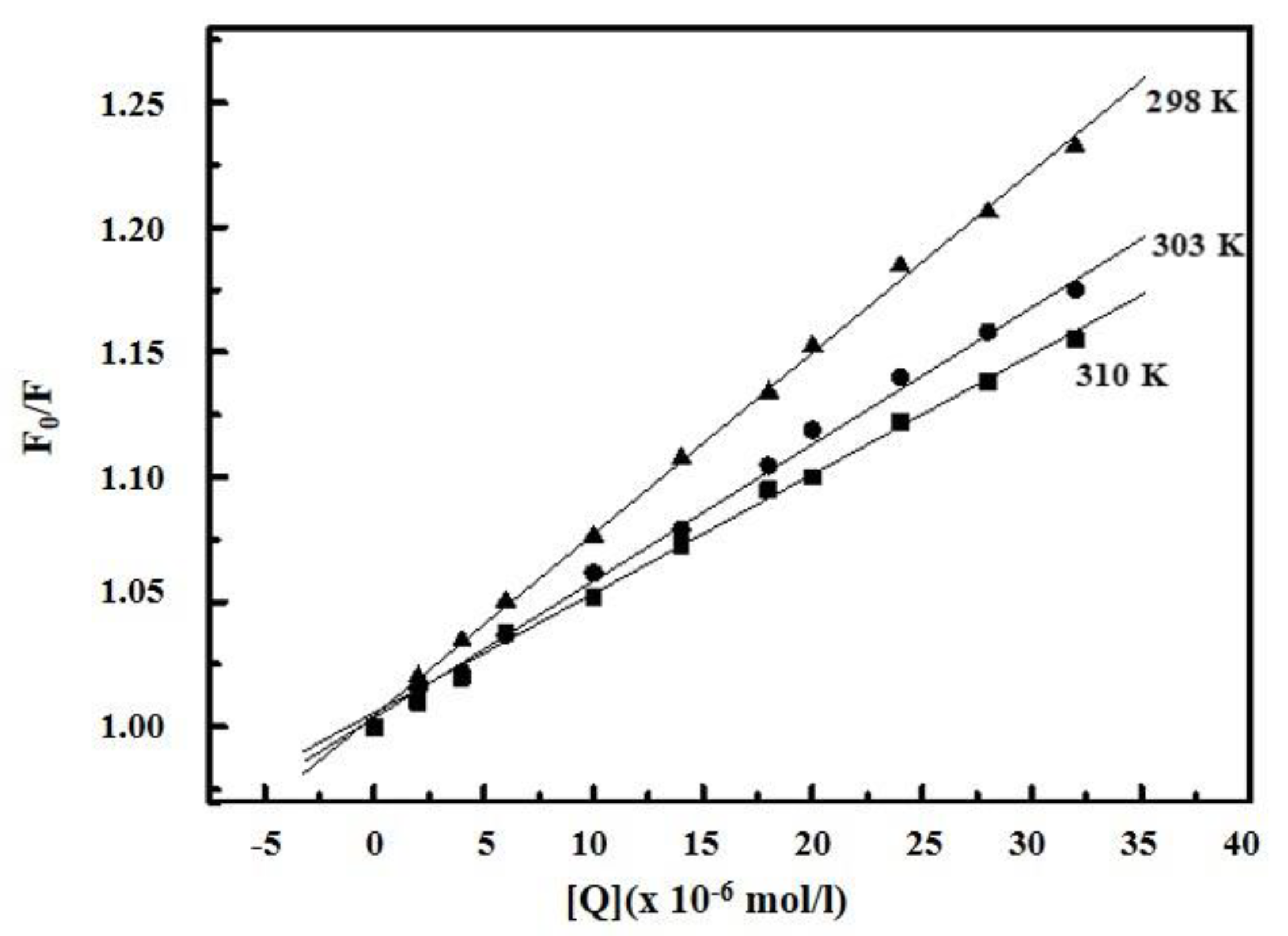
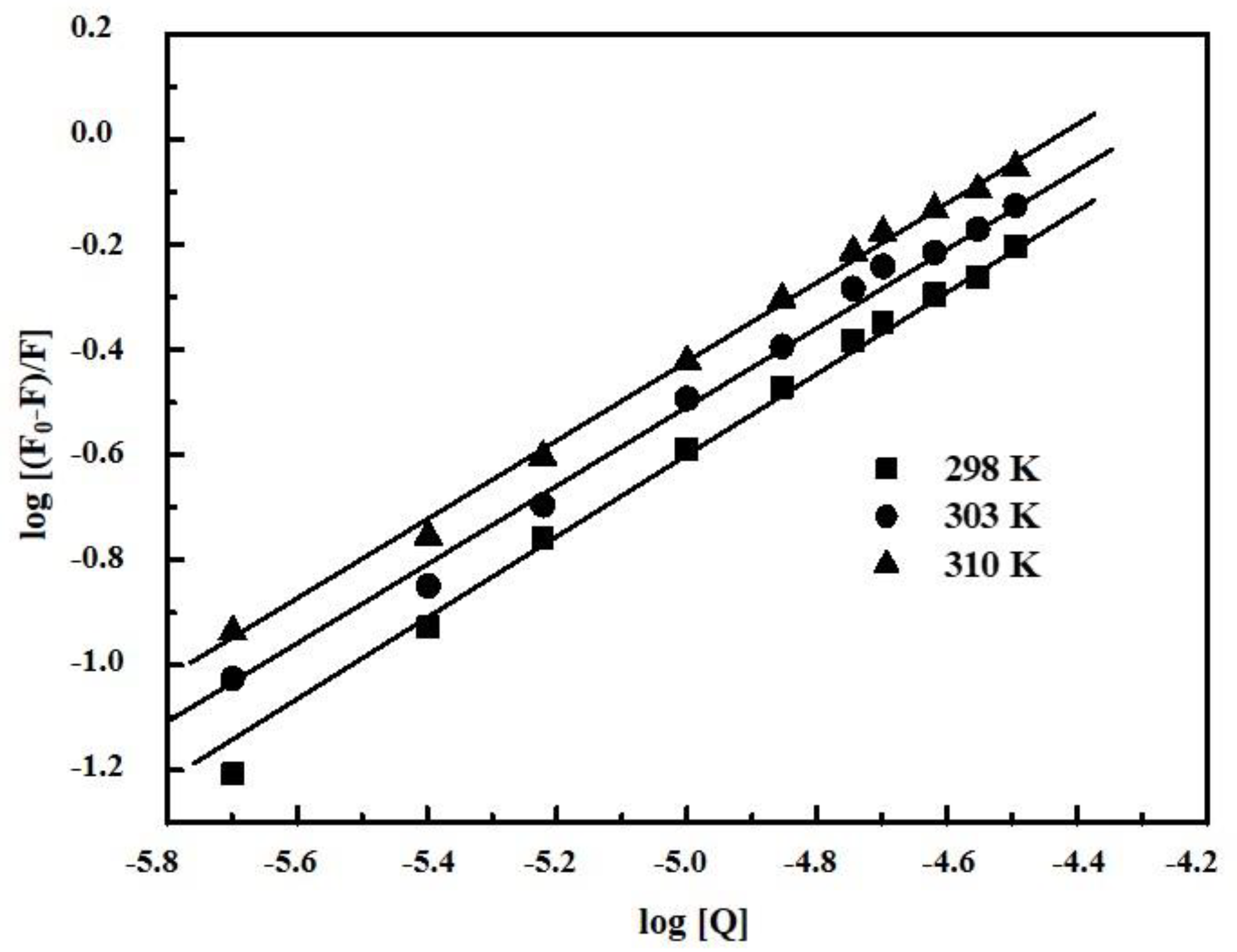




| Complex | T (K) | KA (L/mol) | n | ∆G (kJ/mol) | ∆H (kJ/mol) | ∆S (J/mol.K) |
|---|---|---|---|---|---|---|
| EPX/HSA | 298 | 6.22 × 104 | 0.98 | −26.51 | −105.74 | −265.88 |
| 303 | 2.04 × 104 | 0.95 | −25.18 | |||
| 310 | 8.14 × 103 | 0.91 | −23.32 | |||
| EPX/BSA | 298 | 3.80 × 104 | 0.90 | −22.14 | −81.54 | −199.35 |
| 303 | 1.04 × 104 | 0.96 | −21.13 | |||
| 310 | 7.90 × 103 | 0.95 | −19.73 | |||
| PTC/HSA | 298 | 5.75 × 105 | 1.11 | −32.95 | −64.39 | −105.50 |
| 303 | 3.80 × 105 | 1.07 | −32.42 | |||
| 310 | 2.08 × 105 | 0.98 | −31.68 | |||
| PTC/BSA | 298 | 6.45 × 105 | 1.06 | −32.94 | −84.85 | −174.18 |
| 303 | 2.89 × 105 | 0.99 | −32.07 | |||
| 310 | 1.66 × 105 | 0.95 | −30.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golianová, K.; Havadej, S.; Verebová, V.; Uličný, J.; Holečková, B.; Staničová, J. Interaction of Conazole Pesticides Epoxiconazole and Prothioconazole with Human and Bovine Serum Albumin Studied Using Spectroscopic Methods and Molecular Modeling. Int. J. Mol. Sci. 2021, 22, 1925. https://doi.org/10.3390/ijms22041925
Golianová K, Havadej S, Verebová V, Uličný J, Holečková B, Staničová J. Interaction of Conazole Pesticides Epoxiconazole and Prothioconazole with Human and Bovine Serum Albumin Studied Using Spectroscopic Methods and Molecular Modeling. International Journal of Molecular Sciences. 2021; 22(4):1925. https://doi.org/10.3390/ijms22041925
Chicago/Turabian StyleGolianová, Katarína, Samuel Havadej, Valéria Verebová, Jozef Uličný, Beáta Holečková, and Jana Staničová. 2021. "Interaction of Conazole Pesticides Epoxiconazole and Prothioconazole with Human and Bovine Serum Albumin Studied Using Spectroscopic Methods and Molecular Modeling" International Journal of Molecular Sciences 22, no. 4: 1925. https://doi.org/10.3390/ijms22041925
APA StyleGolianová, K., Havadej, S., Verebová, V., Uličný, J., Holečková, B., & Staničová, J. (2021). Interaction of Conazole Pesticides Epoxiconazole and Prothioconazole with Human and Bovine Serum Albumin Studied Using Spectroscopic Methods and Molecular Modeling. International Journal of Molecular Sciences, 22(4), 1925. https://doi.org/10.3390/ijms22041925








