Qualitative and Quantitative Comparison of Plasma Exosomes from Neonates and Adults
Abstract
1. Introduction
2. Results
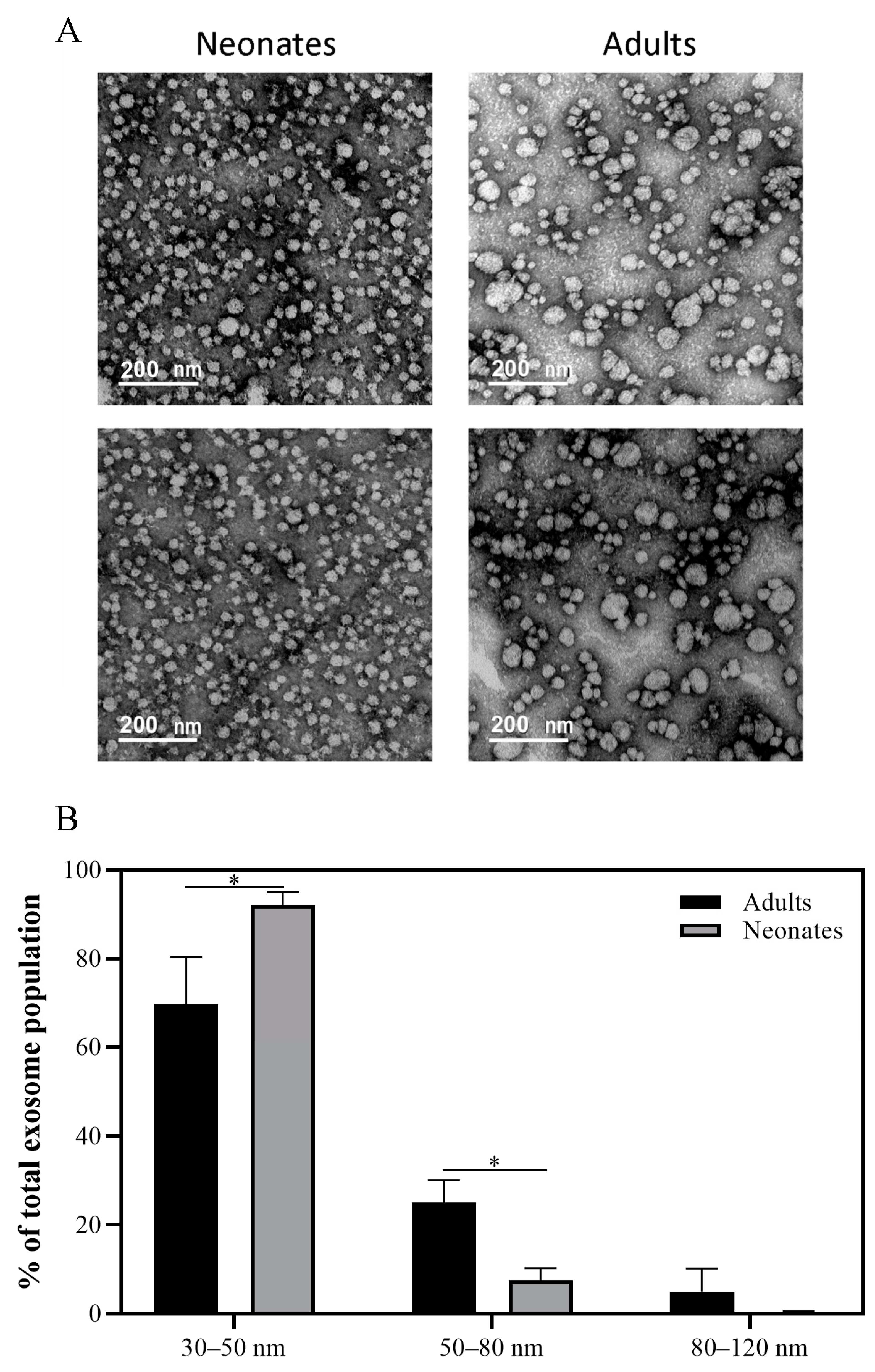
2.1. Characterization of Plasma Exosomes from Neonates and Adults
2.2. Differential Protein Expression
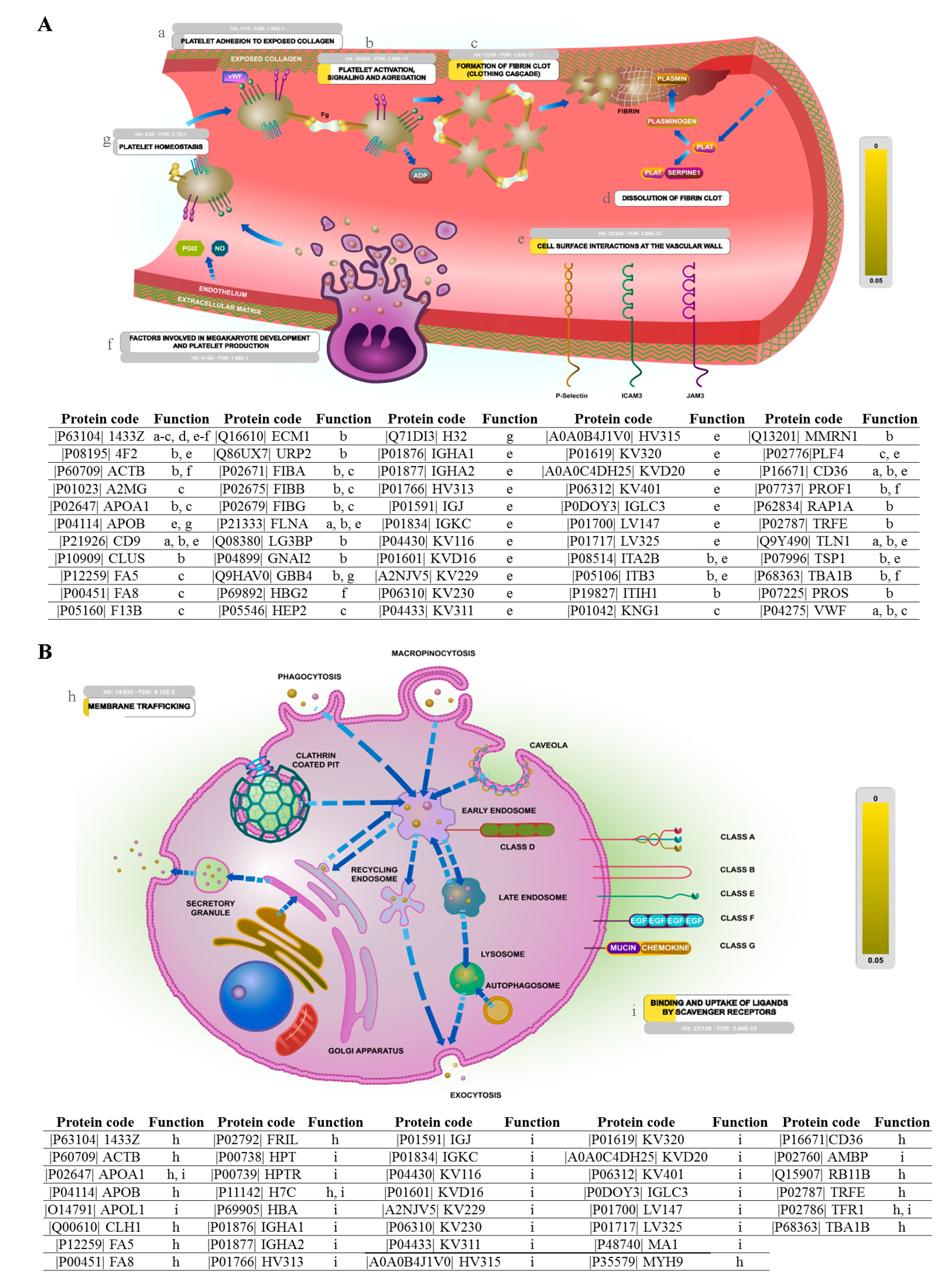
2.3. Biological Pathway Analysis
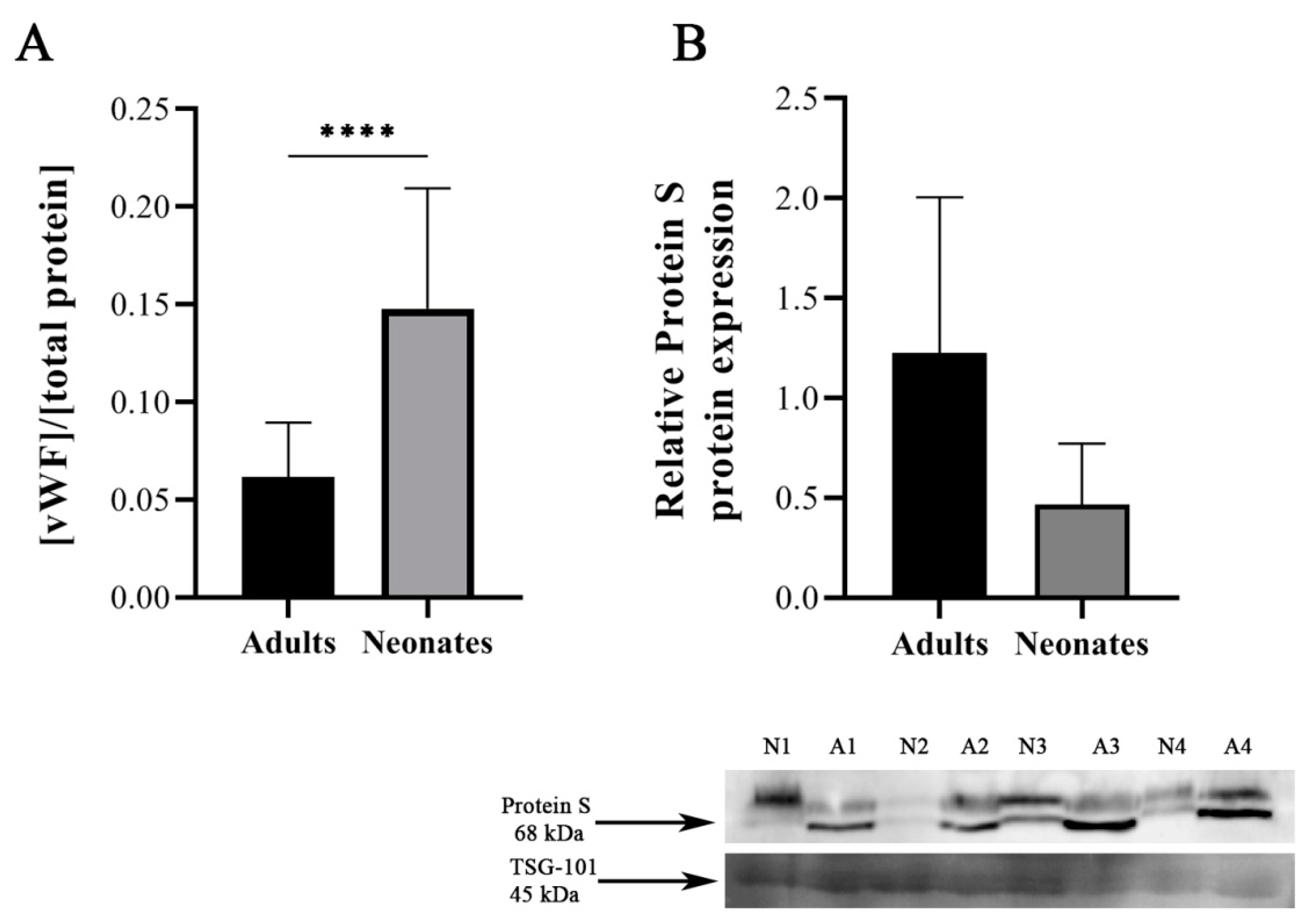
2.4. Validation of Differential Protein Expression in Independent Samples
3. Discussion
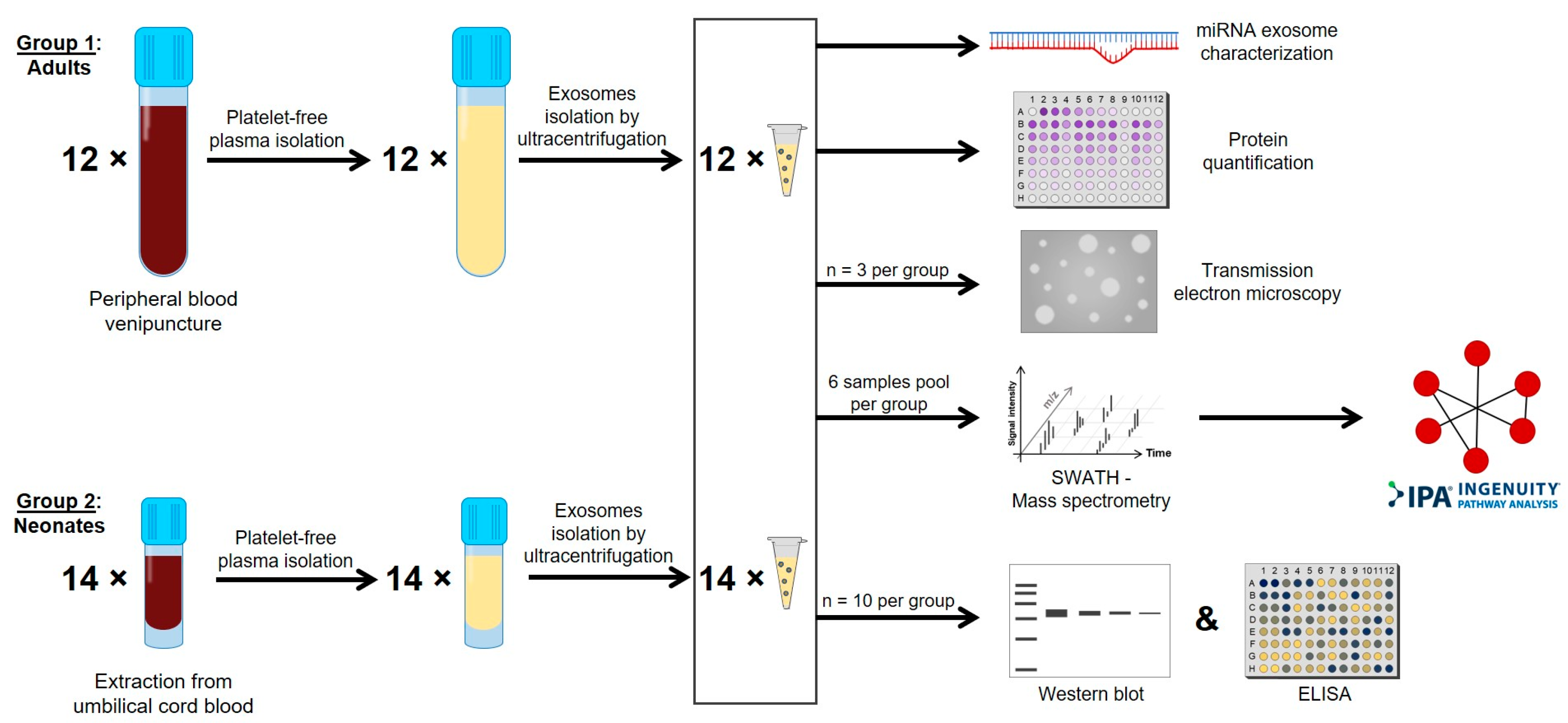
4. Materials and Methods
4.1. Sample Collection
4.2. Platelet-Free Plasma Isolation
4.3. Plasma Exosomes Isolation and Characterization
4.4. Quantitative Proteomic Studies by Liquid Chromatography Mass Spectrometry (LC-MS/MS) Using Sequential Window Acquisition of All Theoretical Mass Spectrometry (SWATH MS) Method
4.4.1. Protein Digestion
4.4.2. Mass Spectrometric Analysis by Sequential Window Acquisition of All Theoretical Mass Spectra (SWATH MS)
4.5. Functional Enrichment and Interaction Network Analysis
4.6. Von Willebrand Factor and Protein S Determination
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A2MG | α2-macroglobulin |
| C4BP | C4b-binding protein |
| CB | Cord blood |
| DDA | Data-dependent acquisition |
| DLS | Dynamic light scattering |
| ELISA | Enzyme-linked immunosorbent assay |
| FDR | False discovery rate |
| FV and FVIII | Factor VIII and Factor V |
| GNAI2 | Guanine nucleotide-binding protein G(i) subunit α-2 |
| MVBs | Multivesicular bodies |
| PBS | Phosphate-buffered saline |
| PCA | Principal components analysis |
| PF4 | Platelet factor 4 |
| RAB11B | Ras-related protein Rab-11B |
| RAP1A | Ras-related protein Rap-1A |
| SCFD2 | Sec 1 family domain containing 2 |
| SNARE | Soluble N-ethylmaleimide-sensitive fusion Attachment protein (SNAP) REceptors |
| TEM | Transmission electron microscopy |
| TLN1 | Talin-1 |
| TSG-101 | Tumor susceptibility gene 101 protein |
| vWF | von Willebrand factor |
References
- Sola-Visner, M. Platelets in the neonatal period: Developmental differences in platelet production, function, and hemostasis and the potential impact of therapies. Hematol. Am. Soc. Hematol. Educ. Program 2012, 2012, 506–511. [Google Scholar] [CrossRef]
- Corby, D.G.; O’Barr, T.P. Decreased alpha-adrenergic receptors in newborn platelets: Cause of abnormal response to epinephrine. Dev. Pharm. 1981, 2, 215–225. [Google Scholar] [CrossRef]
- Gelman, B.; Setty, B.N.Y.; Chen, D.; Amin-Hanjani, S.; Stuart, M.J. Impaired mobilization of intracellular calcium in neonatal platelets. Pediatr. Res. 1996, 39, 692–696. [Google Scholar] [CrossRef]
- Israels, S.J.; Cheang, T.; Roberston, C.; McMillan-Ward, E.M.; McNicol, A. Impaired signal transduction in neonatal platelets. Pediatr. Res. 1999, 45, 687–691. [Google Scholar] [CrossRef][Green Version]
- Schlagenhauf, A.; Schweintzger, S.; Birner-Gruenberger, R.; Leschnik, B.; Muntean, W. Newborn platelets: Lower levels of protease-activated receptors cause hypoaggregability to thrombin. Platelets 2010, 21, 641–647. [Google Scholar] [CrossRef]
- Saxonhouse, M.A.; Garner, R.; Mammel, L.; Li, Q.; Muller, K.E.; Greywoode, J.; Miller, C.; Sola-Visner, M. Closure times measured by the platelet function analyzer PFA-100® Are longer in neonatal blood compared to cord blood samples. Neonatology 2010, 97, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Sitaru, A.G.; Holzhauer, S.; Speer, C.P.; Singer, D.; Obergfell, A.; Walter, U.; Grossmann, R. Neonatal platelets from cord blood and peripheral blood. Platelets 2005, 16, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.T.; Corken, A.; Ware, J. Platelets at the interface of thrombosis, inflammation, and cancer. Blood 2015, 126, 582–588. [Google Scholar] [CrossRef]
- Ferrer-Marin, F.; Stanworth, S.; Josephson, C.; Sola-Visner, M. Distinct differences in platelet production and function between neonates and adults: Implications for platelet transfusion practice. Transfusion 2013, 53, 2814–2821. [Google Scholar] [CrossRef]
- Liu, Z.J.; Hoffmeister, K.M.; Hu, Z.; Mager, D.E.; Ait-Oudhia, S.; Debrincat, M.A.; Pleines, I.; Josefsson, E.C.; Kile, B.T.; Italiano, J.; et al. Expansion of the neonatal platelet mass is achieved via an extension of platelet lifespan. Blood 2014, 123, 3381–3389. [Google Scholar] [CrossRef] [PubMed]
- Baker-Groberg, S.M.; Lattimore, S.; Recht, M.; McCarty, O.J.T.; Haley, K.M. Assessment of neonatal platelet adhesion, activation, and aggregation. J. Thromb. Haemost. 2016, 14, 815–827. [Google Scholar] [CrossRef]
- Margraf, A.; Nussbaum, C.; Sperandio, M. Ontogeny of platelet function. Blood Adv. 2019, 3, 692–703. [Google Scholar] [CrossRef]
- Palma-Barqueros, V.; Torregrosa, J.M.; Caparrós-Pérez, E.; Mota-Pérez, N.; Bohdan, N.; Llanos, M.D.C.; Begonja, A.J.; Sola-Visner, M.; Vicente, V.; Teruel-Montoya, R.; et al. Developmental differences in platelet inhibition response to prostaglandin E1. Neonatology 2019, 117, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Urban, D.; Pluthero, F.G.; Christensen, H.; Baidya, S.; Rand, M.L.; Das, A.; Shah, P.S.; Chitayat, D.; Blanchette, V.S.; Kahr, W.H.A. Decreased numbers of dense granules in fetal and neonatal platelets. Haematologica 2017, 102, e36–e38. [Google Scholar] [CrossRef]
- Caparrós-Pérez, E.; Teruel-Montoya, R.; Palma-Barquero, V.; Torregrosa, J.M.; Blanco, J.E.; Delgado, J.L.; Lozano, M.L.; Vicente, V.; Sola-Visner, M.; Rivera, J.; et al. Down regulation of the Munc18b-syntaxin-11 complex and Β1-Tubulin impairs secretion and spreading in neonatal platelets. Thromb. Haemost. 2017, 117, 2079–2091. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. A Unique SNARE Machinery for Exocytosis of cytotoxic granules and platelets granules. Mol. Membr. Biol. 2015, 32, 120–126. [Google Scholar] [CrossRef]
- Ren, Q.; Ye, S.; Whiteheart, S.W. The platelet release reaction: Just when you thought platelet secretion was simple. Curr. Opin. Hematol. 2008, 15, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Fader, C.M.; Sánchez, D.G.; Mestre, M.B.; Colombo, M.I. TI-VAMP/VAMP7 and VAMP3/Cellubrevin: Two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta 2009, 1793, 1901–1916. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef]
- Marks, M.S. SNARing platelet granule secretion. Blood 2012, 120, 2355–2357. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, N.M.G.; Hackmann, Y.; Aricò, M.; Griffiths, G.M. Munc18-2 is required for syntaxin 11 localization on the plasma membrane in cytotoxic T-lymphocytes. Traffic 2015, 16, 1330–1341. [Google Scholar] [CrossRef] [PubMed]
- Spessott, W.A.; Sanmillan, M.L.; McCormick, M.E.; Kulkarni, V.V.; Giraudo, C.G. SM protein munc18-2 facilitates transition of syntaxin 11-mediated lipid mixing to complete fusion for t-lymphocyte cytotoxicity. Proc. Natl. Acad. Sci. USA 2017, 114, E2176–E2185. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Brisson, A.R.; Tan, S.; Linares, R.; Gounou, C.; Arraud, N. Extracellular vesicles from activated platelets: A semiquantitative cryo-electron microscopy and immuno-gold labeling study. Platelets 2017, 28, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Chim, S.S.C.; Shing, T.K.F.; Hung, E.C.W.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.K.; Lo, Y.M.D. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef]
- Dahlbäck, B. Vitamin K-dependent protein S: Beyond the protein C pathway. Semin. Thromb. Hemost. 2018, 44, 176–184. [Google Scholar] [CrossRef]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2, 1–10. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Teruel-Montoya, R.; Luengo-Gil, G.; Vallejo, F.; Yuste, J.E.; Bohdan, N.; García-Barberá, N.; Espín, S.; Martínez, C.; Espín, J.C.; Vicente, V.; et al. Differential MiRNA expression profile and proteome in plasma exosomes from patients with paroxysmal nocturnal hemoglobinuria. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Vallejo, F.; Yuste, J.E.; Teruel-Montoya, R.; Luengo-Gil, G.; Bohdan, N.; Espín, S.; García-Barberá, N.; Martínez, C.; Vicente, V.; Espín, J.C.; et al. First exploratory study on the metabolome from plasma exosomes in patients with paroxysmal nocturnal hemoglobinuria. Thromb. Res. 2019, 183, 80–85. [Google Scholar] [CrossRef]
- Heijnen, B.H.F.G.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and a-granules. Blood J. 1999, 94, 3791–3799. [Google Scholar] [CrossRef]
- O’Driscoll, L. Expanding on exosomes and ectosomes in cancer. N. Engl. J. Med. 2015, 372, 2359–2362. [Google Scholar] [CrossRef]
- Bock, J.B.; Matern, H.T.; Peden, A.A.; Scheller, R.H. A genomic perspective on membrane compartment organization. Nature 2001, 409, 839–841. [Google Scholar] [CrossRef]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef]
- Caparrós-Pérez, E.; Teruel-Montoya, R.; López-Andreo, M.J.; Llanos, M.C.; Rivera, J.; Palma-Barqueros, V.; Blanco, J.E.; Vicente, V.; Martínez, C.; Ferrer-Marín, F. Comprehensive comparison of neonate and adult human platelet transcriptomes. PLoS ONE 2017, 12, e0183042. [Google Scholar] [CrossRef]
- Hardy, A.T.; Palma-Barqueros, V.; Watson, S.K.; Malcor, J.D.; Eble, J.A.; Gardiner, E.E.; Blanco, J.E.; Guijarro-Campillo, R.; Delgado, J.L.; Lozano, M.L.; et al. Significant hypo-responsiveness to GPVI and CLEC-2 agonists in pre-term and full-term neonatal platelets and following immune thrombocytopenia. Thromb. Haemost. 2018, 118, 1009–1020. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, function, and biomedical applications of exosomes. Science 2020, 367, 1–40. [Google Scholar] [CrossRef]
- Basha, S.; Surendran, N.; Pichichero, M. Immune responses in neonates. Expert Rev. Clin. Immunol. 2014, 10, 1171–1184. [Google Scholar] [CrossRef]
- Kowalska, M.A.; Rauova, L.; Poncz, M. Role of the platelet chemokine platelet factor 4 (PF4) in hemostasis and thrombosis. Thromb. Res. 2010, 125, 292–296. [Google Scholar] [CrossRef]
- Offermanns, S. Activation of platelet function through G protein-coupled receptors. Circ. Res. 2006, 99, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Kuhle, S.; Male, C.; Mitchell, L. Developmental hemostasis: Pro- and anticoagulant systems during childhood. Semin. Thromb. Hemost. 2003, 29, 329–338. [Google Scholar] [CrossRef]
- Levine, J.J.; Udall, J.N., Jr.; Evernden, B.A.; Epstein, M.F.; Bloch, K.J. Elevated levels of A2-Macroglobulin-protease complexes in infants. Biol. Neonate 1987, 51, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Uszyński, M.; Zekanowska, E.; Uszyński, W.; Kuczyński, J.; Zyliński, A. Microparticles (MPs), Tissue Factor (TF) and Tissue Factor Inhibitor (TFPI) in cord blood plasma. A preliminary study and literature survey of procoagulant properties of MPs. Eur. J. Obs. Gynecol. Reprod. Biol. 2011, 158, 37–41. [Google Scholar] [CrossRef]
- Schweintzger, S.; Schlagenhauf, A.; Leschnik, B.; Rinner, B.; Bernhard, H.; Novak, M.; Muntean, W. Microparticles in newborn cord blood: Slight elevation after normal delivery. Thromb. Res. 2011, 128, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Curley, A.; Stanworth, S.J.; Willoughby, K.; Fustolo-Gunnink, S.F.; Venkatesh, V.; Hudson, C.; Deary, A.; Hodge, R.; Hopkins, V.; Lopez Santamaria, B.; et al. Randomized trial of platelet-transfusion thresholds in neonates. N. Engl. J. Med. 2019, 380, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Marin, F.; Chavda, C.; Lampa, M.; Michelson, A.D.; Frelinger, A.L.; Sola-Visner, M. Effects of in vitro adult platelet transfusions on neonatal hemostasis. J. Thromb. Haemost. 2011, 9, 1020–1028. [Google Scholar] [CrossRef]
- Clayton, A.; Boilard, E.; Buzas, E.I.; Cheng, L.; Falcón-Perez, J.M.; Gardiner, C.; Gustafson, D.; Gualerzi, A.; Hendrix, A.; Hoffman, A.; et al. Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. J. Extracell. Vesicles 2019, 8, 1647027. [Google Scholar] [CrossRef] [PubMed]
- Maisano, D.; Mimmi, S.; Russo, R.; Fioravanti, A.; Fiume, G.; Vecchio, E.; Nisticò, N.; Quinto, I.; Iaccino, E. Uncovering the exosomes diversity: A window of opportunity for tumor progression monitoring. Pharmaceuticals 2020, 13, 180. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Jensen, O.N.; Podtelejnikov, A.V.; Neubauer, G.; Mortensen, P.; Mann, M. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 1996, 24, 893–896. [Google Scholar] [CrossRef]
- Couselo-Seijas, M.; López-Canoa, J.N.; Agra-Bermejo, R.M.; Díaz-Rodriguez, E.; Fernandez, A.L.; Martinez-Cereijo, J.M.; Durán-Muñoz, D.; Bravo, S.B.; Velo, A.; González-Melchor, L.; et al. Cholinergic activity regulates the secretome of epicardial adipose tissue: Association with atrial fibrillation. J. Cell. Physiol. 2019, 234, 10512–10522. [Google Scholar] [CrossRef]
- García Vence, M.; del Chantada-Vázquez, M.P.; Vázquez-Estévez, S.; Manuel Cameselle-Teijeiro, J.; Bravo, S.B.; Núñez, C. Potential clinical applications of the personalized, disease-specific protein corona on nanoparticles. Clin. Chim. Acta 2020, 501, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Tamara, C.; Nerea, L.B.; Belén, B.S.; Aurelio, S.; Iván, C.; Fernando, S.; Javier, B.; Felipe, C.F.; María, P. Vesicles Shed by pathological murine adipocytes spread pathology: Characterization and functional role of insulin resistant/hypertrophied adiposomes. Int. J. Mol. Sci. 2020, 21, 2252. [Google Scholar] [CrossRef]
- Izquierdo, I.; Rosa, I.; Bravo, S.B.; Guitián, E.; Pérez-Serra, A.; Campuzano, O.; Brugada, R.; Mangas, A.; García, Á.; Toro, R. Proteomic identification of putative biomarkers for early detection of sudden cardiac death in a family with a LMNA gene mutation causing dilated cardiomyopathy. J. Proteom. 2016, 148, 75–84. [Google Scholar] [CrossRef]
- Del Pilar Chantada-Vázquez, M.; López, A.C.; Vence, M.G.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.D.P.; López, A.C.; García-Vence, M.; Acea-Nebril, B.; Bravo, S.B.; Núñez, C. Protein corona gold nanoparticles fingerprinting reveals a profile of blood coagulation proteins in the serum of Her2-overexpressing breast cancer patients. Int. J. Mol. Sci. 2020, 21, 8449. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.D.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a profile of neutrophil-derived granule proteins in the surface of gold nanoparticles after their interaction with human breast cancer sera. Nanomaterials 2020, 10, 1223. [Google Scholar] [CrossRef]
- Hermida-Nogueira, L.; Barrachina, M.N.; Izquierdo, I.; García-Vence, M.; Lacerenza, S.; Bravo, S.; Castrillo, A.; García, Á. Proteomic Analysis of extracellular vesicles derived from platelet concentrates treated with mirasol® identifies biomarkers of platelet storage lesion. J. Proteom. 2020, 210. [Google Scholar] [CrossRef]
- Lambert, J.; Ivosev, G.; Couzens, A.L.; Larsen, B.; Taipale, M.; Lin, Z.; Zhong, Q.; Lindquist, S.; Vidal, M.; Aebersold, R.; et al. Mapping differential interactomes by affinity purification coupled with data independent mass spectrometry acquisition. Nat. Methods 2014, 10, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- García-Vence, M.; Chantada-Vázquez, M.D.P.; Cameselle-Teijeiro, J.M.; Bravo, S.B.; Núñez, C. A Novel nanoproteomic approach for the identification of molecular targets associated with thyroid tumors. Nanomaterials 2020, 10, 2370. [Google Scholar] [CrossRef] [PubMed]
- Varela-Rodríguez, B.M.; Juiz-Valiña, P.; Varela, L.; Outeiriño-Blanco, E.; Bravo, S.B.; García-Brao, M.J.; Mena, E.; Noguera, J.F.; Valero-Gasalla, J.; Cordido, F.; et al. Beneficial effects of bariatric surgery-induced by weight loss on the proteome of abdominal subcutaneous adipose tissue. J. Clin. Med. 2020, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Redestig, H.; Fukushima, A.; Stenlund, H.; Moritz, T.; Arita, M.; Saito, K.; Kusano, M. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics Data. Anal. Chem. 2009, 81, 7974–7980. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A.; et al. A Novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef] [PubMed]

| Uniprot Code | Gene Name | Protein Name | p-Value | FCh |
|---|---|---|---|---|
| P02786 | TFR1 | Transferrin receptor protein 1 | 0.00104 | 15.32 |
| Q99808 | S29A1 | Equilibrative nucleoside transporter 1 | 0.01424 | 14.01 |
| P11166 | GTR1 | Solute carrier family 2, facilitated glucose transporter member 1 | 0.00106 | 10.15 |
| P69892 | HBG2 | Hemoglobin subunit γ-2 | 0 | 9.43 |
| P02792 | FRIL | Ferritin light chain | 0.00438 | 9.22 |
| Q16819 | MEP1A | Meprin A subunit α | 0.01481 | 7.13 |
| P02776 | PLF4 | Platelet factor 4 | 0.00809 | 7.01 |
| Q02094 | RHAG | Ammonium transporter Rh type A | 0.00772 | 6.71 |
| P27918 | PROP | Properdin | 0 | 6.26 |
| P16671 | CD36 | Platelet glycoprotein 4 | 0.00001 | 5.99 |
| Q9NQ84 | GPC5C | G-protein-coupled receptor family C group 5 member C | 0.00467 | 5.87 |
| P08195 | 4F2 | 4F2 cell surface antigen heavy chain | 0.00224 | 5.77 |
| Q658P3 | STEA3 | Metalloreductase STEAP3 | 0.00262 | 5.34 |
| Q6UX06 | OLFM4 | Olfactomedin-4 | 0.00008 | 5.08 |
| P02787 | TRFE | Serotransferrin | 0.00101 | 5.05 |
| P08514 | ITA2B | Integrin αIIb | 0 | 4.88 |
| P02730 | B3AT | Band 3 anion transport protein | 0.00334 | 4.71 |
| P11142 | HSP7C | Heat shock cognate 71 kDa protein | 0.00239 | 4.7 |
| Q15758 | AAAT | Neutral amino acid transporter B(0) | 0.00829 | 4.56 |
| P05164 | PERM | Myeloperoxidase | 0.00003 | 4.46 |
| P0DMV9 | HS71B | Heat shock 70 kDa protein 1B | 0.0012 | 4.36 |
| P27105 | STOM | Erythrocyte band 7 integral membrane protein | 0.00326 | 4.18 |
| P02743 | SAMP | Serum amyloid P-component | 0 | 4.15 |
| Q13201 | MMRN1 | Multimerin-1 | 0.00228 | 4.1 |
| Q15907 | RB11B | Ras-related protein Rab-11B | 0.00058 | 4.03 |
| Q9HD89 | RETN | Resistin | 0.00002 | 3.79 |
| P01024 | CO3 | Complement C3 | 0 | 3.6 |
| Q86UX7 | URP2 | Fermitin family homolog 3 | 0 | 3.51 |
| P05023 | AT1A1 | Sodium/potassium-transporting ATPase subunit α-1 | 0.00875 | 3.39 |
| P18428 | LBP | Lipopolysaccharide-binding protein | 0.00971 | 3.31 |
| P04275 | VWF | von Willebrand factor | 0 | 3.29 |
| Q00610 | CLH1 | Clathrin heavy chain 1 | 0.00667 | 3.15 |
| P01859 | IGHG2 | Ig heavy constant γ 2 | 0.0001 | 3.06 |
| Q9Y490 | TLN1 | Talin-1 | 0.00002 | 3.02 |
| P00451 | FA8 | Coagulation factor VIII | 0.00055 | 3.01 |
| P05106 | ITB3 | Integrin β3 | 0.00003 | 3 |
| P01042 | KNG1 | Kininogen-1 | 0.00652 | 2.95 |
| O15400 | STX7 | Syntaxin-7 | 0.00007 | 2.89 |
| P12259 | FA5 | Coagulation factor V | 0 | 2.77 |
| P26038 | MOES | Moesin | 0.00065 | 2.74 |
| P10643 | CO7 | Complement component C7 | 0 | 2.73 |
| Q9UP52 | TFR2 | Transferrin receptor protein 2 | 0.03685 | 2.71 |
| P02760 | AMBP | Protein AMBP | 0.00118 | 2.7 |
| P11597 | CETP | Cholesterylester transfer protein | 0.00012 | 2.7 |
| P15144 | AMPN | Aminopeptidase N | 0.00896 | 2.6 |
| P21926 | CD9 | CD9 antigen | 0.00006 | 2.59 |
| P01861 | IGHG4 | Ig heavy constant γ 4 | 0.00009 | 2.58 |
| P0C0L5 | CO4B | Complement C4-B | 0 | 2.54 |
| Q71DI3 | H32 | Histone H3.2 | 0.0305 | 2.49 |
| P05546 | HEP2 | Heparin cofactor 2 | 0.00001 | 2.44 |
| P08603 | CFAH | Complement factor H | 0.0057 | 2.43 |
| P13671 | CO6 | Complement component C6 | 0.00044 | 2.33 |
| P04004 | VTNC | Vitronectin | 0 | 2.3 |
| Q9HAV0 | GBB4 | Guanine nucleotide-binding protein subunit β-4 | 0.00045 | 2.27 |
| P21333 | FLNA | Filamin-A | 0.00019 | 2.23 |
| P02788 | TRFL | Lactotransferrin | 0.01132 | 2.23 |
| P62834 | RAP1A | Ras-related protein Rap-1A | 0.00141 | 2.2 |
| P01717 | LV325 | Ig λ variable 3-25 | 0.04177 | 2.18 |
| P02675 | FIBB | Fibrinogen β chain | 0.00005 | 2.16 |
| P69905 | HBA | Hemoglobin subunit α | 0.00317 | 2.15 |
| P63104 | 1433Z | 14-3-3 protein ζ/δ | 0.00005 | 2.14 |
| P02679 | FIBG | Fibrinogen γ chain | 0 | 2.13 |
| P01857 | IGHG1 | Ig heavy constant γ 1 | 0.00004 | 2.1 |
| P01023 | A2MG | A-2-macroglobulin | 0.00104 | 2.02 |
| P06312 | KV401 | Ig κ variable 4–1 | 0.00234 | 0.48 |
| P06310 | KV230 | Ig κ variable 2–30 | 0.00695 | 0.46 |
| P07225 | PROS | Vitamin K-dependent protein S | 0.0011 | 0.42 |
| P49721 | PSB2 | Proteasome subunit β type-2 | 0.01941 | 0.4 |
| P01834 | IGKC | Ig κ constant | 0.00001 | 0.4 |
| P01766 | HV313 | Ig heavy variable 3–13 | 0.00043 | 0.4 |
| P0DOY3 | IGLC3 | Ig λ constant 3 | 0 | 0.39 |
| A0A0C4DH69 | KV109 | Ig κ variable 1–9 | 0.00017 | 0.39 |
| P04430 | KV116 | Ig κ variable 1–16 | 0.00037 | 0.37 |
| B9A064 | IGLL5 | Ig λ-like polypeptide 5 | 0.00002 | 0.36 |
| P27169 | PON1 | Serum paraoxonase/arylesterase 1 | 0.00304 | 0.36 |
| Q93050 | VPP1 | V-type proton ATPase 116 kDa subunit a isoform 1 | 0.01287 | 0.35 |
| P01700 | LV147 | Ig λ variable 1–47 | 0 | 0.35 |
| P00338 | LDHA | L-lactate dehydrogenase A chain | 0.01646 | 0.34 |
| P04003 | C4BPA | C4b-binding protein α chain | 0 | 0.33 |
| P01601 | KVD16 | Ig κ variable 1D–16 | 0.00931 | 0.32 |
| P02766 | TTHY | Transthyretin | 0.01393 | 0.32 |
| Q6NSI8 | K1841 | Uncharacterized protein KIAA1841 | 0.04514 | 0.31 |
| Q16851 | UGPA | UTP-glucose-1-phosphate uridylyltransferase | 0.04304 | 0.31 |
| A0A0B4J1X5 | HV374 | Ig heavy variable 3–74 | 0.00052 | 0.31 |
| A0A0C4DH25 | KVD20 | Ig κ variable 3D–20 | 0 | 0.31 |
| P0DP03 | HV335 | Ig heavy variable 3–30–5 | 0 | 0.3 |
| A0A0C4DH24 | KV621 | Ig κ variable 6–21 | 0.00154 | 0.28 |
| P08519 | APOA | Apolipoprotein(a) | 0.00237 | 0.28 |
| A2NJV5 | KV229 | Ig κ variable 2–29 | 0.00394 | 0.27 |
| A0A0A0MS15 | HV349 | Ig heavy variable 3–49 | 0 | 0.27 |
| A0A0C4DH68 | KV224 | Ig κ variable 2–24 | 0.00088 | 0.26 |
| P01624 | KV315 | Ig κ variable 3–15 | 0.00002 | 0.25 |
| P28072 | PSB6 | Proteasome subunit β type-6 | 0.02057 | 0.25 |
| P04433 | KV311 | Ig κ variable 3–11 | 0.00002 | 0.24 |
| P01591 | IGJ | Ig J chain | 0.00001 | 0.23 |
| O43866 | CD5L | CD5 antigen-like | 0.00001 | 0.2 |
| A0A0B4J1Y9 | HV372 | Ig heavy variable 3–72 | 0.00097 | 0.17 |
| A0A075B6H9 | LV469 | Ig λ variable 4–69 | 0.00042 | 0.15 |
| P00738 | HPT | Haptoglobin | 0.00134 | 0.13 |
| P00739 | HPTR | Haptoglobin-related protein | 0 | 0.11 |
| O14791 | APOL1 | Apolipoprotein L1 | 0.00002 | 0.08 |
| P01877 | IGHA2 | Ig heavy constant α 2 | 0.00001 | 0.05 |
| Q8TF72 | SHRM3 | Protein Shroom 3 | 0.01003 | 0.05 |
| P01876 | IGHA1 | Ig heavy constant α 1 | 0.00001 | 0.02 |
| Upregulated in Neonates vs. Adults | |||
| Pathway | Protein Count | p Value | Proteins |
| Hemostasis | 23 | <0.001 | RAP1A; YWHAZ; KNG1; FLNA; GNAI2; ITGA2B; ITGB3; A2MG; F5; PF4; PFN1; HIST2H3A; TLN1; FGA; FGB; F8; HIST2H3C; HIST2H3D; TF; FGG; CD36; vWF; HBG2 |
| Formation of fibrin clot (clotting cascade) | 9 | <0.001 | KNG1; A2MG; F5; PF4; FGA; FGB; F8; vWF; FGG; |
| Integrin cell surface interactions | 9 | <0.001 | RAP1A; ITGA2B; ITGB3; THBS1; VTN; TLN1; FGB; FGG; FGA; |
| Complement cascade | 6 | <0.001 | MASP1; C3; C1R; C7; C6; C4B; |
| β3 integrin cell surface interactions | 7 | <0.001 | ITGA2B; ITGB3; THBS1; VTN; FGA; FGB; FGG; |
| Common pathway | 5 | <0.001 | F5; PF4; FGA; FGB; FGG; |
| Platelet activation, signaling and aggregation | 10 | <0.001 | RAP1A; YWHAZ; FLNA; GNAI2; ITGA2B; ITGB3; PFN1; vWF; TF; TLN1; |
| Ephrin B-EPHB pathway | 7 | =0.002 | RAP1A; ITGA2B; ITGB3; FGA; FGB; TF; FGG; |
| p130Cas linkage to MAPK signaling for integrins | 4 | =0.002 | RAP1A; ITGA2B; ITGB3; TLN1; |
| GRB2: SOS provides linkage to MAPK signaling for integrins | 4 | =0.002 | RAP1A; ITGA2B; ITGB3; TLN1; |
| β2 integrin cell surface interactions | 5 | =0.008 | KNG1; FGA; FGB; C3; FGG; |
| Ephrin B reverse signaling | 5 | =0.011 | ITGA2B; ITGB3; FGA; FGB; FGG; |
| Initial triggering of complement | 4 | =0.014 | MASP1; C3; C1R; C4B; |
| Netrin-mediated repulsion signals | 4 | =0.014 | RAP1A; ITGA2B; ITGB3; TLN1; |
| Recycling pathway of L1 | 5 | =0.016 | RAP1A; CLTC; ITGA2B; ITGB3; TLN1; |
| Proteoglycan syndecan-mediated signaling events | 27 | =0.025 | RAP1A; YWHAZ; CLTC; KNG1; FLNA; HSPA1A; GNAI2; SLC3A2; HSPA8; CLU; ITGA2B; ITGB3; A2MG; THBS1; VTN; TLN1; FGA; FGB; HSPA1B; TFRC; TF; FGG; SLC2A1; SDCBP; LBP; STEAP3; HBG2 |
| Intrinsic pathway | 4 | =0.03 | KNG1; A2MG; F8; vWF; |
| Integrin αIIbβ3 signaling | 4 | =0.047 | RAP1A; ITGA2B; ITGB3; TLN1; |
| Downregulated in Neonates vs. Adults | |||
| Pathway | Protein Count | p value | Proteins |
| Chylomicron-mediated lipid transport | 3 | =0.006 | APOA1; APOB; APOC3; |
| Lipoprotein metabolism | 3 | =0.033 | APOA1; APOB; APOC3; |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peñas-Martínez, J.; Barrachina, M.N.; Cuenca-Zamora, E.J.; Luengo-Gil, G.; Bravo, S.B.; Caparrós-Pérez, E.; Teruel-Montoya, R.; Eliseo-Blanco, J.; Vicente, V.; García, Á.; et al. Qualitative and Quantitative Comparison of Plasma Exosomes from Neonates and Adults. Int. J. Mol. Sci. 2021, 22, 1926. https://doi.org/10.3390/ijms22041926
Peñas-Martínez J, Barrachina MN, Cuenca-Zamora EJ, Luengo-Gil G, Bravo SB, Caparrós-Pérez E, Teruel-Montoya R, Eliseo-Blanco J, Vicente V, García Á, et al. Qualitative and Quantitative Comparison of Plasma Exosomes from Neonates and Adults. International Journal of Molecular Sciences. 2021; 22(4):1926. https://doi.org/10.3390/ijms22041926
Chicago/Turabian StylePeñas-Martínez, Julia, María N. Barrachina, Ernesto José Cuenca-Zamora, Ginés Luengo-Gil, Susana Belén Bravo, Eva Caparrós-Pérez, Raúl Teruel-Montoya, José Eliseo-Blanco, Vicente Vicente, Ángel García, and et al. 2021. "Qualitative and Quantitative Comparison of Plasma Exosomes from Neonates and Adults" International Journal of Molecular Sciences 22, no. 4: 1926. https://doi.org/10.3390/ijms22041926
APA StylePeñas-Martínez, J., Barrachina, M. N., Cuenca-Zamora, E. J., Luengo-Gil, G., Bravo, S. B., Caparrós-Pérez, E., Teruel-Montoya, R., Eliseo-Blanco, J., Vicente, V., García, Á., Martínez-Martínez, I., & Ferrer-Marín, F. (2021). Qualitative and Quantitative Comparison of Plasma Exosomes from Neonates and Adults. International Journal of Molecular Sciences, 22(4), 1926. https://doi.org/10.3390/ijms22041926








