Genome-Wide Characterization, Evolution, and Expression Profile Analysis of GATA Transcription Factors in Brachypodium distachyon
Abstract
1. Introduction
2. Results
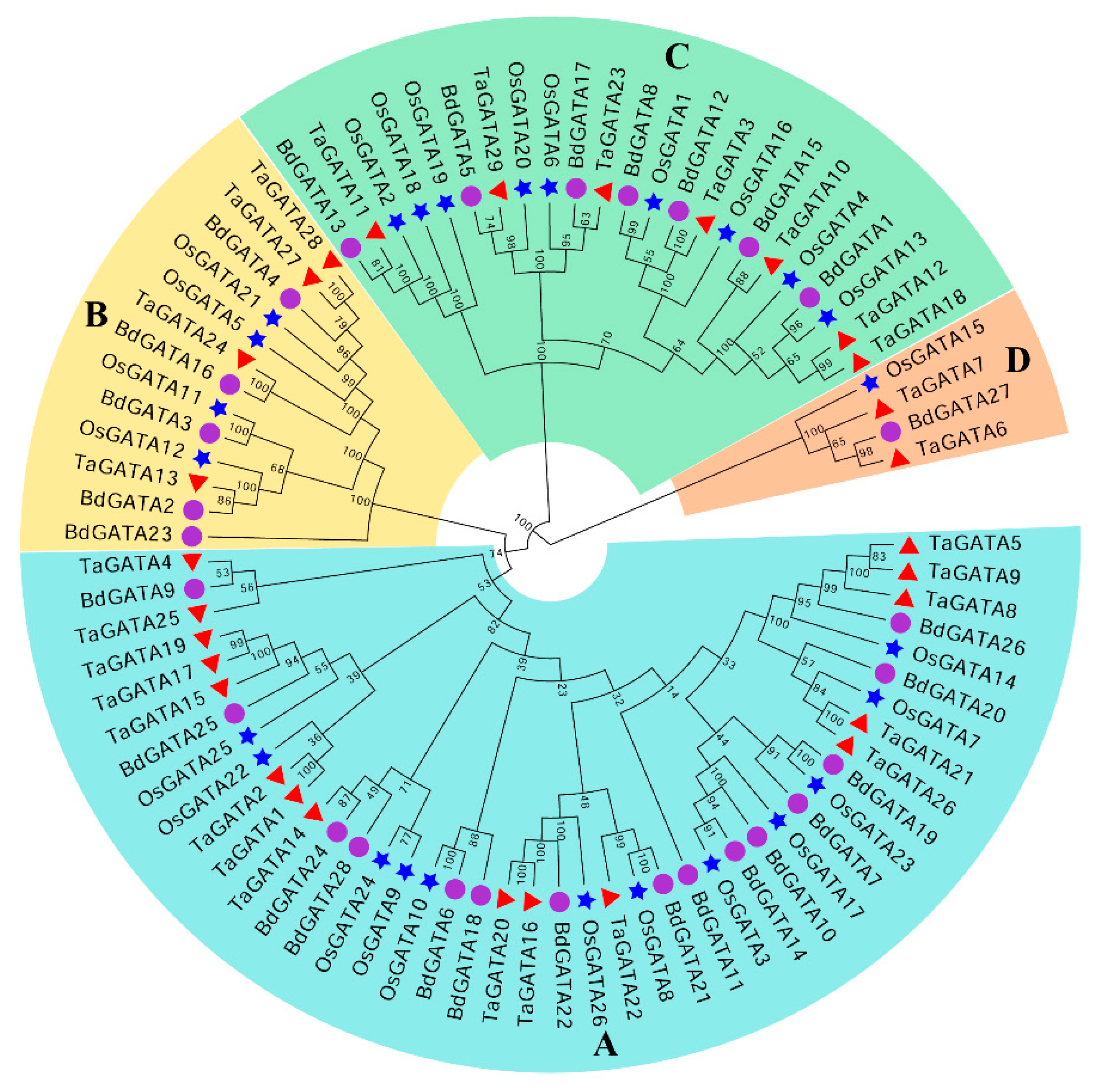
2.1. Identification and Phylogenetic Analysis of GATA Family Proteins in Brachypodium Distachyon
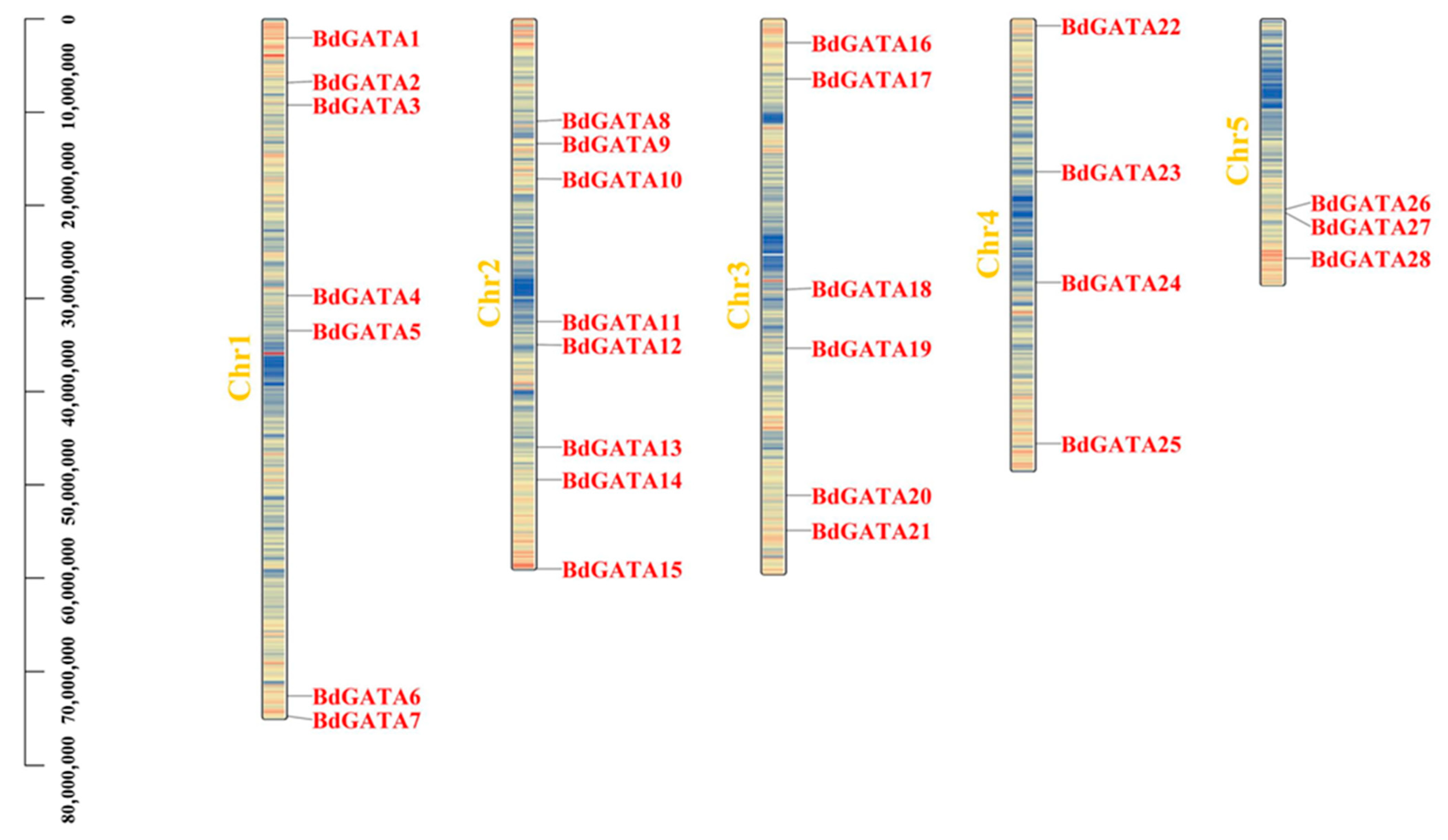
2.2. Chromosome Location and Duplication Analysis of BdGATA Genes
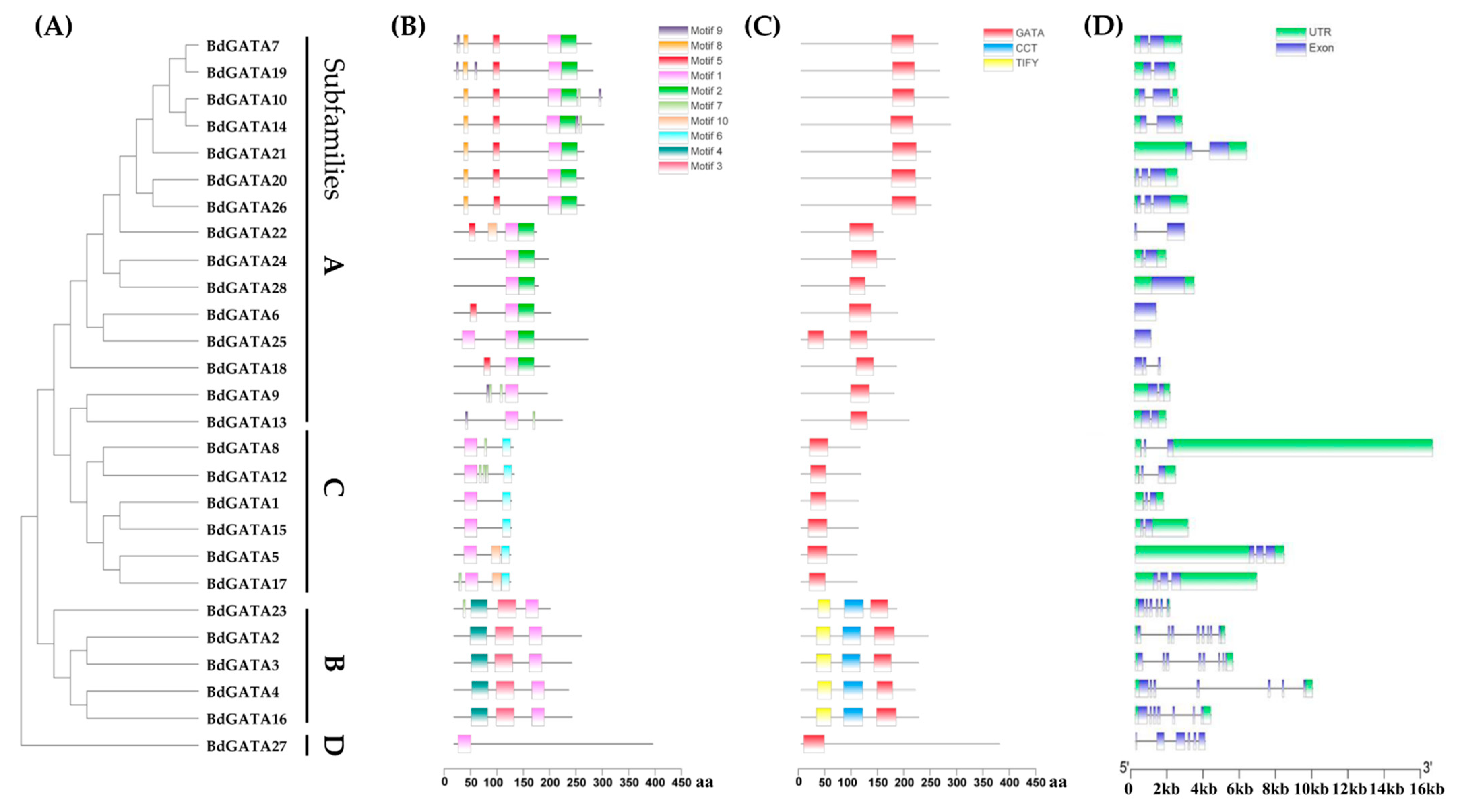
2.3. Structures and Conserved Motifs of BdGATA Genes
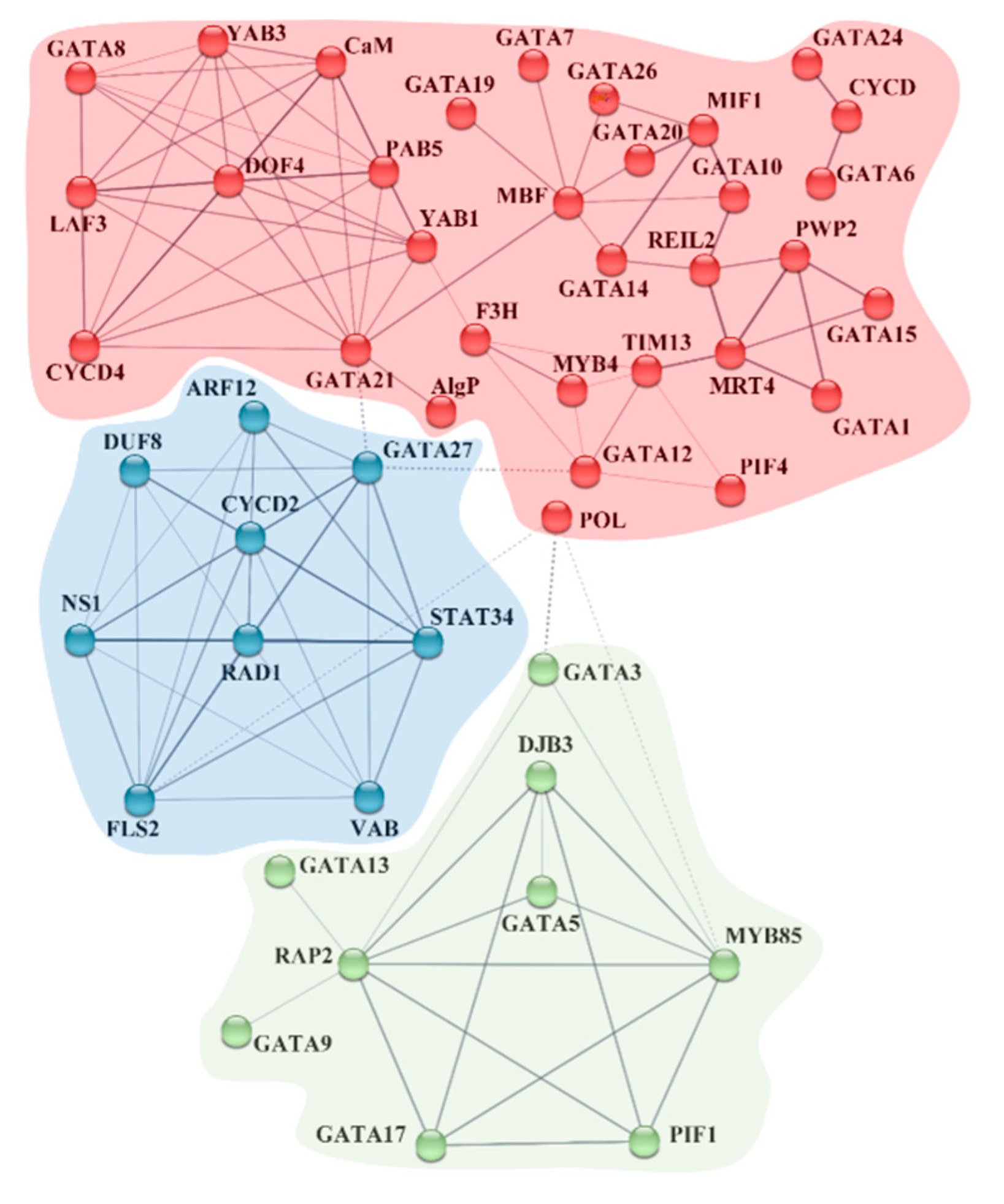
2.4. Prediction of CRE-Involved Pathways, BdGATA Targets of miRNA, and BdGATA Interaction Network
2.5. Expression Profile of BdGATA Genes in Response to Magnaporthe Oryzae and Hormone Treatments
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Hormone Treatments, and Rice Blast Inoculation
4.2. Rice Blast Inoculation
4.3. Identification of Brachypodium Distachyon GATA Gene Family and Phylogenetic Analysis
4.4. Gene Structure, Synteny Analysis, and Bioinformatics Resources
4.5. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA Family of Transcription Factors in Arabidopsis and Rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef]
- Li, Y.; Jia, Z.; Yi, Q.; Song, X.; Liu, Y.; Jia, Y.; Wang, L.; Song, L. A novel GATA-like zinc finger transcription factor involving in hematopoiesis of Eriocheir sinensis. Fish Shellfish Immunol. 2018, 74, 363–371. [Google Scholar] [CrossRef]
- Daniel-Vedele, F.; Caboche, M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. MGG 1993, 240, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hou, Y.; Hao, Q.; Chen, H.; Chen, L.; Yuan, S.; Shan, Z.; Zhang, X.; Yang, Z.; Qiu, D.; et al. Genome-Wide Survey of the Soybean GATA Transcription Factor Gene Family and Expression Analysis under Low Nitrogen Stress. PLoS ONE 2015, 10, e0125174. [Google Scholar] [CrossRef]
- Ranftl, Q.L.; Bastakis, E.; Klermund, C.; Schwechheimer, C. LLM-Domain Containing B-GATA Factors Control Different Aspects of Cytokinin-Regulated Development in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2295–2311. [Google Scholar] [CrossRef]
- An, Y.; Zhou, Y.; Han, X.; Shen, C.; Wang, S.; Liu, C.; Yin, W.; Xia, X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2020, 71, 1969–1984. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Bi, Y.-M.; Rothstein, S.J. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 2017, 94, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Bastakis, E.; Hedtke, B.; Klermund, C.; Grimm, B.; Schwechheimer, C. LLM-Domain B-GATA Transcription Factors Play Multifaceted Roles in Controlling Greening in Arabidopsis. Plant Cell 2018, 30, 582–599. [Google Scholar] [CrossRef]
- Stamm, P.; Ravindran, P.; Mohanty, B.; Tan, E.; Yu, H.; Kumar, P.P. Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 179. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, P.; Verma, V.; Stamm, P.; Kumar, P.P. A Novel RGL2–DOF6 Complex Contributes to Primary Seed Dormancy in Arabidopsis thaliana by Regulating a GATA Transcription Factor. Mol. Plant 2017, 10, 1307–1320. [Google Scholar] [CrossRef]
- Sen, S.; Kundu, S.; Dutta, S.K. Proteomic analysis of JAZ interacting proteins under methyl jasmonate treatment in finger millet. Plant Physiol. Biochem. 2016, 108, 79–89. [Google Scholar] [CrossRef]
- Pant, S.R.; Krishnavajhala, A.; McNeece, B.T.; Lawrence, G.W.; Klink, V.P. The syntaxin 31-induced gene, LESION SIMULATING DISEASE1 (LSD1), functions in Glycine max defense to the root parasite Heterodera glycines. Plant Signal. Behav. 2015, 10, e977737. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.R.; Joshi, G.; Kukreja, B.; Malik, V.; Arora, P.; Pandey, R.; Shukla, R.N.; Bankar, K.G.; Katiyar-Agarwal, S.; Goel, S.; et al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015, 15, 9. [Google Scholar] [CrossRef]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic Stresses Cause Differential Regulation of Alternative Splice Forms of GATA Transcription Factor in Rice. Front. Plant Sci. 2017, 8, 1944. [Google Scholar] [CrossRef]
- Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J. Exp. Bot. 2019, erz368. [Google Scholar] [CrossRef]
- Bi, Y.-M.; Zhang, Y.; Signorelli, T.; Zhao, R.; Zhu, T.; Rothstein, S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity: A GATA gene for chlorophyll and glucose sensitivity. Plant J. 2005, 44, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.-M.; Rothstein, S.J. Rice Cytokinin GATA Transcription Factor1 Regulates Chloroplast Development and Plant Architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef]
- Li, W.-T.; Chen, W.-L.; Yang, C.; Wang, J.; Yang, L.; He, M.; Wang, J.-C.; Qin, P.; Wang, Y.-P.; Ma, B.-T.; et al. Identification and network construction of zinc finger protein (ZFP) genes involved in the rice-Magnaporthe oryzae interaction. Plant Omics 2014, 7, 540–548. [Google Scholar]
- He, S.; Tan, G.; Liu, Q.; Huang, K.; Ren, J.; Zhang, X.; Yu, X.; Huang, P.; An, C. The LSD1-Interacting Protein GILP Is a LITAF Domain Protein That Negatively Regulates Hypersensitive Cell Death in Arabidopsis. PLoS ONE 2011, 6, e18750. [Google Scholar] [CrossRef]
- Deb, A.; Kundu, S. Deciphering Cis-Regulatory Element Mediated Combinatorial Regulation in Rice under Blast Infected Condition. PLoS ONE 2015, 21. [Google Scholar] [CrossRef]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2009, 10, 729–731. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.-Z. Computational detection of microRNAs targeting transcription factor genes in Arabidopsis thaliana. Comput. Biol. Chem. 2005, 29, 360–367. [Google Scholar] [CrossRef]
- Singh, R.; Tiwari, J.K.; Rawat, S.; Sharma, V.; Singh, B.P. In silico identification of candidate microRNAs and their targets in potato somatic hybrid Solanum tuberosum (+) S. pinnatisectum for late blight resistance. Plant Omics 2016, 9, 159–164. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, N.; Ma, C.; Qu, Y.; Si, H.; Wang, D. Prediction and verification of microRNAs related to proline accumulation under drought stress in potato. Comput. Biol. Chem. 2013, 46, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Zhang, C.-L.; Zhao, T.-T.; Xu,, X.Y. Research advances of GATA transcription factor in plant. Mol. Plant Breed. 2017, 15, 1702–1707. [Google Scholar] [CrossRef]
- Chen, H.; Shao, H.; Li, K.; Zhang, D.; Fan, S.; Li, Y.; Han, M. Genome-wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus × domestica Borkh.). Gene 2017, 627, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, Y.-C.; Wang, W.-P.; Zhang, C. Bioinformatics Analysis of GATA Gene Family in Rice. Mol. Plant Breed. 2018, 16, 5514–5522. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, X.; Huang, Z.; Fan, S.; Qun, G.; Liu, A.; Gong, J.; Li, J.; Gong, W.; Shi, Y.; et al. Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene 2019, 680, 72–83. [Google Scholar] [CrossRef]
- Zhu, W.; Guo, Y.; Chen, Y.; Wu, D.; Jiang, L. Genome-Wide Identification, Phylogenetic and Expression Pattern Analysis of GATA Family Genes in Brassica Napus. BMC Plant Biol. 2020, 20, 543. [Google Scholar] [CrossRef]
- Jiang, L.; Yu, X.; Chen, D.; Feng, H.; Li, J. Identification, Phylogenetic Evolution and Expression Analysis of GATA Transcription Factor Family in Maize (Zea Mays). Int. J. Agric. Bio. 2020, 23, 7. [Google Scholar] [CrossRef]
- Jin, J.-H.; Zhang, H.-X.; Ali, M.; Wei, A.-M.; Luo, D.-X.; Gong, Z.-H. The CaAP2/ERF064 Regulates Dual Functions in Pepper: Plant Cell Death and Resistance to Phytophthora capsici. Genes 2019, 10, 541. [Google Scholar] [CrossRef]
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.P.; Caracuel-Rios, Z.; Overy, D.P.; Snowdon, S.; Talbot, N.J.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–737. [Google Scholar] [CrossRef]
- Routledge, A.P.M.; Shelley, G.; Smith, J.V.; Talbot, N.J.; Draper, J.; Mur, L.A.J. Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 2004, 5, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Ping, H.-T.; Guo, P.-C.; Yu, P.; Zhang, Y.-J.; Zhang, B.; Hu, Z.-L. Cloning and Analysis of Pathogen Resistance of Multiprotein Bridging Factor Gene LeMBF1 in Tomato. Life Sci. Res. 2012, 16, 138–143. [Google Scholar]
- Zhang, Y.; Zhang, G.; Dong, Y.-L.; Guo, J.; Huang, L.-L.; Kang, Z.-S. Cloning and Characterization of a MBF1 Transcriptional Coactivator Factor in Wheat Induced by Stripe Rust Pathogen: Cloning and Characterization of a MBF1 Transcriptional Coactivator Factor in Wheat Induced by Stripe Rust Pathogen. ACTA Agron. Sin. 2009, 35, 11–17. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, T.; Yin, K.-Q.; Chen, Z.; Gu, H.; Qu, L.-J.; Qin, G. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 2012, 195, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Fu, M.; Li, H.; Chen, Y.; Wang, L.; Liu, R. Systematic analysis of NAC transcription factors in Gossypium barbadense uncovers their roles in response to Verticillium wilt. PeerJ 2019, 7, e7995. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Chen, S.; Li, J.; Liu, Y.; Ji, W.; Li, H. Genome-wide identification of GRAS genes in Brachypodium distachyon and functional characterization of BdSLR1 and BdSLRL1. BMC Genom. 2019, 20, 635. [Google Scholar] [CrossRef]
- Ebel, C.; BenFeki, A.; Hanin, M.; Solano, R.; Chini, A. Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum Durum TdTIFY11a in salt stress tolerance. PLoS ONE 2018, 13, e0200566. [Google Scholar] [CrossRef]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, M.A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae Chitinase MoChia1 by a Rice Tetratricopeptide Repeat Protein Allows Free Chitin to Trigger Immune Responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, L.; Zhu, Y.; Li, Y.; Yan, H.; Xiang, Y. Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol. Direct 2015, 10, 48. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, W.; Li, W.; Song, N.; Tang, Z.; Liu, J.; Wang, Y.; Pan, S.; Dai, L.; Wang, B. Genome-Wide Characterization, Evolution, and Expression Profile Analysis of GATA Transcription Factors in Brachypodium distachyon. Int. J. Mol. Sci. 2021, 22, 2026. https://doi.org/10.3390/ijms22042026
Peng W, Li W, Song N, Tang Z, Liu J, Wang Y, Pan S, Dai L, Wang B. Genome-Wide Characterization, Evolution, and Expression Profile Analysis of GATA Transcription Factors in Brachypodium distachyon. International Journal of Molecular Sciences. 2021; 22(4):2026. https://doi.org/10.3390/ijms22042026
Chicago/Turabian StylePeng, Weiye, Wei Li, Na Song, Zejun Tang, Jing Liu, Yunsheng Wang, Sujun Pan, Liangying Dai, and Bing Wang. 2021. "Genome-Wide Characterization, Evolution, and Expression Profile Analysis of GATA Transcription Factors in Brachypodium distachyon" International Journal of Molecular Sciences 22, no. 4: 2026. https://doi.org/10.3390/ijms22042026
APA StylePeng, W., Li, W., Song, N., Tang, Z., Liu, J., Wang, Y., Pan, S., Dai, L., & Wang, B. (2021). Genome-Wide Characterization, Evolution, and Expression Profile Analysis of GATA Transcription Factors in Brachypodium distachyon. International Journal of Molecular Sciences, 22(4), 2026. https://doi.org/10.3390/ijms22042026




