A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1
Abstract
:1. Introduction
2. Results
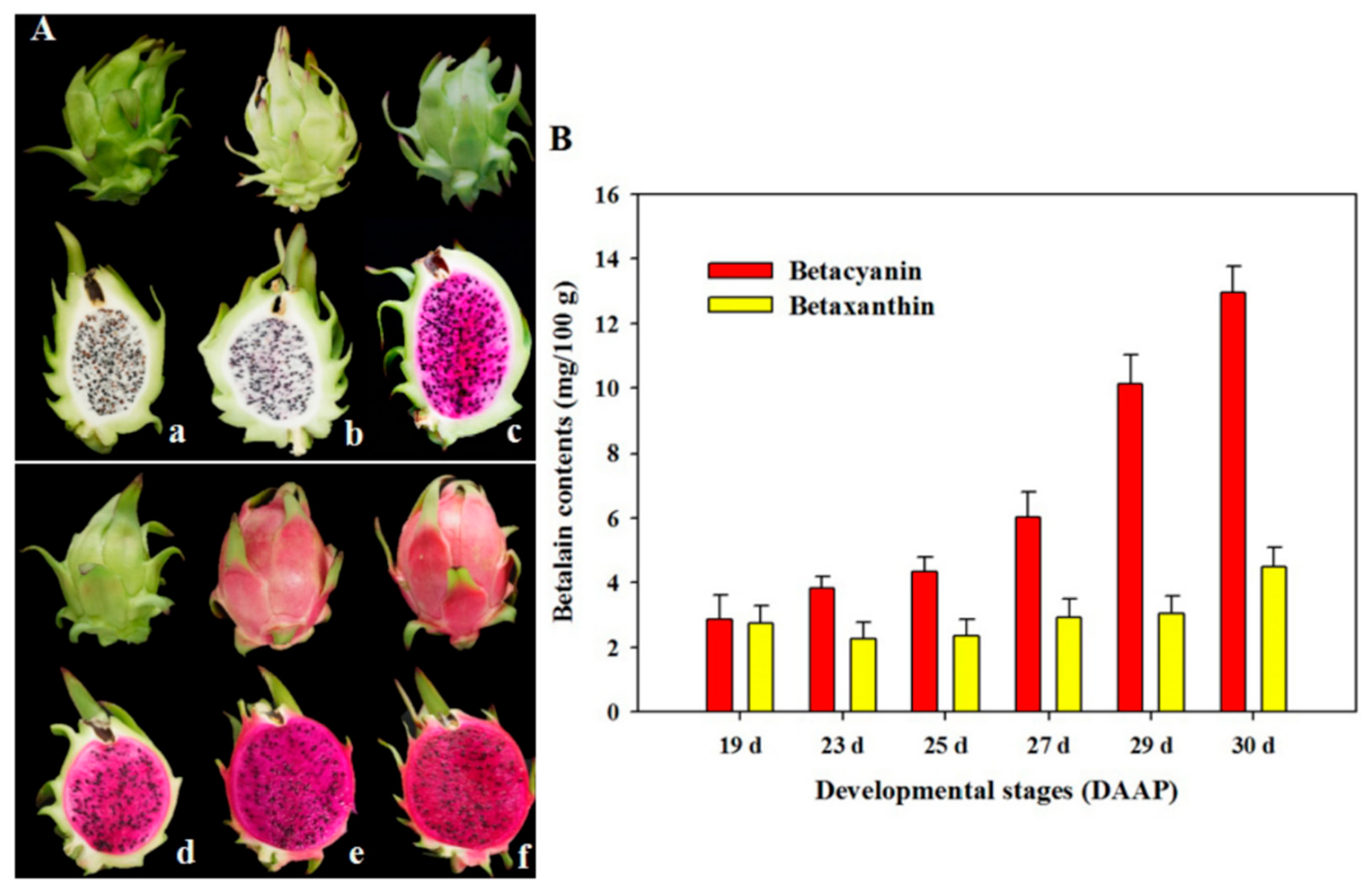
2.1. Changes in Betalain Contents at All Coloration Stages of Pitaya Pulp
2.2. Promoter Analyses of HmoCYP76AD1
2.3. Cloning and Sequence Analyses of HmoWRKY40
2.4. Expression Analyses of HmoWRKY40 and HmoCYP76AD1
2.5. Nuclear Localization of HmoWRKY40
2.6. Analyses of Transcriptional Activities of HmoWRKY40
2.7. Virus-Induced Gene Silencing (VIGS) Analyses of HmoWRKY40
2.8. Interaction of HmoWRKY40 with the W-Box in the Promoter of HmoCYP76AD1
2.9. HmoWRKY40 Activates the Transcription of Promoter of HmoCYP76AD1
3. Discussion and Conclusions
4. Materials and Methods
4.1. Materials
4.2. Measurement of Betalain Contents
4.3. Gene Cloning and Sequence Analyses
4.4. RT-qPCR Analyses
4.5. Subcellular Localization of HmoWRKY40
4.6. Gene Silencing of HmoWRKY40
4.7. Promoter Analysis of HmoCYP76AD1
4.8. Yeast One-Hybrid Experiment
4.9. Transcriptional Activity
4.10. Dual-Luciferase Reporter Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| cDNA | Complementary DNA |
| CTAB | Hexadecyltrimethylammonium bromide |
| CYP | Cytochrome P450 |
| DAAP | Day after artificial pollination |
| DOD | Dioxygenase |
| DOPA | 4,5-dihydroxy-phenylalanine |
| GFP | Green fluorescent protein |
| GTs | Glucosyltransferases |
| L-DOPA | L-3,4-dihydroxyphenylalanine |
| MES | (2-Nmorpholino) ethanesulfonic acid |
| MMA | MES+MgCl2+acetosyringone |
| MYB | Myb proto-oncogene protein |
| NCBI | National Center for Biotechnology Information |
| ORF | Open reading frame |
| pI | Theoretical isoelectric point |
| RFP | Red fluorescent protein |
| RNA-Seq | RNA sequencing |
| RT-qPCR | Reverse transcription quantitative real-time polymerase chain reaction |
| TFs | Transcription factors |
| TYR | Tyrosinase |
| VIGS | Virus-induced gene silencing |
References
- Fernando, G.H.; Francisco, G.C. Characterization of recombinant Beta vulgaris 4,5-DOPA-extradiol-dioxygenase active in the biosynthesis of betalains. Planta 2012, 236, 91–100. [Google Scholar]
- Stafford, H.A. Anthocyanins and betalains: Evolution of the mutually exclusive pathways. Plant Sci. 1994, 101, 91–98. [Google Scholar] [CrossRef]
- Jain, G.; Gould, K.S. Functional significance of betalain biosynthesis in leaves of Disphyma australe under salinity stress. Environ. Exp. Bot. 2015, 109, 131–140. [Google Scholar] [CrossRef]
- Jain, G.; Schwinn, K.E.; Gould, K.S. Betalain induction by l-DOPA application confers photoprotection to saline-exposed leaves of Disphyma australe. New Phytol. 2015, 207, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, P.; Singh, K.P.; Singh, S.K.; Singh, M.C.; Swaroop, K. Influence of biotic and abiotic elicitors on production of betalain pigments in bougainvillea callus cultures. Indian J. Hortic. 2014, 71, 373–378. [Google Scholar]
- Gengatharan, A.; Dykes, G.A.; Choo, W.S. Betalains: Natural plant pigments with potential application in functional foods. LWT Food Sci. Technol. 2015, 64, 645–649. [Google Scholar] [CrossRef]
- Clifford, T.; Howatson, G.; West, D.J.; Stevenson, E.J. The potential benefits of red beetroot supplementation in health and disease. Nutrients 2015, 7, 2801–2822. [Google Scholar] [CrossRef]
- Da Silva, D.V.T.; dos Santos Baião, D.; de Oliveira Silva, F.; Alves, G.; Perrone, D.; Del Aguila, E.M.; Paschoalin, V.M.F. Betanin, a natural food additive: Stability, bioavailability, antioxidant and preservative ability assessments. Molecules 2019, 24, 458. [Google Scholar] [CrossRef] [Green Version]
- Hatlestad, G.J.; Sunnadeniya, R.M.; Akhavan, N.A.; Gonzalez, A.; Goldman, I.L.; McGrath, J.M.; Lloyd, A.M. The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat. Genet. 2012, 44, 816–820. [Google Scholar] [CrossRef]
- Hua, Q.Z.; Chen, C.J.; Chen, Z.; Chen, P.K.; Ma, Y.W.; Wu, J.Y.; Zheng, J.; Hu, G.B.; Zhao, J.T.; Qin, Y.H. Transcriptomic analysis reveals key genes related to betalain biosynthesis in pulp coloration of Hylocereus polyrhizus. Front. Plant Sci. 2016, 6, 1179. [Google Scholar]
- Polturak, G.; Breitel, D.; Grossman, N.; Sarrion-Perdigones, A.; Weithorn, E.; Pliner, M.; Orzaez, D.; Granell, A.; Rogachev, I.; Aharoni, A. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 2016, 210, 269–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polturak, G.; Aharoni, A. “La Vie en Rose”: Biosynthesis, sources, and applications of betalain pigments. Mol. Plant 2018, 11, 7–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.F.; Hua, Q.Z.; Chen, C.B.; Zhang, L.L.; Zhang, Z.K.; Chen, J.Y.; Zhang, R.; Zhao, J.S.; Hu, G.B.; Zhao, J.T.; et al. Transcriptomics-based identification and characterization of glucosyltransferases involved in betalain biosynthesis in Hylocereus megalanthus. Plant Physiol. Bioch. 2020, 152, 112–124. [Google Scholar] [CrossRef]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB, genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Hatlestad, G.J.; Akhavan, N.A.; Sunnadeniya, R.M.; Elam, L.; Cargile, S.; Hembd, A.; Gonzalez, A.; McGrath, J.M.; Lloyd, A.M. The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 2014, 47, 92–96. [Google Scholar] [CrossRef]
- Cheng, M.N.; Huang, Z.J.; Hua, Q.Z.; Shan, W.; Kuang, J.F.; Lu, W.J.; Qin, Y.H.; Chen, J.Y. The WRKY transcription factor HpWRKY44 regulates CytP450-like1 expression in red pitaya fruit (Hylocereus polyrhizus). Hortic. Res. 2017, 4, 17039. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, S.R.M.; Mohamed, G.A.; Khedr, A.I.M.; Zayed, M.F.; EI-Kholy, A.A.E.S. Genus Hylocereus: Beneficial phytochemicals, nutritional importance, and biological relevance-A review. J. Food Biochem. 2018, 42, e12491. [Google Scholar] [CrossRef]
- Sunnadeniya, R.; Bean, A.; Brown, M.; Akhavan, N.; Hatlestad, G.; Gonzalez, A.; Symonds, V.V.; Lloyd, A. Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS ONE 2016, 11, e0149417. [Google Scholar]
- Chen, L.G.; Song, Y.; Li, S.J.; Zhang, L.P.; Zou, C.S.; Yu, D.Q. The role of WRKY transcription factors in plant abiotic stresses. BBA 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, J.C. Identification and characterization of the grape WRKY family. Biomed Res. Int. 2014, 2014, 787680. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.B.; Yu, D.Q. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Jadaun, J.S.; Kushwaha, A.K.; Sangwan, N.S.; Narnoliya, L.K.; Mishra, S.; Sangwan, R.S. WRKY1-mediated regulation of tryptophan decarboxylase in tryptamine generation for withanamide production in Withania somnifera (Ashwagandha). Plant Cell Rep. 2020, 39, 1443–1465. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Kumar, S.R.; Dwivedi, V.; Rai, A.; Pal, S.; Shasany, A.K.; Nagegowda, D.A. A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 2017, 215, 1115–1131. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Zhang, Z.L.; Zou, X.L.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.X.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Gong, X.Q.; Zhang, J.Y.; Hu, J.B.; Wang, W.; Wu, H.; Zhang, Q.H.; Liu, J.H. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015, 38, 2248–2262. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.J.; Xiao, Y.Y.; Han, Y.C.; Shan, W.; Fan, Z.Q.; Xu, Q.G.; Kuang, J.F.; Lu, W.J.; Lakshmanan, P.; Chen, J.Y. Banana fruit VQ motif-containing protein5 represses cold-responsive transcription factor MaWRKY26 involved in the regulation of JA biosynthetic genes. Sci. Rep. 2016, 6, 23632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lee, L.Y.C.; Hou, X.L.; Fang, L.; Fan, S.G.; Kumar, P.P.; Yu, H. STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 2012, 139, 1568–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouwerkerk, P.B.; Meijer, A.H. Yeast one-hybrid screening for DNA-protein interactions. Curr. Protoc. Mol. Biol. 2001, 12, 12. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [Green Version]







| Site Names | Positions | Sequences | Functions |
|---|---|---|---|
| AAGAA-motif | 353 (+) | GAAAGAA | |
| ABRE | 162 (−); 771 (−) | ACGTG/CGTACGTGCA | Cis-acting element involved in the abscisic acid responsiveness |
| ABRE3a | 162 (−) | TACGTG | |
| ABRE4 | 162 (+) | CACGTA | |
| ARE | 659 (−) | AAACCA | Cis-acting regulatory element essential for the anaerobic induction |
| AT~TATA-box | 310 (+) | TATATA | |
| Box III | 628 (+) | ATCATTTTCACT | Protein binding site |
| CAAT-box | 11 (+); 60 (−); 179372 (−); 191 (−); 203 (+); 228 (+); 372 (+); 378 (+); 403 (−); 451 (−); 512 (−); 527 (−); 640 (−); 645 (+); 746 (+); 781 (+); 782 (+) | CCAAT/CAAAT | Common cis-acting element in promoter and enhancer regions |
| G-box | 162 (−); | TACGTG | Cis-acting regulatory element involved in light responsiveness |
| GARE-motif | 591 (+); | TCTGTTG | Gibberellin-responsive element |
| GCN4_motif | 570 (−); | TGAGTCA | Cis-regulatory element involved in endosperm expression |
| MRE | 538 (+);543 (+); | AACCTAA | MYB binding site involved in light responsiveness |
| MYB-binding site | 471 (+); 592 (−) | TAACCA/CAACAG | |
| MYC | 190 (+); 718 (−); 720 (+); 746 (−) | CATTTG/CATGTG | |
| MYB | 566 (+) | TAACTG | |
| STRE | 873 (−) | AGGGG | stress response element |
| TATA | 550 (−) | TATAAAAT | |
| TATA-box | 135 (−); 136 (+); 166 (+); 174 (−); 194 (−); 196 (+); 213 (−); 245 (+); 246 (−); 247 (+); 262 (+); 263 (−)264 (+); 309 (+); 310 (+); 312 (+); 343 (+); 551 (−); 552 (−); 553 (−); 554 (−); 728 (−); 729 (−); 730 (−) | TATAA/TATA/TATAAA/TATAAAA/TATATA/ATATAT/ATTATA/TACAAAA/TATACA/CCTATAAAAA/TATTTAAA | Core promoter element around -30 of transcription start |
| WRE3 | 487 (+) | CCACCT | Wnt-responsive element |
| W-box | 120 (−) | TTGACT | WRKY binding site |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Chen, C.; Xie, F.; Hua, Q.; Zhang, Z.; Zhang, R.; Chen, J.; Zhao, J.; Hu, G.; Qin, Y. A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. Int. J. Mol. Sci. 2021, 22, 2171. https://doi.org/10.3390/ijms22042171
Zhang L, Chen C, Xie F, Hua Q, Zhang Z, Zhang R, Chen J, Zhao J, Hu G, Qin Y. A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. International Journal of Molecular Sciences. 2021; 22(4):2171. https://doi.org/10.3390/ijms22042171
Chicago/Turabian StyleZhang, Lulu, Canbin Chen, Fangfang Xie, Qingzhu Hua, Zhike Zhang, Rong Zhang, Jianye Chen, Jietang Zhao, Guibing Hu, and Yonghua Qin. 2021. "A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1" International Journal of Molecular Sciences 22, no. 4: 2171. https://doi.org/10.3390/ijms22042171
APA StyleZhang, L., Chen, C., Xie, F., Hua, Q., Zhang, Z., Zhang, R., Chen, J., Zhao, J., Hu, G., & Qin, Y. (2021). A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. International Journal of Molecular Sciences, 22(4), 2171. https://doi.org/10.3390/ijms22042171









