New Insights Into Oral Squamous Cell Carcinoma: From Clinical Aspects to Molecular Tumorigenesis
Abstract
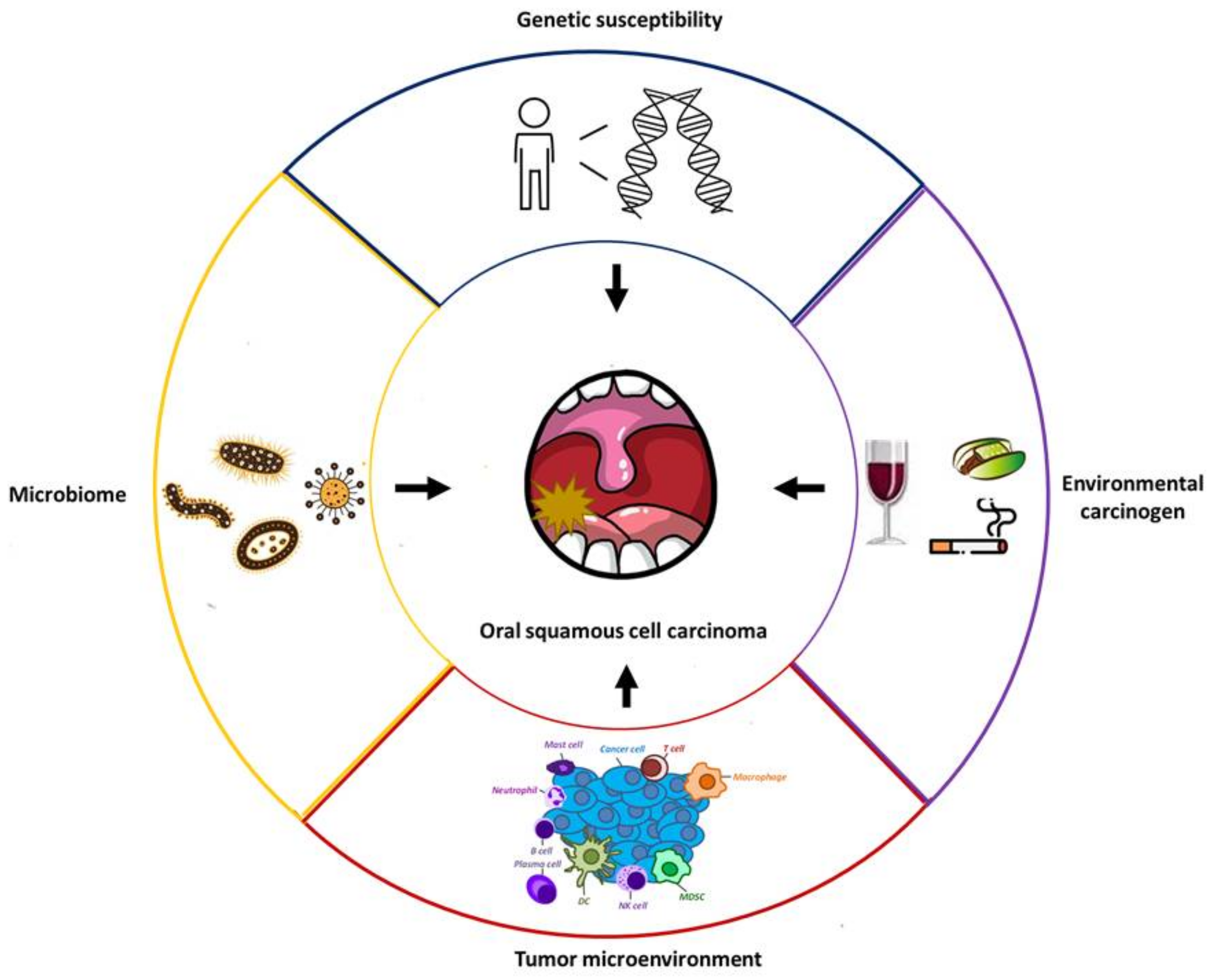
:1. Introduction
2. Perspectives from Clinicopathological Risk Factors
2.1. Conventional Overview of Clinicopathological Risk Factors
2.2. Oral Hygiene and Microbiota
2.3. Gene Susceptibility
3. Perspectives on Molecular Tumorigenesis
3.1. General Concepts of Molecular Tumorigenesis Inside Oral Cancer Cells
3.2. Molecular Overview of Lymph Node Metastases of Oral Cancer
3.3. Novel Investigations of Lymphovascular Invasion in Oral Cancer
4. Overview of the Impact of the TME on Oral SCC
4.1. Current Achievements and Limitations of Immunotherapy
4.2. Outlook on the Immunosuppressive Network in the TME
4.3. Role of IL-1β in Immunomodulation of Oral Cancer
4.4. Metabolism Reprogramming and Antitumor Immunity in Oral Cancer
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CAF | cancer-associated fibroblast |
| CPS | combined positive score |
| CSF1 | colony stimulating factor 1 |
| CSF1R | colony stimulating factor 1 receptor |
| ECM | extracellular matrix |
| EGFR | epidermal growth factor receptor |
| EMT | epithelial–mesenchymal transition |
| EV | extracellular vesicle |
| FAP | fibroblast activation protein |
| HNC | head and neck cancer |
| HPV | human papillomavirus |
| ICI | immune checkpoint inhibitor |
| IFN | interferon |
| IL | interleukin |
| ISG15 | interferon-stimulated gene 15 |
| LC–MS | liquid chromatography–tandem mass spectrometry |
| MAPK | mitogen-activated protein kinase |
| MDSC | myeloid-derived suppressor cell |
| miRNA | microRNA |
| mtDNA | mitochondrial DNA |
| PD-1 | programmed cell death 1 |
| ROS | reactive oxygen species |
| SCC | squamous cell carcinoma |
| scRNA-seq | single-cell RNA-seq |
| SNP | single nucleotide polymorphism |
| TGF-β | transforming growth factor-β |
| TLR | toll-like receptor |
| TME | tumor microenvironment |
| TMEM | transmembrane protein |
| TNF-α | tumor necrosis factor-α |
| VEGF-C | vascular endothelial growth factor C |
References
- Dhanuthai, K.; Rojanawatsirivej, S.; Thosaporn, W.; Kintarak, S.; Subarnbhesaj, A.; Darling, M.; Kryshtalskyj, E.; Chiang, C.P.; Shin, H.I.; Choi, S.Y.; et al. Oral cancer: A multicenter study. Med. Oral Patol. Oral Cir. Bucal 2018, 23, e23–e29. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. Available online: http://gco.iarc.fr/ (accessed on 10 October 2020).
- Cancer Registry Annual Report, 2016 (Taiwan). Available online: https://www.hpa.gov.tw/Pages/Detail.aspx?nodeid=269&pid=12235 (accessed on 20 January 2020).
- Chuang, S.L.; Su, W.W.; Chen, S.L.; Yen, A.M.; Wang, C.P.; Fann, J.C.; Chiu, S.Y.; Lee, Y.C.; Chiu, H.M.; Chang, D.C.; et al. Population-based screening program for reducing oral cancer mortality in 2,334,299 Taiwanese cigarette smokers and/or betel quid chewers. Cancer 2017, 123, 1597–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerawala, C.; Roques, T.; Jeannon, J.P.; Bisase, B. Oral cavity and lip cancer: United Kingdom National Multidisciplinary Guidelines. J. Laryngol. Otol. 2016, 130, S83–S89. [Google Scholar] [CrossRef]
- Montero, P.H.; Patel, S.G. Cancer of the oral cavity. Surg. Oncol. Clin. N. Am. 2015, 24, 491–508. [Google Scholar] [CrossRef] [Green Version]
- Mohan, M.; Jagannathan, N. Oral field cancerization: An update on current concepts. Oncol. Rev. 2014, 30, 244. [Google Scholar] [CrossRef] [Green Version]
- Hashim, D.; Genden, E.; Posner, M.; Hashibe, M.; Boffetta, P. Head and neck cancer prevention: From primary prevention to impact of clinicians on reducing burden. Ann. Oncol. 2019, 30, 744–756. [Google Scholar] [CrossRef] [Green Version]
- Koyfman, S.A.; Ismaila, N.; Crook, D.; D’Cruz, A.; Rodriguez, C.P.; Sher, D.J.; Silbermins, D.; Sturgis, E.M.; Tsue, T.T.; Weiss, J.; et al. Management of the Neck in Squamous Cell Carcinoma of the Oral Cavity and Oropharynx: ASCO Clinical Practice Guideline. J. Clin. Oncol. 2019, 37, 1753–1774. [Google Scholar] [CrossRef]
- Oosting, S.F.; Haddad, R.I. Best Practice in Systemic Therapy for Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2019, 9, 815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasco, A.J.; Secretan, M.B.; Straif, K. Tobacco smoking and cancer: A brief review of recent epidemiological evidence. Lung Cancer 2004, 45, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, K.A.; Ralhan, R. Clinical, pathological, cellular and molecular lesions caused by oral smokeless tobacco--a review. J. Oral Pathol. Med. 2007, 36, 63–77. [Google Scholar] [CrossRef]
- Hernandez, B.Y.; Zhu, X.; Goodman, M.T.; Gatewood, R.; Mendiola, P.; Quinata, K.; Paulino, Y.C. Betel nut chewing, oral premalignant lesions, and the oral microbiome. PLoS ONE 2017, 12, e0172196. [Google Scholar] [CrossRef]
- Li, Y.C.; Cheng, A.J.; Lee, L.Y.; Huang, Y.C.; Chang, J.T. Multifaceted Mechanisms of Areca Nuts in Oral Carcinogenesis: The Molecular Pathology from Precancerous Condition to Malignant Transformation. J. Cancer 2019, 10, 4054–4062. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H.; Chang, J.S.; Syu, S.H.; Wong, T.S.; Chan, J.Y.; Tang, Y.C.; Yang, Z.P.; Yang, W.C.; Chen, C.T.; Lu, S.C.; et al. IL-1β promotes malignant transformation and tumor aggressiveness in oral cancer. J. Cell Physiol. 2015, 230, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Altieri, A.; Cogliano, V. WHO International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007, 8, 292–293. [Google Scholar] [CrossRef]
- Stornetta, A.; Guidolin, V.; Balbo, S. Alcohol-Derived Acetaldehyde Exposure in the Oral Cavity. Cancers 2018, 10, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Williams, V.; Filippova, M.; Filippov, V.; Duerksen-Hughes, P. Viral carcinogenesis: Factors inducing DNA damage and virus integration. Cancers 2014, 6, 2155. [Google Scholar] [CrossRef] [PubMed]
- Ha, P.K.; Califano, J.A. The role of human papillomavirus in oral carcinogenesis. Crit. Rev. Oral Biol. Med. 2004, 15, 188–196. [Google Scholar] [CrossRef] [Green Version]
- Hübbers, C.U.; Akgül, B. HPV and cancer of the oral cavity. Virulence 2015, 6, 244–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, H.; Donovan, B.; Wand, H.; Read, T.R.; Regan, D.G.; Grulich, A.E.; Fairley, C.K.; Guy, R.J. Genital warts in young Australians five years into national human papillomavirus vaccination programme: National surveillance data. BMJ 2013, 346, f2032. [Google Scholar] [CrossRef] [Green Version]
- Rosenquist, K.; Wennerberg, J.; Schildt, E.B.; Bladström, A.; Göran Hansson, B.; Andersson, G. Oral status, oral infections and some lifestyle factors as risk factors for oral and oropharyngeal squamous cell carcinoma. A population-based case-control study in southern Sweden. Acta Otolaryngol. 2005, 125, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Slot, D.E.; Van der Weijden, F.; Ciancio, S.G. Oral health, dental care and mouthwash associated with upper aerodigestive tract cancer risk in Europe: The ARCAGE study. Oral Oncol. 2014, 50, e57. [Google Scholar] [CrossRef] [PubMed]
- Hashim, D.; Sartori, S.; Brennan, P.; Curado, M.P.; Wünsch-Filho, V.; Divaris, K.; Olshan, A.F.; Zevallos, J.P.; Winn, D.M.; Franceschi, S.; et al. The role of oral hygiene in head and neck cancer: Results from International Head and Neck Cancer Epidemiology (INHANCE) consortium. Ann. Oncol. 2016, 27, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Eslami, H.; Yousefi, M.; Asgharzadeh, M.; Aghazadeh, M.; Kafil, H.S. Role of oral microbiome on oral cancers, a review. Biomed. Pharmacother. 2016, 84, 552–558. [Google Scholar] [CrossRef]
- Karpiński, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, J.R.; Chang, C.C.; Lee, W.T.; Huang, C.C.; Ou, C.Y.; Tsai, S.T.; Chen, K.C.; Huang, J.S.; Wong, T.Y.; Lai, Y.H.; et al. The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogenesis 2018, 39, 778–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Zhang, Q.; Hua, H.; Chen, F. Changes in the salivary microbiota of oral leukoplakia and oral cancer. Oral Oncol. 2016, 56, e6–e8. [Google Scholar] [CrossRef]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, Y.; Fukuma, N.; Totsika, M.; Kenny, L.; Morrison, M.; Punyadeera, C. The Performance of an Oral Microbiome Biomarker Panel in Predicting Oral Cavity and Oropharyngeal Cancers. Front. Cell Infect. Microbiol. 2018, 8, 267. [Google Scholar] [CrossRef] [Green Version]
- Mager, D.L.; Haffajee, A.D.; Devlin, P.M.; Norris, C.M.; Posner, M.R.; Goodson, J.M. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.Y.; Yeh, Y.M.; Yu, H.Y.; Chin, C.Y.; Hsu, C.W.; Liu, H.; Huang, P.J.; Hu, S.N.; Liao, C.T.; Chang, K.P.; et al. Oral Microbiota Community Dynamics Associated With Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef] [Green Version]
- Katz, J.; Onate, M.D.; Pauley, K.M.; Bhattacharyya, I.; Cha, S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int. J. Oral Sci. 2011, 3, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Ramadas, K.; Hariharan, R.; Rejnish Kumar, R.; Radhakrishna Pillai, M. Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncol. 2006, 42, 350–362. [Google Scholar] [CrossRef]
- Fan, J.; Liu, W.; Zhang, M.; Xing, C. A literature review and systematic meta-analysis on XRCC3 Thr241Met polymorphism associating with susceptibility of oral cancer. Oncol. Lett. 2019, 18, 3265–3273. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Li, C.Y.; Kong, A.N. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 2005, 28, 249–268. [Google Scholar] [CrossRef]
- Ghosh, T.; Gupta, S.; Bajpai, P.; Agarwal, D.; Agarwal, M.; Gupta, O.P.; Agrawal, D. Association of CYP1A1, GSTM1, and GSTT1 gene polymorphism with risk of oral submucous fibrosis in a section of North Indian population. Mol. Biol. Rep. 2012, 39, 9383–9389. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, D.; Gupta, S.; Agarwal, D.; Gupta, O.P.; Agarwal, M. Role of GSTM1 and GSTT1 polymorphism: Susceptibility to oral submucous fibrosis in the North Indian population. Oncology 2010, 79, 181–186. [Google Scholar] [CrossRef]
- Yadav, B.K.; Kaur, J.; Srivastava, A.; Ralhan, R. Effect of polymorphisms in XRCC1, CCND1 and GSTM1 and tobacco exposure as risk modifier for oral leukoplakia. Int. J. Biol. Markers 2009, 24, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2005, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kluwe, J.; Mencin, A.; Schwabe, R.F. Toll-like receptors, wound healing, and carcinogenesis. J. Mol. Med. 2009, 87, 125–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisani, L.P.; Estadella, D.; Ribeiro, D.A. The Role of Toll Like Receptors (TLRs) in Oral Carcinogenesis. Anticancer Res. 2017, 37, 5389–5394. [Google Scholar]
- Kauppila, J.H.; Mattila, A.E.; Karttunen, T.J.; Salo, T. Toll-like receptor 5 and the emerging role of bacteria in carcinogenesis. Oncoimmunology 2013, 2, e23620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.C.; Zhang, F.; Zhang, Z.J.; Meng, S.Y.; Wang, Y.; Xiang, X.R.; Wang, C.; Tang, Y.Y. Tumor necrosis factor-α gene polymorphisms and risk of oral cancer: Evidence from a meta-analysis. Asian Pac. J. Cancer Prev. 2013, 14, 7243–7249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Hao, S.H.; Sun, Y.; Hu, C.M.; Ma, Z.H.; Wang, Z.M.; Liu, J.; Liu, H.B.; Ye, M.; Zhang, Y.F.; et al. Functional Polymorphisms in COX-2 Gene Are Correlated with the Risk of Oral Cancer. Biomed. Res. Int. 2015, 2015, 580652. [Google Scholar] [PubMed]
- Yang, W.H.; Wang, S.J.; Chang, Y.S.; Su, C.M.; Yang, S.F.; Tang, C.H. Association of Resistin Gene Polymorphisms with Oral Squamous Cell Carcinoma Progression and Development. Biomed. Res. Int. 2018, 2018, 9531315. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Mastronikolis, N.; Ragos, V.; Kyrodimos, E.; Chrysovergis, A.; Papanikolaou, V.; Mastronikolis, S.; Stamatelopoulos, A.; Tsiambas, E. Mechanisms of C-myc oncogenic activity in head and neck squamous cell carcinoma. J. BUON 2019, 24, 2242–2244. [Google Scholar] [PubMed]
- Marquard, F.E.; Jücker, M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem. Pharmacol. 2020, 172, 113729. [Google Scholar] [CrossRef]
- Hsieh, J.C.; Wang, H.M.; Wu, M.H.; Chang, K.P.; Chang, P.H.; Liao, C.T.; Liau, C.T. Review of emerging biomarkers in head and neck squamous cell carcinoma in the era of immunotherapy and targeted therapy. Head Neck. 2019, 41 (Suppl. 1), 19–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picon, H.; Guddati, A.K. Mechanisms of resistance in head and neck cancer. Am. J. Cancer Res. 2020, 10, 2742–2751. [Google Scholar]
- Barnes, P.; Yeboah, F.A.; Zhu, J.; Saahene, R.O.; Obirikorang, C.; Adinortey, M.B.; Amoani, B.; Kyei, F.; Akakpo, P.; Awuku, Y.A. Prognostic Worth of Epidermal Growth Factor Receptor (EGFR) in Patients with Head and Neck Tumors. J. Cancer Epidemiol. 2020, 2020, 5615303. [Google Scholar] [CrossRef]
- Vermorken, J.B.; Mesia, R.; Rivera, F.; Remenar, E.; Kawecki, A.; Rottey, S.; Erfan, J.; Zabolotnyy, D.; Kienzer, H.R.; Cupissol, D.; et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 2008, 359, 1116–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCCN Guidelines for Head and Neck Cancers. Available online: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf (accessed on 10 October 2020).
- Zhu, G.; Pan, C.; Bei, J.X.; Li, B.; Liang, C.; Xu, Y.; Fu, X. Mutant p53 in Cancer Progression and Targeted Therapies. Front. Oncol. 2020, 10, 595187. [Google Scholar] [CrossRef] [PubMed]
- Ragos, V.; Mastronikolis, N.S.; Tsiambas, E.; Baliou, E.; Mastronikolis, S.N.; Tsoukalas, N.; Patsouri, E.E.; Fotiades, P.P. p53 mutations in oral cavity carcinoma. J. BUON 2018, 23, 1569–1572. [Google Scholar] [PubMed]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [Green Version]
- Lim, A.M.; Do, H.; Young, R.J.; Wong, S.Q.; Angel, C.; Collins, M.; Takano, E.A.; Corry, J.; Wiesenfeld, D.; Kleid, S.; et al. Differential mechanisms of CDKN2A (p16) alteration in oral tongue squamous cell carcinomas and correlation with patient outcome. Int. J. Cancer 2014, 135, 887–895. [Google Scholar] [CrossRef]
- Adkins, D.; Ley, J.; Neupane, P.; Worden, F.; Sacco, A.G.; Palka, K.; Grilley-Olson, J.E.; Maggiore, R.; Salama, N.N.; Trinkaus, K.; et al. Palbociclib and cetuximab in platinum-resistant and in cetuximab-resistant human papillomavirus-unrelated head and neck cancer: A multicentre, multigroup, phase 2 trial. Lancet Oncol. 2019, 20, 1295–1305. [Google Scholar] [CrossRef]
- Massano, J.; Regateiro, F.S.; Januário, G.; Ferreira, A. Oral squamous cell carcinoma: Review of prognostic and predictive factors. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2006, 102, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, C.; Vaidya, S.; Wadhwan, V.; Malik, S. Lymph node metastasis: A bearing on prognosis in squamous cell carcinoma. Indian J. Cancer 2015, 52, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Baik, S.H.; Seo, J.W.; Kim, J.H.; Lee, S.K.; Choi, E.C.; Kim, J. Prognostic Value of Cervical Nodal Necrosis Observed in Preoperative CT and MRI of Patients With Tongue Squamous Cell Carcinoma and Cervical Node Metastases: A Retrospective Study. AJR Am. J. Roentgenol. 2019, 213, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Wissmann, C.; Detmar, M. Pathways targeting tumor lymphangiogenesis. Clin. Cancer Res. 2006, 12, 6865–6868. [Google Scholar] [CrossRef] [Green Version]
- Yanase, M.; Kato, K.; Yoshizawa, K.; Noguchi, N.; Kitahara, H.; Nakamura, H. Prognostic value of vascular endothelial growth factors A and C in oral squamous cell carcinoma. J. Oral Pathol. Med. 2014, 43, 514–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naruse, T.; Yanamoto, S.; Yamada, S.I.; Takahashi, H.; Matsushita, Y.; Imayama, N.; Ikeda, H.; Shiraishi, T.; Fujita, S.; Ikeda, T.; et al. Immunohistochemical study of vascular endothelial growth factor-C/vascular endothelial growth factor receptor-3 expression in oral tongue squamous cell carcinoma: Correlation with the induction of lymphangiogenesis. Oncol Lett. 2015, 10, 2027–2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasahira, T.; Ueda, N.; Yamamoto, K.; Kurihara, M.; Matsushima, S.; Bhawal, U.K.; Kirita, T.; Kuniyasu, H. Prox1 and FOXC2 act as regulators of lymphangiogenesis and angiogenesis in oral squamous cell carcinoma. PLoS ONE 2014, 9, e92534. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, L.H.; Niu, Y.H.; Geng, N.B.; Feng, C.J. AEG-1 promotes angiogenesis and may be a novel treatment target for tongue squamous cell carcinoma. Oral Dis. 2020, 26, 876–884. [Google Scholar] [CrossRef]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Ota, I.; Masui, T.; Kurihara, M.; Yook, J.I.; Mikami, S.; Kimura, T.; Shimada, K.; Konishi, N.; Yane, K.; Yamanaka, T.; et al. Snail-induced EMT promotes cancer stem cell-like properties in head and neck cancer cells. Oncol. Rep. 2016, 35, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Hsu, D.S.; Hwang, W.L.; Yuh, C.H.; Chu, C.H.; Ho, Y.H.; Chen, P.B.; Lin, H.S.; Lin, H.K.; Wu, S.P.; Lin, C.Y.; et al. Lymphotoxin-beta interacts with methylated EGFR to mediate acquired resistance to cetuximab in head and neck cancer. Clin. Cancer Res. 2017, 23, 4388–4401. [Google Scholar] [CrossRef] [Green Version]
- Goppel, J.; Mockelmann, N.; Munscher, A.; Sauter, G.; Schumacher, U. Expression of epithelial-mesenchymal transition regulating transcription factors in head and neck squamous cell carcinomas. Anticancer Res. 2017, 37, 5435–5440. [Google Scholar] [PubMed]
- Zhou, Y.; Zhang, H.; Zhuo, X.; Liu, Y.; Zhang, G.; Tan, Y. Over-expression of TWIST, an epithelial-mesenchymal transition inducer, predicts poor survival in patients with oral carcinoma. Int. J. Clin. Exp. Med. 2015, 8, 9239–9247. [Google Scholar]
- Seyedmajidi, M.; Seifi, S.; Moslemi, D.; Mozaffari, S.F.; Gholinia, H.; Zolfaghari, Z. Immunohistochemical expression of TWIST in oral squamous cell carcinoma and its correlation with clinicopathologic factors. J. Cancer Res. Ther. 2018, 14, 964–969. [Google Scholar] [PubMed]
- Bai, Y.; Sha, J.; Kanno, T. The Role of Carcinogenesis-Related Biomarkers in the Wnt Pathway and Their Effects on Epithelial-Mesenchymal Transition (EMT) in Oral Squamous Cell Carcinoma. Cancers 2020, 12, 555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, M.; Flores, T.; Betancur, D.; Peña-Oyarzún, D.; Torres, V.A. Wnt/β-Catenin Signaling in Oral Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 4682. [Google Scholar] [CrossRef]
- Mikels, A.J.; Nusse, R. Wnts as ligands: Processing, secretion and reception. Oncogene 2006, 25, 7461–7468. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.H.; Chang, J.S.; Hsiao, J.R.; Yen, Y.C.; Jiang, S.S.; Liu, S.H.; Chen, Y.L.; Shen, Y.Y.; Chang, J.Y.; Chen, Y.W. Tumour cell-derived WNT5B modulates in vitro lymphangiogenesis via induction of partial endothelial-mesenchymal transition of lymphatic endothelial cells. Oncogene 2017, 36, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Wu, W.L.; Jang, C.W.; Yen, Y.C.; Wang, S.H.; Tsai, F.Y.; Shen, Y.Y.; Chen, Y.W. Interferon-stimulated gene 15 modulates cell migration by interacting with Rac1 and contributes to lymph node metastasis of oral squamous cell carcinoma cells. Oncogene 2019, 38, 4480–4495. [Google Scholar] [CrossRef] [PubMed]
- Cajee, U.F.; Hull, R.; Ntwasa, M. Modification by ubiquitin-like proteins: Significance in apoptosis and autophagy pathways. Int. J. Mol. Sci. 2012, 13, 1804. [Google Scholar] [CrossRef]
- Kazanietz, M.G.; Caloca, M.J. The Rac GTPase in Cancer: From Old Concepts to New Paradigms. Cancer Res. 2017, 77, 5445–5451. [Google Scholar] [CrossRef] [Green Version]
- Choi, D.; Spinelli, C.; Montermini, L.; Rak, J. Oncogenic Regulation of Extracellular Vesicle Proteome and Heterogeneity. Proteomics 2019, 19, e1800169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, A.; Boutros, M. Extracellular vesicles and oncogenic signaling. Mol. Oncol. 2021, 15, 3–26. [Google Scholar] [CrossRef]
- Wang, S.H.; Liou, G.G.; Liu, S.H.; Chang, J.S.; Hsiao, J.R.; Yen, Y.C.; Chen, Y.L.; Wu, W.L.; Chang, J.Y.; Chen, Y.W. Laminin γ2-enriched extracellular vesicles of oral squamous cell carcinoma cells enhance in vitro lymphangiogenesis via integrin α3-dependent uptake by lymphatic endothelial cells. Int. J. Cancer 2019, 144, 2795–2810. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.C.; Gomez, R.S. MicroRNA and oral cancer: Future perspectives. Oral Oncol. 2008, 44, 910–914. [Google Scholar] [CrossRef]
- Min, A.; Zhu, C.; Peng, S.; Rajthala, S.; Costea, D.E.; Sapkota, D. MicroRNAs as Important Players and Biomarkers in Oral Carcinogenesis. Biomed. Res. Int. 2015, 2015, 186904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, C.; Li, Y. Prospective applications of microRNAs in oral cancer. Oncol. Lett. 2019, 18, 3974–3984. [Google Scholar] [PubMed]
- Shiah, S.G.; Hsiao, J.R.; Chang, W.M.; Chen, Y.W.; Jin, Y.T.; Wong, T.Y.; Huang, J.S.; Tsai, S.T.; Hsu, Y.M.; Chou, S.T.; et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014, 74, 7560–7572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.M.; Shiah, S.G.; Huang, C.C.; Hsiao, J.-R.; Chang, J.-Y. Up-regulation of miR-455-5p by the TGF-β-SMAD signalling axis promotes the proliferation of oral squamous cancer cells by targeting, U.B.E.2.B. J. Pathol. 2016, 240, 38–49. [Google Scholar] [CrossRef]
- Chang, W.M.; Lin, Y.F.; Su, C.Y.; Peng, H.Y.; Chang, Y.C.; Lai, T.C.; Wu, G.H.; Hsu, Y.M.; Chi, L.H.; Hsiao, J.R.; et al. Dysregulation of RUNX2/Activin-A Axis upon miR-376c Downregulation Promotes Lymph Node Metastasis in Head and Neck Squamous Cell Carcinoma. Cancer Res. 2016, 76, 7140–7150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsing, E.W.; Shiah, S.G.; Peng, H.Y.; Chen, Y.W.; Chuu, C.P.; Hsiao, J.R.; Lyu, P.C.; Chang, J.Y. TNF-α-induced miR-450a mediates TMEM182 expression to promote oral squamous cell carcinoma motility. PLoS ONE 2019, 14, e0213463. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Mishra, L.; Deng, C.X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Pang, X.; Tang, Y.L.; Liang, X.H. Transforming growth factor-β signaling in head and neck squamous cell carcinoma: Insights into cellular responses. Oncol. Lett. 2018, 16, 4799–4806. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, C.A.; Yamamoto, H.; Fujita, T.; Furuichi, T.; Ito, K.; Inoue, K.; Yamana, K.; Zanma, A.; Takada, K.; Ito, Y.; et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004, 18, 952–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wajant, H. The role of TNF in cancer. Results Probl. Cell Differ. 2009, 49, 1–15. [Google Scholar] [PubMed]
- Josephs, S.F.; Ichim, T.E.; Prince, S.M.; Kesari, S.; Marincola, F.M.; Escobedo, A.R.; Jafri, A. Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med. 2018, 16, 242. [Google Scholar] [CrossRef]
- Wrzesiński, T.; Szelag, M.; Cieślikowski, W.A.; Ida, A.; Giles, R.; Zodro, E.; Szumska, J.; Poźniak, J.; Kwias, Z.; Bluyssen, H.A.; et al. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer 2015, 15, 518. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Guo, J.; Chen, L.; Luo, N.; Yang, W.; Qu, X. Overexpression of TMEM158 contributes to ovarian carcinogenesis. J. Exp. Clin. Cancer Res. 2015, 34, 75. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.C.; Shen, B.Y.; Deng, X.X.; Chen, H.; Zhu, Z.G.; Peng, C.H. TMEM45B promotes proliferation, invasion and migration and inhibits apoptosis in pancreatic cancer cells. Mol. Biosyst. 2016, 12, 1860–1870. [Google Scholar] [CrossRef]
- Boxberg, M.; Leising, L.; Steiger, K.; Jesinghaus, M.; Alkhamas, A.; Mielke, M.; Pfarr, N.; Götz, C.; Wolff, K.D.; Weichert, W.; et al. Composition and Clinical Impact of the Immunologic Tumor Microenvironment in Oral Squamous Cell Carcinoma. J. Immunol. 2019, 202, 278–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peltanova, B.; Raudenska, M.; Masarik, M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: A systematic review. Mol. Cancer 2019, 18, 63. [Google Scholar] [CrossRef]
- Mohan, S.P.; Bhaskaran, M.K.; George, A.L.; Thirutheri, A.; Somasundaran, M.; Pavithran, A. Immunotherapy in Oral Cancer. J. Pharm. Bioallied. Sci. 2019, 11 (Suppl. 2), S107–S111. [Google Scholar] [CrossRef]
- Kujan, O.; van Schaijik, B.; Farah, C.S. Immune Checkpoint Inhibitors in Oral Cavity Squamous Cell Carcinoma and Oral Potentially Malignant Disorders: A Systematic Review. Cancers 2020, 12, 1937. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Hum. Vaccin Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Yan, Y.Y.; Li, J.J.; Adhikari, R.; Fu, L.W. PD-1/PD-L1 Based Combinational Cancer Therapy: Icing on the Cake. Front. Pharmacol. 2020, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Soulieres, D.; Le Tourneau, C.; Dinis, J.; Licitra, L.; Ahn, M.J.; Soria, A.; Machiels, J.P.; Mach, N.; Mehra, R.; et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 2019, 393, 156–167. [Google Scholar] [CrossRef]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Zandberg, D.P.; Strome, S.E. The role of the PD-L1: PD-1 pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2014, 50, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Hua, Y.; Yu, B.; Ye, X.; He, Z.; Li, C.; Wang, J.; Mo, Y.; Wei, X.; Chen, Y.; et al. Predictive biomarkers and mechanisms underlying resistance to PD1/PD-L1 blockade cancer immunotherapy. Mol. Cancer. 2020, 19, 19. [Google Scholar] [CrossRef]
- Chen, Y.P.; Wang, Y.Q.; Lv, J.W.; Li, Y.Q.; Chua, M.L.K.; Le, Q.T.; Lee, N.; Colevas, A.D.; Seiwert, T.; Hayes, D.N.; et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: Implications for immunotherapy. Ann. Oncol. 2019, 30, 68–75. [Google Scholar] [CrossRef]
- Qin, X.; Yan, M.; Wang, X.; Xu, Q.; Wang, X.; Zhu, X.; Shi, J.; Li, Z.; Zhang, J.; Chen, W. Cancer-associated Fibroblast-derived IL-6 Promotes Head and Neck Cancer Progression via the Osteopontin-NF-kappa B Signaling Pathway. Theranostics 2018, 8, 921–940. [Google Scholar] [CrossRef]
- Tsai, M.S.; Chen, W.C.; Lu, C.H.; Chen, M.F. The prognosis of head and neck squamous cell carcinoma related to immunosuppressive tumor microenvironment regulated by IL-6 signaling. Oral Oncol. 2019, 91, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Punyani, S.R.; Sathawane, R.S. Salivary level of interleukin-8 in oral precancer and oral squamous cell carcinoma. Clin. Oral Investig. 2013, 17, 517–524. [Google Scholar] [CrossRef]
- Nishio, Y.; Gojoubori, T.; Kaneko, Y.; Shimizu, N.; Asano, M. Cancer cell-derived IL-8 induces monocytic THP1 cells to secrete IL-8 via the mitogen-activated protein kinase pathway. Tumour Biol. 2015, 36, 9171–9177. [Google Scholar] [CrossRef]
- Chuang, C.-Y.; Sung, W.-W.; Wang, L.; Lin, W.L.; Yeh, K.T.; Su, M.C.; Hsin, C.H.; Lee, S.Y.; Wu, B.C.; Cheng, Y.W.; et al. Differential impact of IL-10 expression on survival and relapse between HPV16-positive and -negative oral squamous cell carcinomas. PLoS ONE 2012, 7, e47541. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Sun, M.; Gu, C.; Wang, X.; Wang, X.; Chen, D.; Zhao, E.; Jiao, X.; Zheng, J. Expression of CD163, interleukin-10, and interferon-gamma in oral squamous cell carcinoma: Mutual relationships and prognostic implications. Eur. J. Oral Sci. 2014, 122, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.S.; Arantes, D.A.; Bernardes, V.F.; Jaeger, F.; Silva, J.M.; Silva, T.A.; Aguiar, M.C.; Batista, C. Immunosuppressive mediators of oral squamous cell carcinoma in tumour samples and saliva. Hum. Immunol. 2015, 76, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Watari, K.; Shibata, T.; Kawahara, A.; Sata, K.; Nabeshima, H.; Shinoda, A.; Abe, H.; Azuma, K.; Murakami, Y.; Izumi, H.; et al. Tumor-derived interleukin-1 promotes lymphangiogenesis and lymph node metastasis through M2-type macrophages. PLoS ONE 2014, 9, e99568. [Google Scholar] [CrossRef] [PubMed]
- Shchors, K.; Shchors, E.; Rostker, F.; Lawlor, E.R.; Brown-Swigart, L.; Evan, G.I. The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1beta. Genes Dev. 2006, 20, 2527–2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.H.; Chang, C.Y.; Kuo, Y.Z.; Fang, W.Y.; Kao, H.Y.; Tsai, S.T.; Wu, L.W. Cancer-associated fibroblast-derived interleukin-1β activates protumor C-C motif chemokine ligand 22 signaling in head and neck cancer. Cancer Sci. 2019, 110, 2783–2793. [Google Scholar] [CrossRef] [Green Version]
- Astradsson, T.; Sellberg, F.; Berglund, D.; Ehrsson, Y.T.; Laurell, G.F.E. Systemic Inflammatory Reaction in Patients with Head and Neck Cancer-An Explorative Study. Front. Oncol. 2019, 9, 1177. [Google Scholar] [CrossRef] [Green Version]
- Allison, K.E.; Coomber, B.L.; Bridle, B.W. Metabolic reprogramming in the tumour microenvironment: A hallmark shared by cancer cells and T lymphocytes. Immunology 2017, 152, 175–184. [Google Scholar] [CrossRef]
- Cerezo, M.; Rocchi, S. Cancer cell metabolic reprogramming: A keystone for the response to immunotherapy. Cell Death Dis. 2020, 11, 964. [Google Scholar] [CrossRef]
- Kumar, D.; New, J.; Vishwakarma, V.; Joshi, R.; Enders, J.; Lin, F.; Dasari, S.; Gutierrez, W.R.; Leef, G.; Ponnurangam, S.; et al. Cancer-Associated Fibroblasts Drive Glycolysis in a Targetable Signaling Loop Implicated in Head and Neck Squamous Cell Carcinoma Progression. Cancer Res. 2018, 78, 3769–3782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colegio, O.R.; Chu, N.Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Jha, A.K.; Huang, S.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [Green Version]
- El Kasmi, K.C.; Stenmark, K.R. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin Immunol. 2015, 27, 267–275. [Google Scholar] [CrossRef] [Green Version]
- Cemerski, S.; Cantagrel, A.; Van Meerwijk, J.P.; Romagnoli, P. Reactive oxygen species differentially affect T cell receptor-signaling pathways. J. Biol. Chem. 2002, 277, 19585–19593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamoto, K.; Chowdhury, P.S.; Kumar, A.; Sonomura, K.; Matsuda, F.; Fagarasan, S.; Honjo, T. Mitochondrial activation chemicals synergize with surface receptor PD-1 blockade for T cell-dependent antitumor activity. Proc. Natl. Acad. Sci. USA 2017, 114, E761–E770. [Google Scholar] [CrossRef] [Green Version]
- Maj, T.; Wang, W.; Crespo, J.; Zhang, H.; Wang, W.; Wei, S.; Zhao, L.; Vatan, L.; Shao, I.; Szeliga, W.; et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017, 18, 1332–1341. [Google Scholar] [CrossRef]
- Sung, Y.J.; Kao, T.Y.; Kuo, C.L.; Fan, C.C.; Cheng, A.N.; Fang, W.C.; Chou, H.Y.; Lo, Y.K.; Chen, C.H.; Jiang, S.S.; et al. Mitochondrial Lon sequesters and stabilizes p53 in the matrix to restrain apoptosis under oxidative stress via its chaperone activity. Cell Death Dis. 2018, 9, 697. [Google Scholar] [CrossRef]
- Voos, W.; Pollecker, K. The Mitochondrial Lon Protease: Novel Functions off the Beaten Track? Biomolecules 2020, 10, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.L.; Chou, H.Y.; Chiu, Y.C.; Cheng, A.N.; Fan, C.C.; Chang, Y.N.; Chen, C.H.; Jiang, S.S.; Chen, N.J.; Lee, A.Y. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett. 2020, 474, 138–150. [Google Scholar] [CrossRef]
- Cheng, A.N.; Cheng, L.C.; Kuo, C.L.; Lo, Y.K.; Chou, H.Y.; Chen, C.H.; Wang, Y.H.; Chuang, T.H.; Cheng, S.J.; Lee, A.Y. Mitochondrial Lon-induced mtDNA leakage contributes to PD-L1-mediated immunoescape via STING-IFN signaling and extracellular vesicles. J. Immunother. Cancer. 2020, 8, e001372. [Google Scholar] [CrossRef]
- Qi, Z.; Barrett, T.; Parikh, A.S.; Tirosh, I.; Puram, S.V. Single-cell sequencing and its applications in head and neck cancer. Oral Oncol. 2019, 99, 104441. [Google Scholar] [CrossRef]
- Huang, L.Y.; Hsieh, Y.P.; Wang, Y.Y.; Hwang, D.Y.; Jiang, S.S.; Huang, W.T.; Chiang, W.F.; Liu, K.J.; Huang, T.T. Single-Cell Analysis of Different Stages of Oral Cancer Carcinogenesis in a Mouse Model. Int. J. Mol. Sci. 2020, 21, 8171. [Google Scholar] [CrossRef] [PubMed]
- Evrard, D.; Szturz, P.; Tijeras-Raballand, A.; Astorgues-Xerri, L.; Abitbol, C.; Paradis, V.; Raymond, E.; Albert, S.; Barry, B.; Faivre, S. Macrophages in the microenvironment of head and neck cancer: Potential targets for cancer therapy. Oral Oncol. 2019, 88, 29–38. [Google Scholar] [CrossRef]
- Pathria, P.; Louis, T.L.; Varner, J.A. Targeting Tumor-Associated Macrophages in Cancer. Trends Immunol. 2019, 40, 310–327. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Nabell, L.; Wong, D.J.L.; Day, T.A.; Daniels, G.A.; Milhem, M.M.; Deva, S.; Jameson, M.B.; Guntinas-Lichius, O.; Almubarak, M.; et al. Phase 1b/2, open label, multicenter study of intratumoral SD-101 in combination with pembrolizumab in anti-PD-1 treatment naïve patients with recurrent or metastatic head and neck squamous cell carcinoma (HNSCC). J. Clin. Oncol. 2019, 37, 6039. [Google Scholar] [CrossRef]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef]
- Huber, M.A.; Kraut, N.; Park, J.E.; Schubert, R.D.; Rettig, W.J.; Peter, R.U.; Garin-Chesa, P. Fibroblast activation protein: Differential expression and serine protease activity in reactive stromal fibroblasts of melanocytic skin tumors. J. Investig. Dermatol. 2003, 120, 182–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soerensen, M.M.; Ros, W.; Rodriguez-Ruiz, M.E.; Robbrecht, D.; Rohrberg, K.; Martin-Liberal, J.; Lassen, U.; Bermejo, I.M.; Lolkema, M.P.; Tabernero, J.; et al. Safety, PK/PD, and anti-tumor activity of RO6874281, an engineered variant of interleukin-2 (IL-2v) targeted to tumor-associated fibroblasts via binding to fibroblast activation protein (FAP). J. Clin. Oncol. 2018, 36, e15155. [Google Scholar] [CrossRef]

| Bacterial Species | Characteristic | Sample Type | Reference |
|---|---|---|---|
| Actinomyces spp. | Gram-positive anaerobe | Saliva | [28] |
| Clostridium spp. | Gram-positive anaerobe | Saliva | [28] |
| Capnocytophaga spp. | Gram-negative bacteria | Premalignant lesion, tumor, and saliva | [29,30,31] |
| Enterobacteriaceae spp. | Gram-negative bacteria | Saliva | [28] |
| Fusobacterium nucleatum | Gram-negative anaerobe | Saliva and oral rinse | [27,32] |
| Porphyromonas gingivalis | Gram-negative anaerobe | Saliva and tumor | [31,33] |
| Prevotella intermedia | Gram-negative anaerobe | Saliva and oral rinse | [27,32] |
| Prevotella melaninogenica | Gram-negative anaerobe | Saliva | [31] |
| Prevotella tannerae | Gram-negative anaerobe | Saliva | [27] |
| Streptococcus spp. | Gram-positive anaerobe | Premalignant lesion, tumor, and saliva | [29,30,31] |
| Veillonella spp. | Gram-negative anaerobe | Oral rinse | [32] |
| Gene Name | Polymorphism | Protein Function | Reference |
|---|---|---|---|
| XRCC1 | Arg399Gln | DNA damage repair | [34] |
| XRCC3 | Thr241Met | DNA damage repair | [35] |
| CYP1A1 | MspI site | Phase I enzyme | [37] |
| GSTM1 | Null genotype | Phase II enzyme | [37] |
| GSTT1 | Null genotype | Phase II enzyme | [37] |
| TNF-α | −238G > A | Inflammation | [44] |
| COX-2 | −765G > C, +837 T > C | Inflammation | [45] |
| RETN | A/T/G/G haplotype | Inflammation and metabolism | [46] |
| Molecule Name | Biological Function | Potential Mechanisms Regulating Lymph Node Metastasis | Reference |
|---|---|---|---|
| Wnt5B | Activator of the Wnt pathway | Promotes lymphangiogenesis and endothelial–mesenchymal transition by regulating the expression of Snail and Slug proteins | [77] |
| ISG15 | Ubiquitin-like protein | Induces lymphovascular invasion by targeting Rac1 activity | [78] |
| Laminin γ2 | Basement membrane protein | Enhances lymphangiogenesis through the uptake of laminin γ2-enriched EVs by lymphatic endothelial cells | [83] |
| miRNA Name | Biological Function | Potential Mechanisms Regulating Lymph Node Metastasis | Reference |
|---|---|---|---|
| miR329 and miR410 | Tumor suppressor microRNA | Reduced expression of miR329 and miR410 activates the Wnt pathway by targeting Wnt7B | [89] |
| miR-455-5p | Oncogenic microRNA |
| [90] |
| miR-376c | Tumor suppressor microRNA | Downregulated miR-376c promotes lymph node metastasis by targeting the RUNX2/Activin-A axis | [91] |
| miR-450a | Oncogenic microRNA |
| [92] |
| Drug | Action Mechanism | Clinical Benefit | Reference |
|---|---|---|---|
| Cetuximab | Antiepidermal growth factor receptor antibody | In the EXTREME study, the addition of cetuximab to platinum-based chemotherapy in first-line therapy extended the overall survival of patients with recurrent or metastatic disease (hazard ratio: 0.80; 95% confidence interval: 0.64–0.99; p = 0.04). | [53] |
| Nivolumab | Antiprogrammed cell death protein 1 antibody | In the CheckMate 141 study, nivolumab monotherapy prolonged overall survival in patients with advanced disease refractory to a platinum-based regimen (hazard ratio: 0.70; 95% confidence interval: 0.51–0.96; p = 0.01). | [107] |
| Pembrolizumab | Antiprogrammed cell death protein 1 antibody |
| [108,109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.-H.; Hsiao, S.-Y.; Chang, K.-Y.; Chang, J.-Y. New Insights Into Oral Squamous Cell Carcinoma: From Clinical Aspects to Molecular Tumorigenesis. Int. J. Mol. Sci. 2021, 22, 2252. https://doi.org/10.3390/ijms22052252
Chen S-H, Hsiao S-Y, Chang K-Y, Chang J-Y. New Insights Into Oral Squamous Cell Carcinoma: From Clinical Aspects to Molecular Tumorigenesis. International Journal of Molecular Sciences. 2021; 22(5):2252. https://doi.org/10.3390/ijms22052252
Chicago/Turabian StyleChen, Shang-Hung, Sheng-Yen Hsiao, Kwang-Yu Chang, and Jang-Yang Chang. 2021. "New Insights Into Oral Squamous Cell Carcinoma: From Clinical Aspects to Molecular Tumorigenesis" International Journal of Molecular Sciences 22, no. 5: 2252. https://doi.org/10.3390/ijms22052252
APA StyleChen, S.-H., Hsiao, S.-Y., Chang, K.-Y., & Chang, J.-Y. (2021). New Insights Into Oral Squamous Cell Carcinoma: From Clinical Aspects to Molecular Tumorigenesis. International Journal of Molecular Sciences, 22(5), 2252. https://doi.org/10.3390/ijms22052252





