LONP1 Regulates Mitochondrial Accumulations of HMGB1 and Caspase-3 in CA1 and PV Neurons Following Status Epilepticus
Abstract
1. Introduction
2. Results
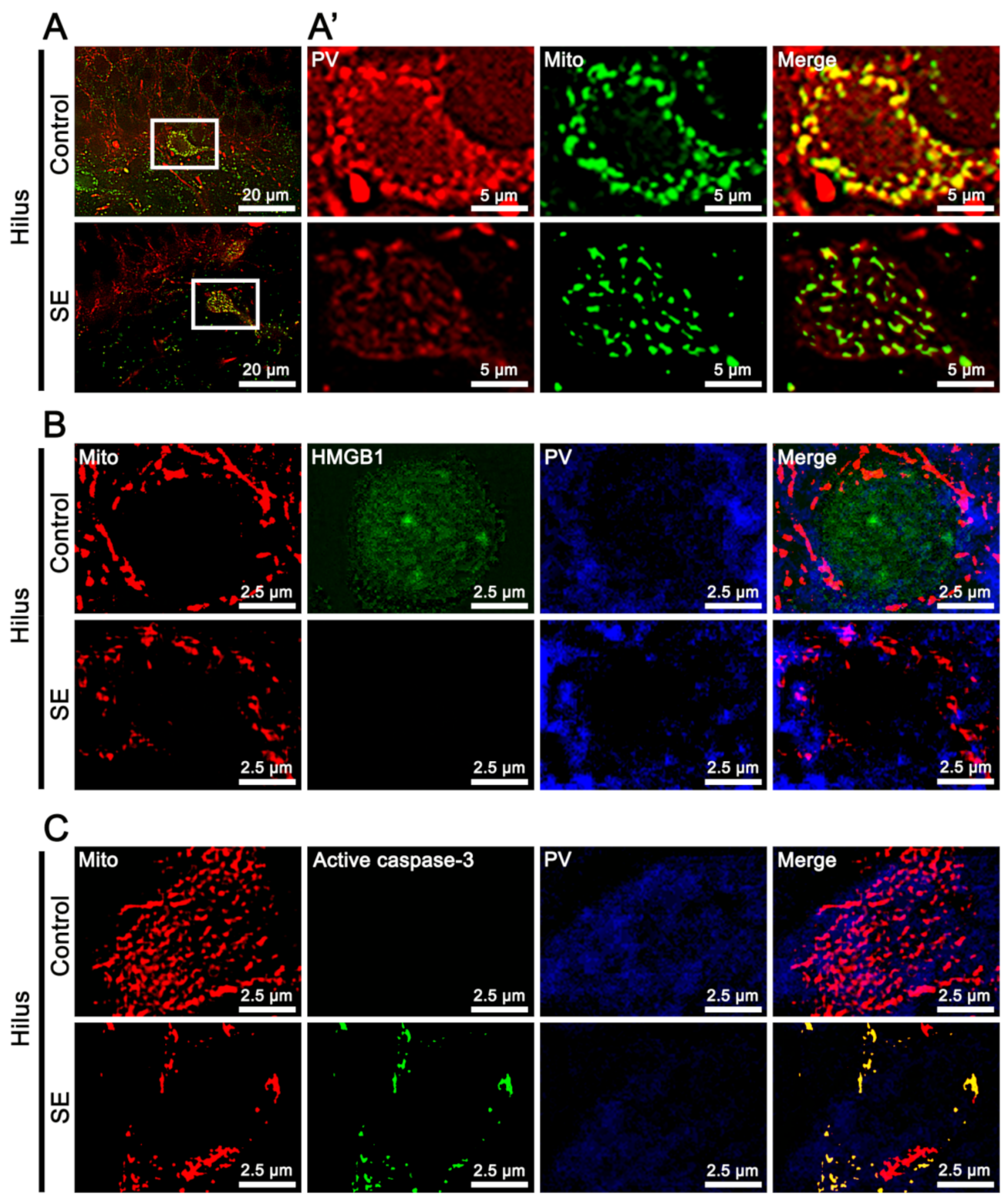
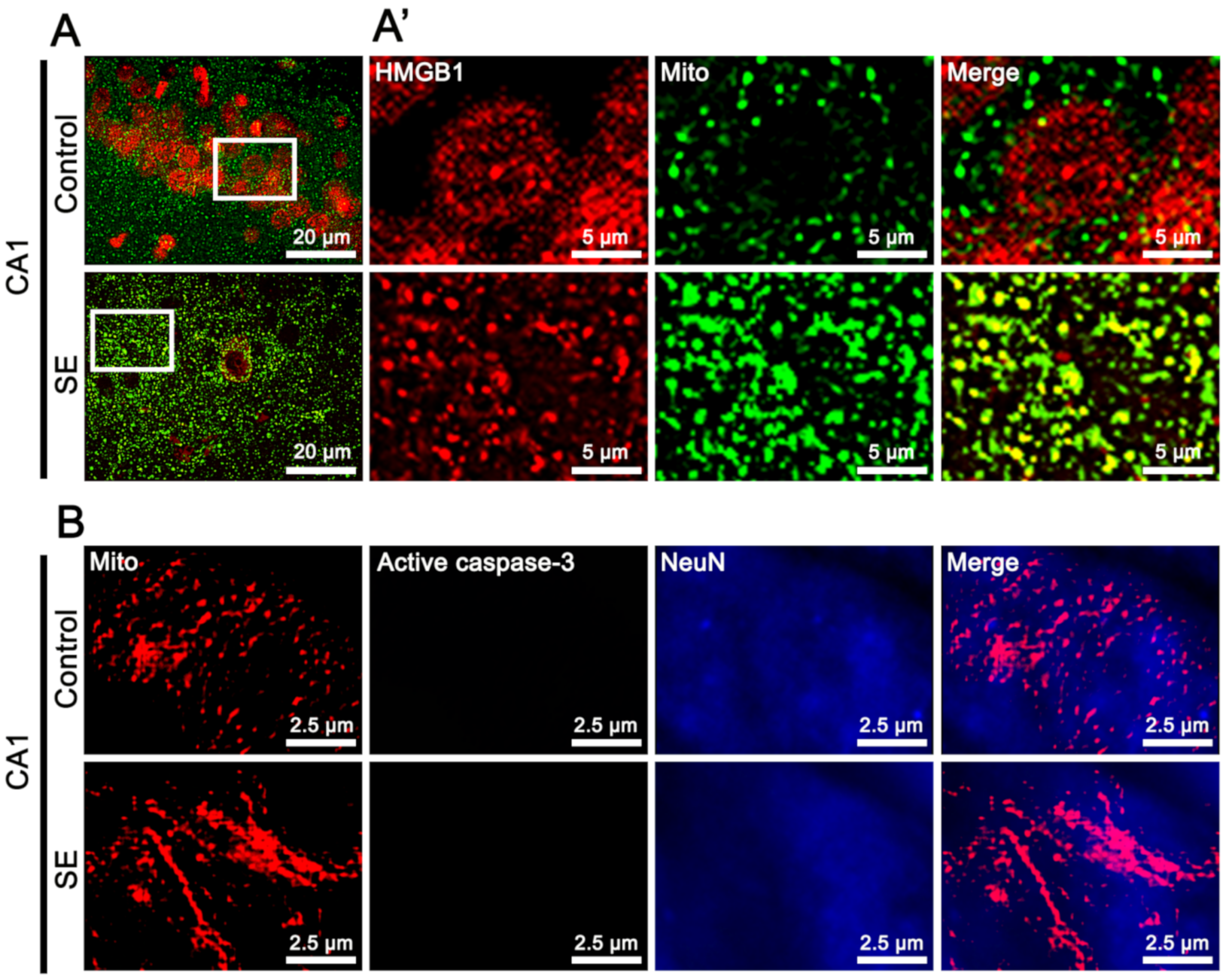
2.1. SE-Induced Mitochondrial Caspase-3 and HMGB1 Translocations in PV and CA1 Neurons, Respectively
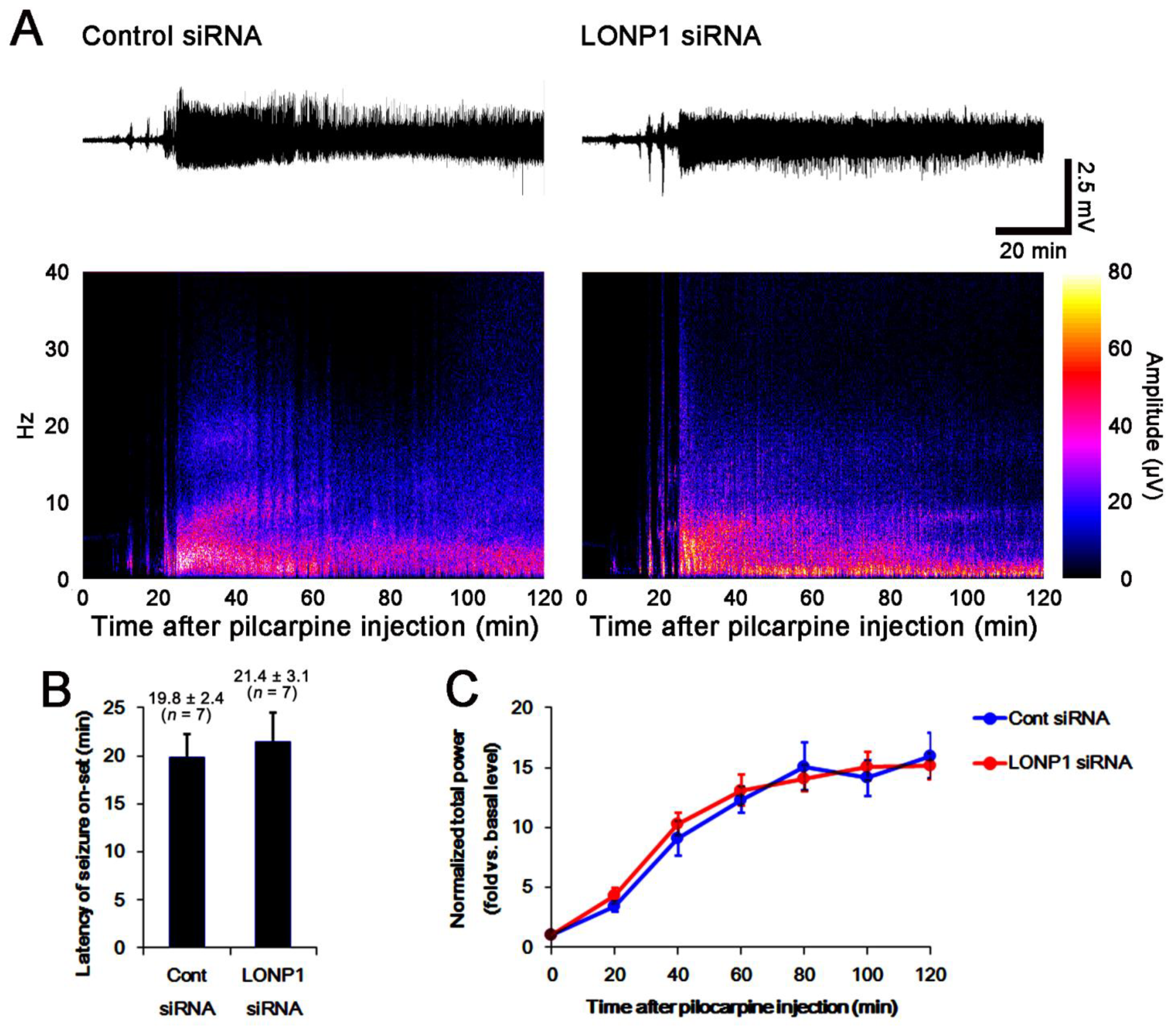
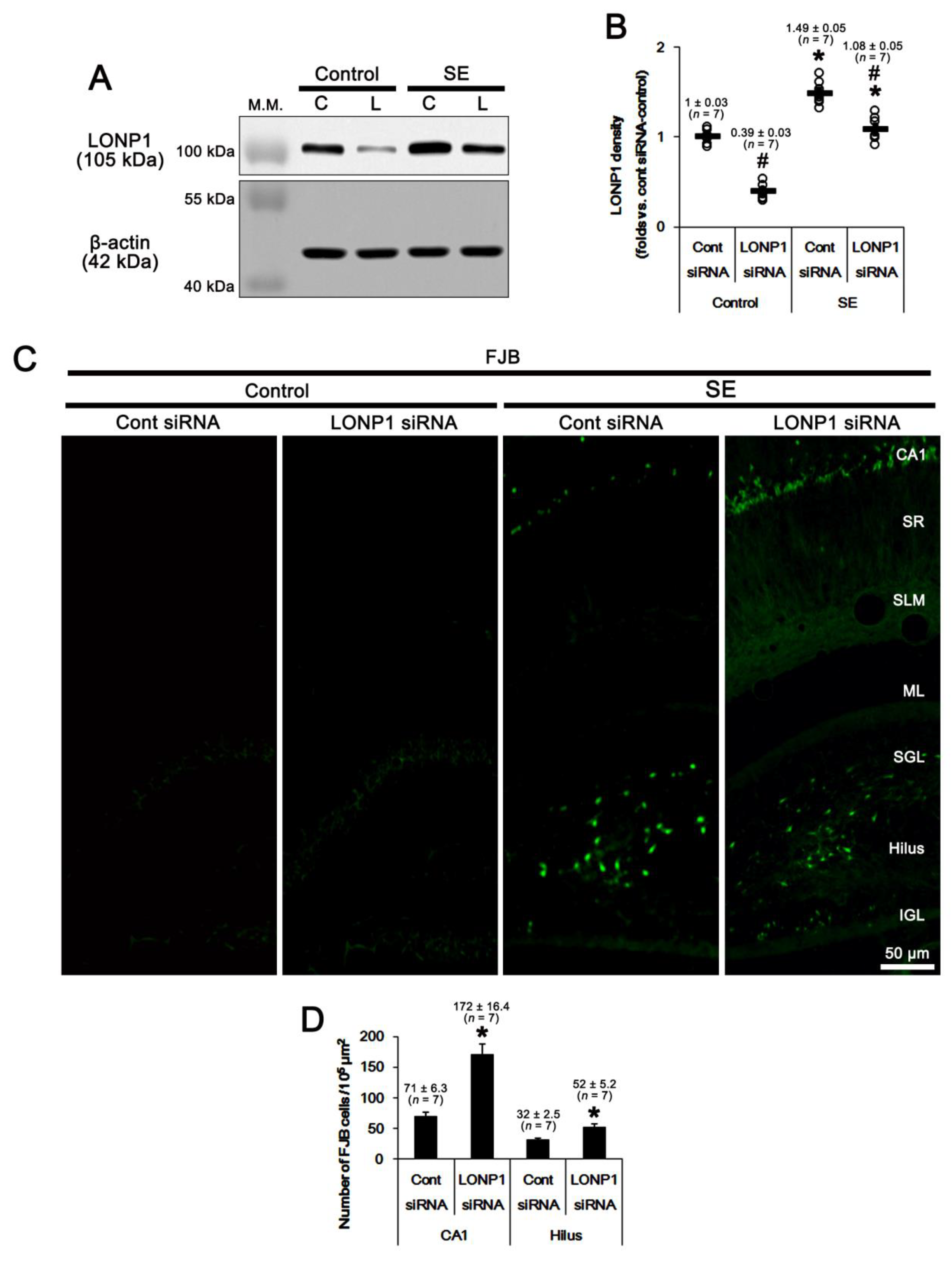
2.2. Effects of LONP1 Knockdown on SE-Induced Neuronal Death
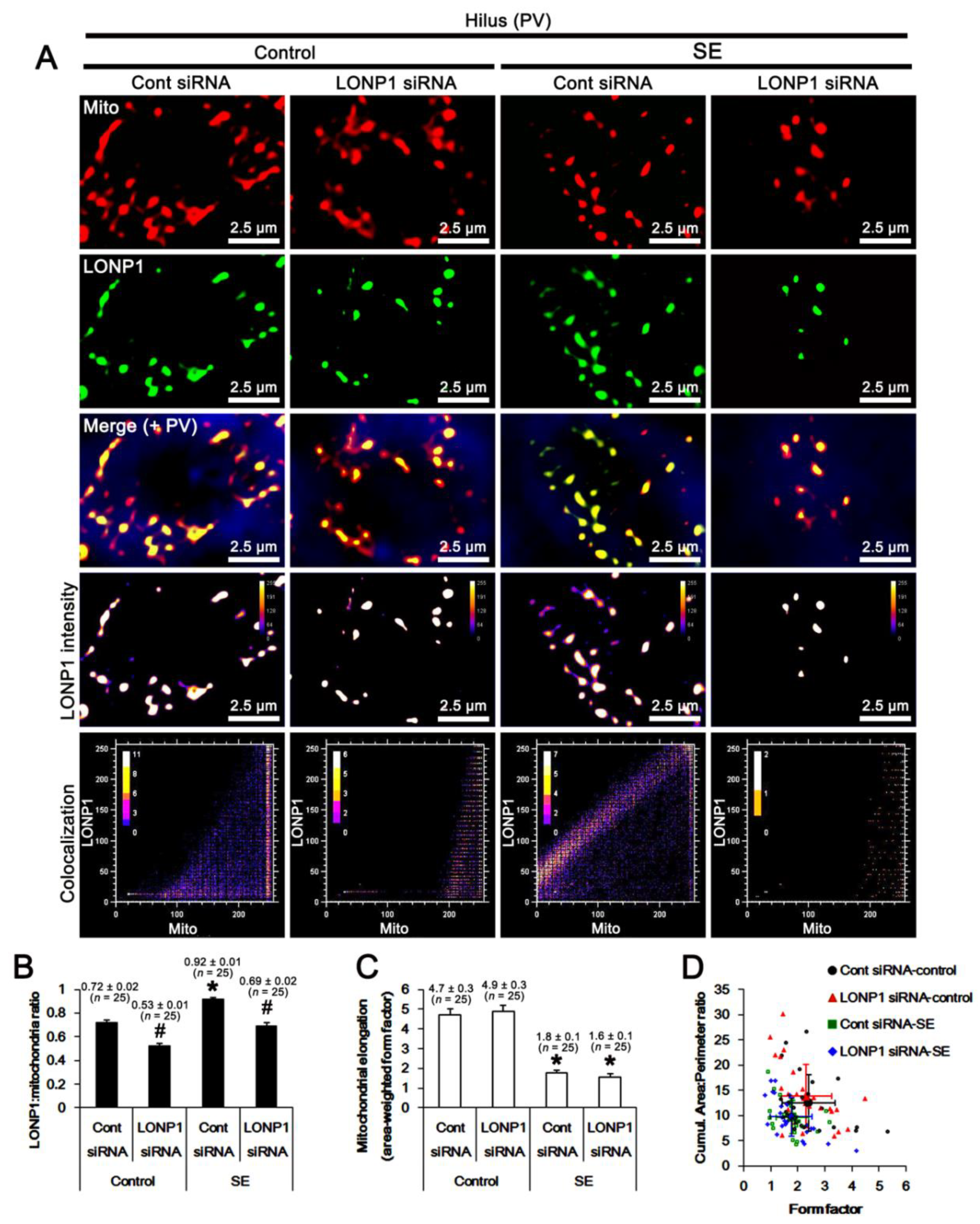
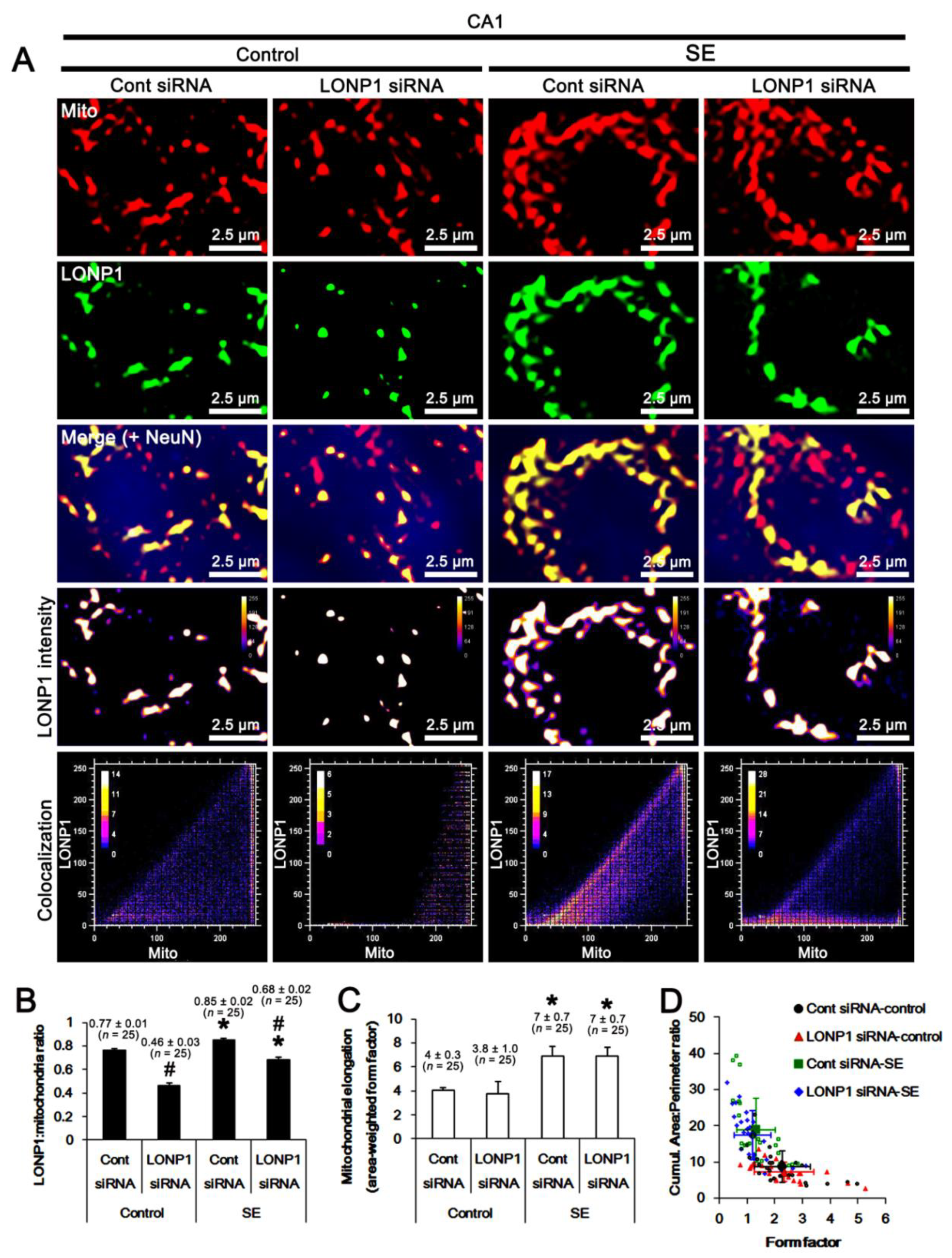
2.3. Effects of LONP1 Knockdown on Mitochondrial LONP1 Level and Mitochondrial Dynamics Following SE
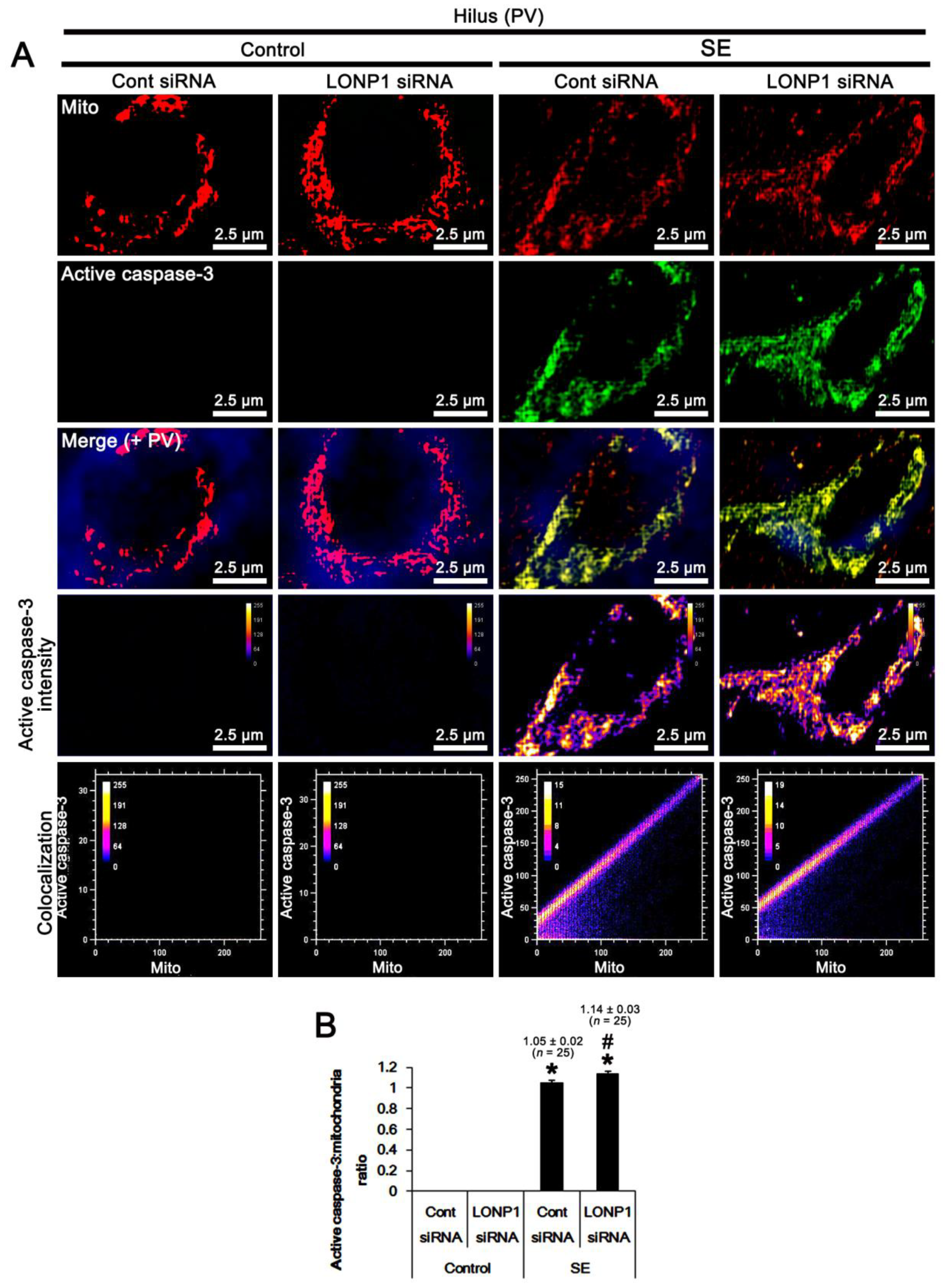
2.4. Effects of LONP1 Knockdown in Mitochondrial Accumulation of Active Caspase-3 in PV Cells Following SE
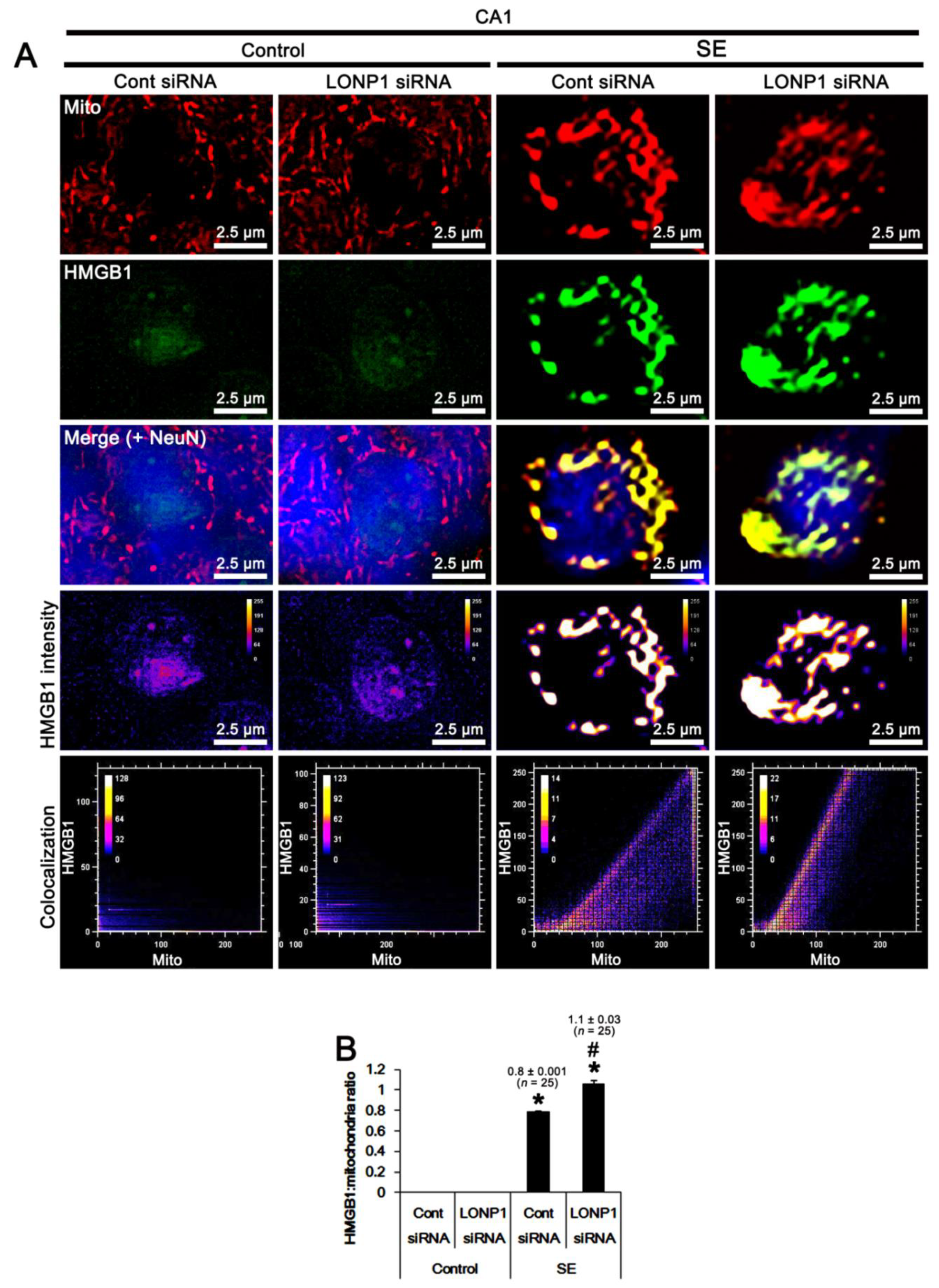
2.5. Effects of LONP1 Knockdown on Mitochondrial HMGB1 Accumulation in CA1 Neurons Following SE
3. Discussion
4. Materials and Methods
4.1. Experimental Animals and Chemicals
4.2. LONP1 Knockdown and Electrode Implantation
4.3. SE Induction and EEG Analysis
4.4. Tissue Processing
4.5. Western Blot
4.6. Immunohistochemistry and FJB Staining
4.7. Cell Count and Mitochondrial Morphometry
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HMGB1 | High mobility group box 1 |
| LONP1 | Lon protease 1 |
| PV | Parvalbumin |
| SE | Status epilepticus |
References
- Pandya, J.D.; Nukala, V.N.; Sullivan, P.G. Concentration dependent effect of calcium on brain mitochondrial bioenergetics and oxidative stress parameters. Front. Neuroenerget. 2013, 5, 10. [Google Scholar] [CrossRef]
- Lezi, E.; Swerdlow, R.H. Mitochondria in neurodegeneration. Adv. Exp. Med. Biol. 2012, 942, 269–286. [Google Scholar] [PubMed]
- Venkatesh, S.; Lee, J.; Singh, K.; Lee, I.; Suzuki, C.K. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim. Biophys. Acta 2012, 1823, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Bulteau, A.L.; Mena, N.P.; Auchère, F.; Lee, I.; Prigent, A.; Lobsiger, C.S.; Camadro, J.M.; Hirsch, E.C. Dysfunction of mitochondrial Lon protease and identification of oxidized protein in mouse brain following exposure to MPTP: Implications for Parkinson disease. Free Radic. Biol. Med. 2017, 108, 236–246. [Google Scholar] [CrossRef]
- Ngo, J.K.; Davies, K.J. Mitochondrial Lon protease is a human stress protein. Free Radic. Biol. Med. 2009, 46, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Bota, D.A.; Ngo, J.K.; Davies, K.J. Downregulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic. Biol. Med. 2005, 38, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Fedoce, A.D.G.; Ferreira, F.; Bota, R.G.; Bonet-Costa, V.; Sun, P.Y.; Davies, K.J.A. The role of oxidative stress in anxiety disorder: Cause or consequence? Free Radic. Res. 2018, 52, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 2006, 125, 1241–1252. [Google Scholar] [CrossRef]
- Detmer, S.A.; Chan, D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell. Biol. 2007, 8, 870–879. [Google Scholar] [CrossRef]
- Terman, A.; Gustafsson, B.; Brunk, U.T. Mitochondrial damage and intralysosomal degradation in cellular aging. Mol. Aspects. Med. 2006, 27, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Barsoum, M.J.; Yuan, H.; Gerencser, A.A.; Liot, G.; Kushnareva, Y.; Gräber, S.; Kovacs, I.; Lee, W.D.; Waggoner, J.; Cui, J.; et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006, 25, 3900–3911. [Google Scholar] [CrossRef]
- Chandra, D.; Tang, D.G. Mitochondrially localized active caspase-9 and caspase-3 result mostly from translocation from the cytosol and partly from caspase-mediated activation in the organelle. Lack of evidence for Apaf-1-mediated procaspase-9 activation in the mitochondria. J. Biol. Chem. 2003, 278, 17408–17420. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, C.; Fulda, S.; Srinivasan, A.; Friesen, C.; Li, F.; Tomaselli, K.J.; Debatin, K.M.; Krammer, P.H.; Peter, M.E. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998, 17, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Ryu, H.J.; Kim, M.J.; Kang, T.C. LIM kinase-2 induces programmed necrotic neuronal death via dysfunction of DRP1-mediated mitochondrial fission. Cell Death Differ. 2014, 21, 1036–1049. [Google Scholar] [CrossRef]
- Ko, A.R.; Hyun, H.W.; Min, S.J.; Kim, J.E.; Kang, T.C. Endothelin-1 induces LIMK2-mediated programmed necrotic neuronal death independent of NOS activity. Mol. Brain 2015, 8, 58. [Google Scholar] [CrossRef][Green Version]
- Hyun, H.W.; Ko, A.R.; Kang, T.C. Mitochondrial translocation of high mobility group box 1 facilitates LIM kinase 2-mediated programmed necrotic neuronal death. Front. Cell. Neurosci. 2016, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Kang, T.C. p47Phox/CDK5/DRP1-mediated mitochondrial fission evokes PV cell degeneration in the rat dentate gyrus following status epilepticus. Front. Cell. Neurosci. 2017, 11, 267. [Google Scholar] [CrossRef]
- Kim, J.E.; Kang, T.C. Differential roles of mitochondrial translocation of active caspase-3 and HMGB1 in neuronal death induced by status epilepticus. Front. Cell. Neurosci. 2018, 12, 301. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Dixit, V.M. Caspases: Intracellular signaling by proteolysis. Cell 1997, 91, 443–446. [Google Scholar] [CrossRef]
- Campello, S.; Scorrano, L. Mitochondrial shape changes: Orchestrating cell pathophysiology. EMBO Rep. 2010, 11, 678–684. [Google Scholar] [CrossRef]
- Verrijdt, G.; Haelens, A.; Schoenmakers, E.; Rombauts, W.; Claessens, F. Comparative analysis of the influence of the high-mobility group box 1 protein on DNA binding and transcriptional activation by the androgen, glucocorticoid, progesterone and mineralocorticoid receptors. Biochem. J. 2002, 361, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef]
- Gdynia, G.; Keith, M.; Kopitz, J.; Bergmann, M.; Fassl, A.; Weber, A.N.; George, J.; Kees, T.; Zentgraf, H.W.; Wiestler, O.D.; et al. Danger signaling protein HMGB1 induces a distinct form of cell death accompanied by formation of giant mitochondria. Cancer Res. 2010, 70, 8558–8568. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Park, H.; Choi, S.H.; Kong, M.J.; Kang, T.C. TRPC6-mediated ERK1/2 activation increases dentate granule cell resistance to status epilepticus via regulating Lon protease-1 expression and mitochondrial dynamics. Cells 2019, 8, 1376. [Google Scholar] [CrossRef]
- Lu, B.; Lee, J.; Nie, X.; Li, M.; Morozov, Y.I.; Venkatesh, S.; Bogenhagen, D.F.; Temiakov, D.; Suzuki, C.K. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell. 2013, 49, 121–132. [Google Scholar] [CrossRef]
- Cribbs, J.T.; Strack, S. Functional characterization of phosphorylation sites in dynamin-related protein 1. Methods Enzymol. 2009, 457, 231–253. [Google Scholar] [PubMed]
- Merrill, R.A.; Dagda, R.K.; Dickey, A.S.; Cribbs, J.T.; Green, S.H.; Usachev, Y.M.; Strack, S. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 2011, 9, e1000612. [Google Scholar] [CrossRef]
- Kim, J.E.; Park, H.; Choi, S.H.; Kong, M.J.; Kang, T.C. CDDO-Me selectively attenuates CA1 neuronal death induced by status epilepticus via facilitating mitochondrial fission independent of LONP1. Cells 2019, 8, 833. [Google Scholar] [CrossRef]
- Fukuda, R.; Zhang, H.; Kim, J.W.; Shimoda, L.; Dang, C.V.; Semenza, G.L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007, 129, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Bota, D.A.; Davies, K.J. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell. Biol. 2002, 4, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Bota, D.A.; Davies, K.J. Mitochondrial Lon protease in human disease and aging: Including an etiologic classification of Lon-related diseases and disorders. Free Radic. Biol. Med. 2016, 100, 188–198. [Google Scholar] [CrossRef]
- Pomatto, L.C.D.; Carney, C.; Shen, B.; Wong, S.; Halaszynski, K.; Salomon, M.P.; Davies, K.J.A.; Tower, J. The mitochondrial Lon protease is required for age-specific and sex-specific adaptation to oxidative stress. Curr. Biol. 2017, 27, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Faraco, G.; Fossati, S.; Bianchi, M.E.; Patrone, M.; Pedrazzi, M.; Sparatore, B.; Moroni, F.; Chiarugi, A. High mobility group box 1 protein is released by neural cells upon different stresses and worsens ischemic neurodegeneration in vitro and in vivo. J. Neurochem. 2007, 103, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Nishimura, M.; Wang, Y.; Sims, J.R.; Qiu, S.; Savitz, S.I.; Salomone, S.; Moskowitz, M.A. Early release of HMGB-1 from neurons after the onset of brain ischemia. J. Cereb. Blood Flow Metab. 2008, 28, 927–938. [Google Scholar] [CrossRef]
- Maroso, M.; Balosso, S.; Ravizza, T.; Liu, J.; Aronica, E.; Iyer, A.M.; Rossetti, C.; Molteni, M.; Casalgrandi, M.; Manfredi, A.A.; et al. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat. Med. 2010, 16, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, K.; Fukuyasu, Y.; Teshigawara, K.; Fu, L.; Wake, H.; Ohtsuka, A.; Nishibori, M. HMGB1 translocation in neurons after ischemic insult: Subcellular localization in mitochondria and peroxisomes. Cells 2020, 9, 643. [Google Scholar] [CrossRef] [PubMed]
- Stumbo, A.C.; Cortez, E.; Rodrigues, C.A.; Henriques, M.; Porto, L.C.; Barbosa, H.S.; Carvalho, L. Mitochondrial localization of non-histone protein HMGB1 during human endothelial cell-Toxoplasma gondii infection. Cell. Biol. Int. 2008, 32, 235–238. [Google Scholar] [CrossRef]
- Ito, H.; Fujita, K.; Tagawa, K.; Chen, X.; Homma, H.; Sasabe, T.; Shimizu, J.; Shimizu, S.; Tamura, T.; Muramatsu, S.; et al. HMGB1 facilitates repair of mitochondrial DNA damage and extends the lifespan of mutant ataxin-1 knock-in mice. EMBO Mol. Med. 2015, 7, 78–101. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.W.; Liu, Y.J.; Lv, Z.; Mao, Y.F.; Wang, Y.W.; Zhu, X.Y.; Jiang, L. Lung endothelial barrier protection by resveratrol involves inhibition of HMGB1 release and HMGB1-induced mitochondrial oxidative damage via an Nrf2-dependent mechanism. Free Radic. Biol. Med. 2015, 88, 404–416. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, W.; Tohme, S.; Chen, M.; Fu, Y.; Tian, D.; Lotze, M.; Tang, D.; Tsung, A. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. J. Hepatol. 2015, 63, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hu, L.; Jiang, J.; Li, H.; Wu, Q.; Ooi, K.; Wang, J.; Feng, Y.; Zhu, D.; Xia, C. HMGB1/RAGE axis mediates stress-induced RVLM neuroinflammation in mice via impairing mitophagy flux in microglia. J. Neuroinflammat. 2020, 17, 15. [Google Scholar] [CrossRef] [PubMed]
- Parkkinen, J.; Raulo, E.; Merenmies, J.; Nolo, R.; Kajander, E.O.; Baumann, M.; Rauvala, H. Amphoterin, the 30-kDa protein in a family of HMG1-type polypeptides. Enhanced expression in transformed cells, leading edge localization, and interactions with plasminogen activation. J. Biol. Chem. 1993, 268, 19726–19738. [Google Scholar] [CrossRef]
- Louneva, N.; Cohen, J.W.; Han, L.Y.; Talbot, K.; Wilson, R.S.; Bennett, D.A.; Trojanowski, J.Q.; Arnold, S.E. Caspase-3 is enriched in postsynaptic densities and increased in Alzheimer’s disease. Am. J. Pathol. 2008, 173, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.J.; Kao, T.Y.; Kuo, C.L.; Fan, C.C.; Cheng, A.N.; Fang, W.C.; Chou, H.Y.; Lo, Y.K.; Chen, C.H.; Jiang, S.S.; et al. Mitochondrial Lon sequesters and stabilizes p53 in the matrix to restrain apoptosis under oxidative stress via its chaperone activity. Cell Death Dis. 2018, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Zamzami, N.; Hirsch, T.; Dallaporta, B.; Petit, P.X.; Kroemer, G. Mitochondrial implication in accidental and programmed cell death: Apoptosis and necrosis. J. Bioenerg. Biomembr. 1997, 29, 185–193. [Google Scholar] [CrossRef]
| Antigen | Host | Manufacturer (Catalog Number) | Dilution Used |
|---|---|---|---|
| Active caspase-3 | Rabbit | Cell signaling (#9664) | 1:400 (IH) |
| High mobility group box 1 (HMGB1) | Rabbit | Abcam (#ab18256) | 1:100 (IH) |
| Lon protease 1 (LONP1) | Rabbit | Proteintech (15440-1-AP) | 1:100 (IH) 1:1000 (WB) |
| Mitochondrial marker (Mitochondrial complex IV subunit 1, MTCO1) | Mouse | Abcam (#ab14705) | 1:500 (IH) |
| NeuN | Guinea pig | Millipore (#3238431) | 1:1000 (IH) |
| Parvalbumin (PV) | Goat | Swant (#PVG213) | 1:100,000 (IH) |
| β-actin | Mouse | Sigma (A5316) | 1:5000 (WB) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-E.; Park, H.; Kim, T.-H.; Kang, T.-C. LONP1 Regulates Mitochondrial Accumulations of HMGB1 and Caspase-3 in CA1 and PV Neurons Following Status Epilepticus. Int. J. Mol. Sci. 2021, 22, 2275. https://doi.org/10.3390/ijms22052275
Kim J-E, Park H, Kim T-H, Kang T-C. LONP1 Regulates Mitochondrial Accumulations of HMGB1 and Caspase-3 in CA1 and PV Neurons Following Status Epilepticus. International Journal of Molecular Sciences. 2021; 22(5):2275. https://doi.org/10.3390/ijms22052275
Chicago/Turabian StyleKim, Ji-Eun, Hana Park, Tae-Hyun Kim, and Tae-Cheon Kang. 2021. "LONP1 Regulates Mitochondrial Accumulations of HMGB1 and Caspase-3 in CA1 and PV Neurons Following Status Epilepticus" International Journal of Molecular Sciences 22, no. 5: 2275. https://doi.org/10.3390/ijms22052275
APA StyleKim, J.-E., Park, H., Kim, T.-H., & Kang, T.-C. (2021). LONP1 Regulates Mitochondrial Accumulations of HMGB1 and Caspase-3 in CA1 and PV Neurons Following Status Epilepticus. International Journal of Molecular Sciences, 22(5), 2275. https://doi.org/10.3390/ijms22052275






