Analysis of ADAM12-Mediated Ephrin-A1 Cleavage and Its Biological Functions
Abstract
1. Introduction
2. Results
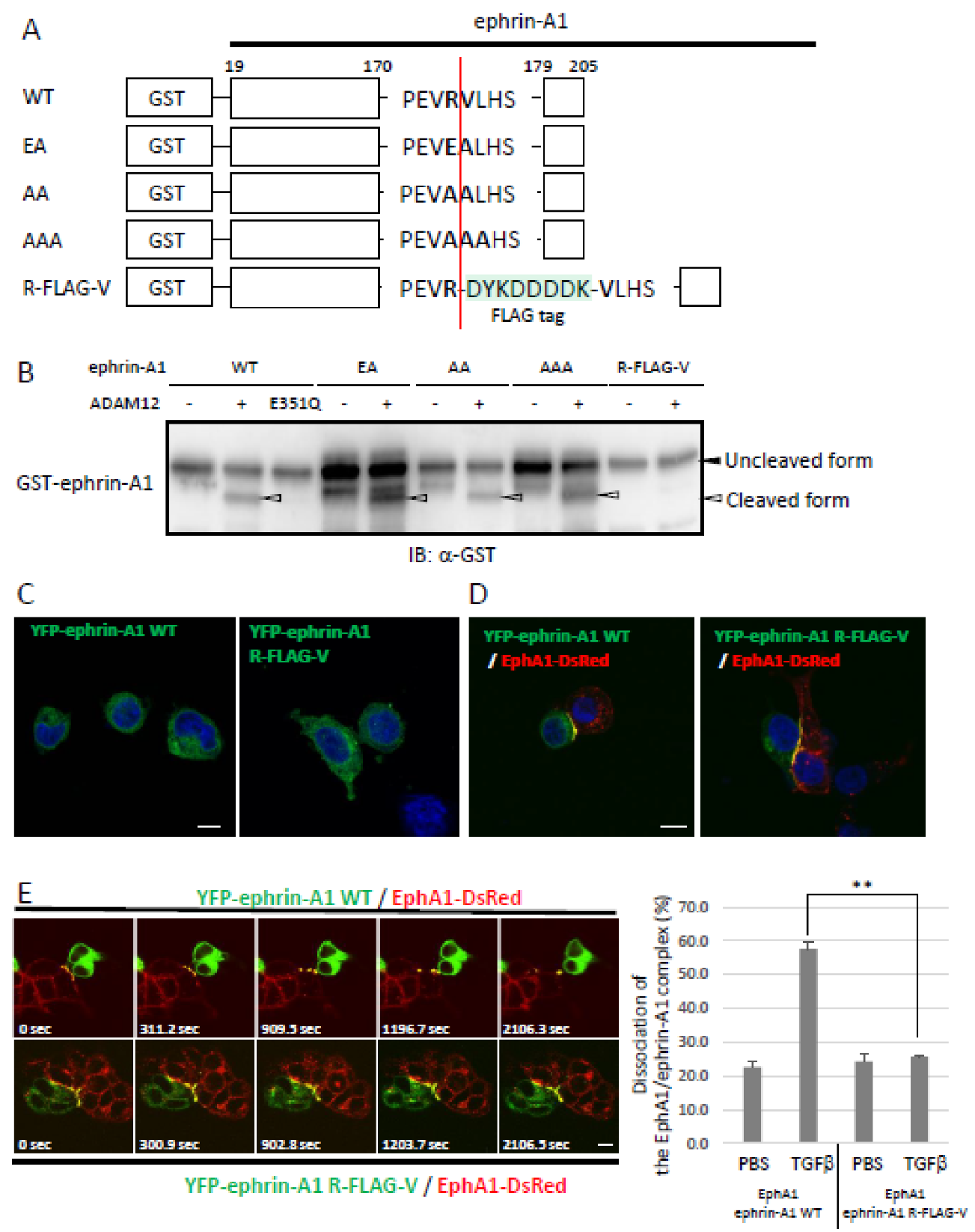
2.1. Identification of an ADAM12-Mediated Cleavage Site in Ephrin-A1 by Mass Spectrometry
2.2. Bioactivity of ADAM12-Cleaved Ephrin-A1 on the Eph/Ephrin Signal
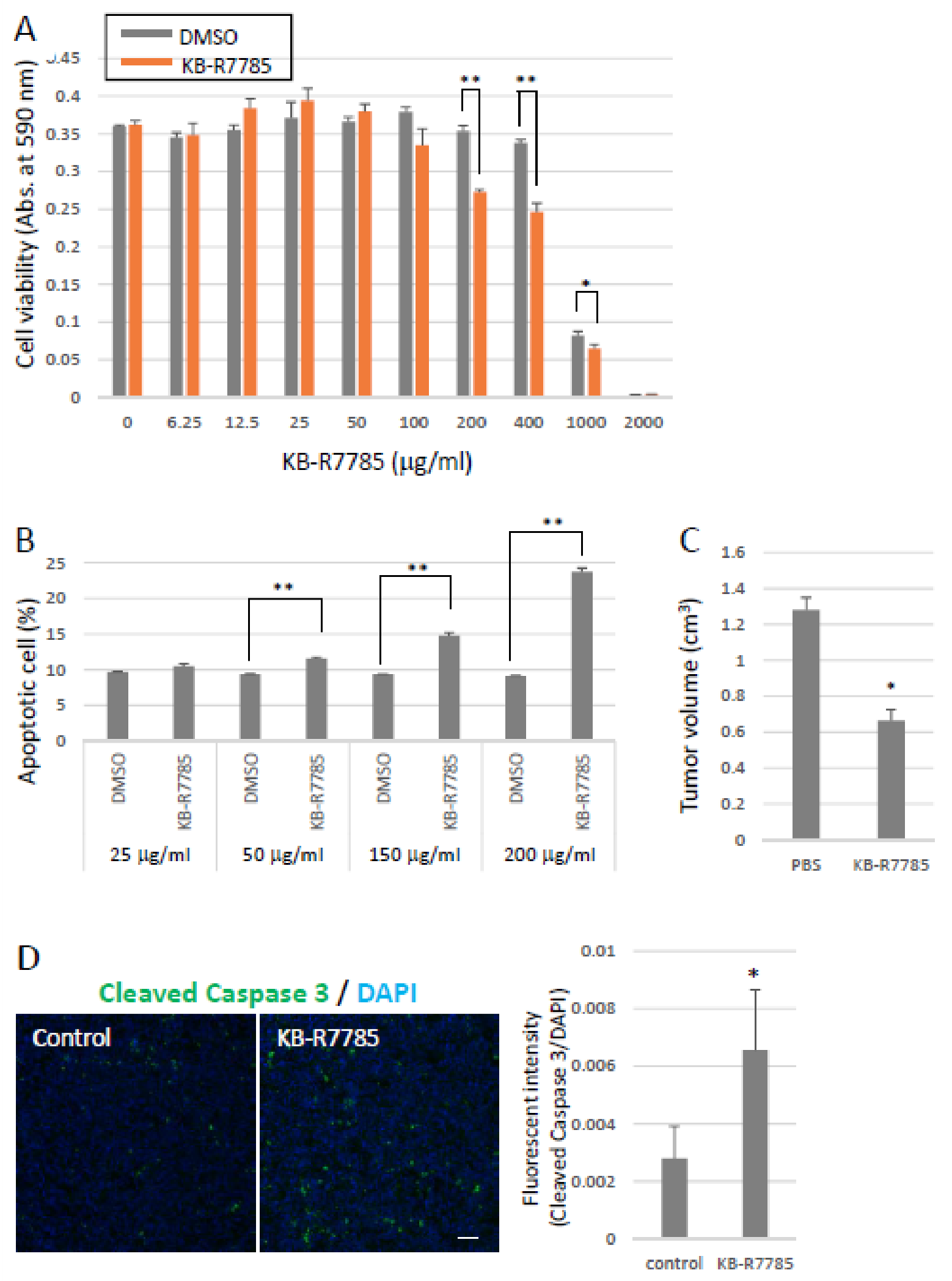
2.3. Effects of Ephrin-A1 Cleavage on Tumor Growth and Metastasis
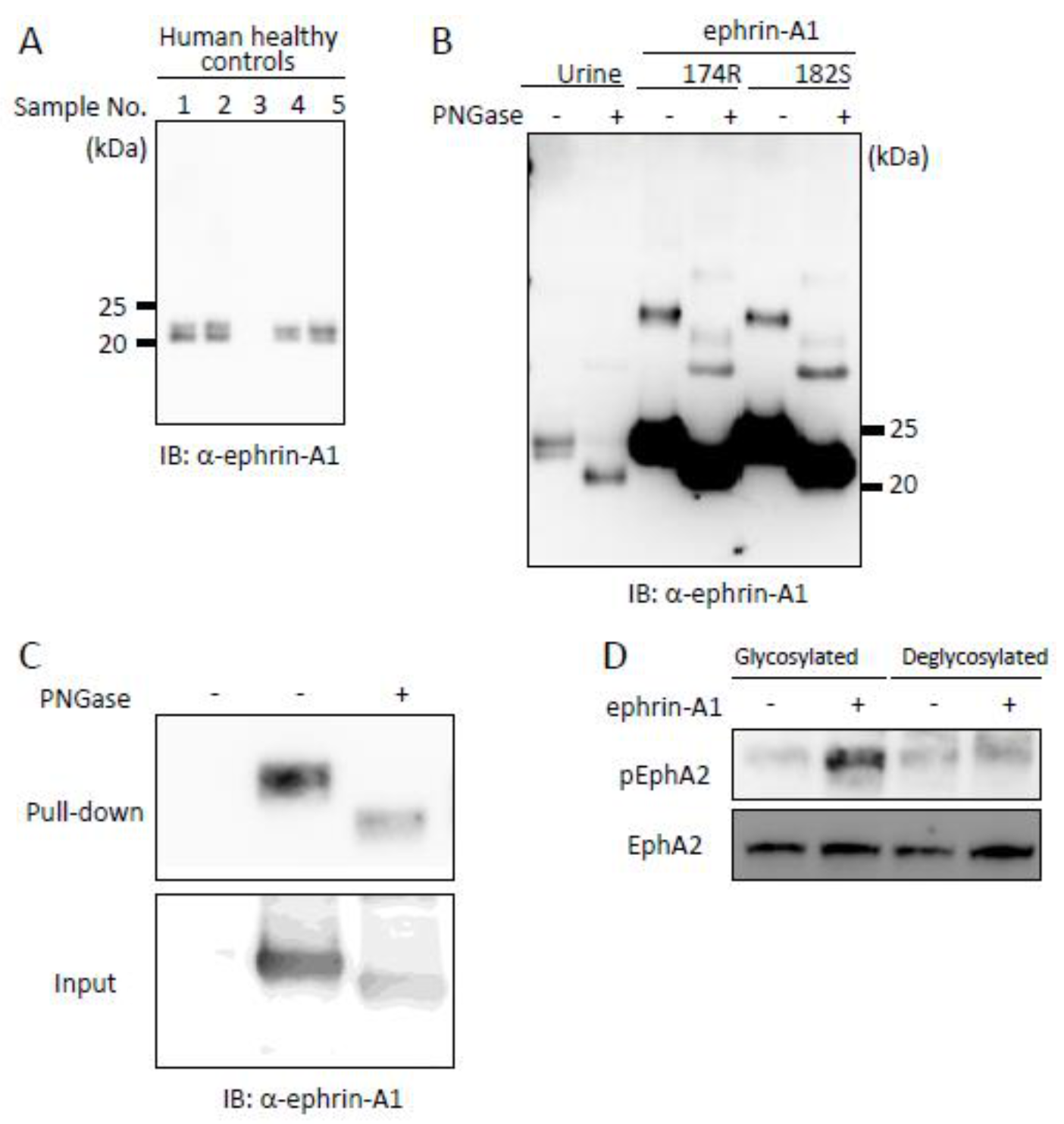
2.4. Tumor-Derived Ephrin-A1 as a Putative Biomarker for Metastasis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Plasmids
4.3. Preparation of Recombinant Protein
4.4. Cell Culture
4.5. Electrophoresis and in-Gel Digestion
4.6. Μatrix-Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
4.7. Synthesis of KB-R7785 and UniPR1331
4.8. Hibit Luminescence Assay
4.9. Cell-Spreading Assay
4.10. Dissociation Assay of the Epha1/Ephrin-A1 Complex
4.11. Animal Study
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Mullooly, M.; McGowan, P.M.; Crown, J.; Duffy, M.J. The ADAMs family of proteases as targets for the treatment of cancer. Cancer Biol. Ther. 2016, 17, 870–880. [Google Scholar] [CrossRef]
- Mochizuki, S.; Okada, Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007, 98, 621–628. [Google Scholar] [CrossRef]
- Singh, D.; Srivastava, S.K.; Chaudhuri, T.K.; Upadhyay, G. Multifaceted role of matrix metalloproteinases (MMPs). Front. Mol. Biosci. 2015, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Diaz, B.; Yuen, A.; Iizuka, S.; Higashiyama, S.; Courtneidge, S.A. Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J. Cell Biol. 2013, 201, 279–292. [Google Scholar] [CrossRef]
- Tien, W.S.; Chen, J.H.; Wu, K.P. SheddomeDB: The ectodomain shedding database for membrane-bound shed markers. BMC Bioinform. 2017, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Mullooly, M.; McGowan, P.M.; Kennedy, S.A.; Madden, S.F.; Crown, J.; O’Donovan, N.; Duffy, M.J. ADAM10: A new player in breast cancer progression? Br. J. Cancer 2015, 113, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Kodama, T.; Ikeda, E.; Okada, A.; Ohtsuka, T.; Shimoda, M.; Shiomi, T.; Yoshida, K.; Nakada, M.; Ohuchi, E.; Okada, Y. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am. J. Pathol. 2004, 165, 1743–1753. [Google Scholar] [CrossRef]
- Kuefer, R.; Day, K.C.; Kleer, C.G.; Sabel, M.S.; Hofer, M.D.; Varambally, S.; Zorn, C.S.; Chinnaiyan, A.M.; Rubin, M.A.; Day, M.L. ADAM15 disintegrin is associated with aggressive prostate and breast cancer disease. Neoplasia 2006, 8, 319–329. [Google Scholar] [CrossRef]
- Ieguchi, K.; Maru, Y. Savior or not: ADAM17 inhibitors overcome radiotherapy-resistance in non-small cell lung cancer. J. Thorac. Dis. 2016, 8, E813–E815. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ieguchi, K. Eph as a target in inflammation. Endocr. Metab. Immune Disord. Drug Targets 2015, 15, 119–128. [Google Scholar] [CrossRef]
- Ieguchi, K.; Maru, Y. Roles of EphA1/A2 and ephrin-A1 in cancer. Cancer Sci. 2019, 110, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Holzman, L.B.; Marks, R.M.; Dixit, V.M. A novel immediate-early response gene of endothelium is induced by cytokines and encodes a secreted protein. Mol. Cell Biol. 1990, 10, 5830–5838. [Google Scholar] [CrossRef]
- Foo, S.S.; Turner, C.J.; Adams, S.; Compagni, A.; Aubyn, D.; Kogata, N.; Lindblom, P.; Shani, M.; Zicha, D.; Adams, R.H. Ephrin-B2 controls cell motility and adhesion during blood-vessel-wall assembly. Cell 2006, 124, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nakayama, M.; Pitulescu, M.E.; Schmidt, T.S.; Bochenek, M.L.; Sakakibara, A.; Adams, S.; Davy, A.; Deutsch, U.; Luthi, U.; et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 2010, 465, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Nettiksimmons, J.; Tranah, G.; Evans, D.S.; Yokoyama, J.S.; Yaffe, K. Gene-based aggregate SNP associations between candidate AD genes and cognitive decline. Age 2016, 38, 41. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Yamamoto, H.; Kim, C.; Uemura, M.; Akita, H.; Tomimaru, Y.; Hama, N.; Kawamoto, K.; Kobayashi, S.; Eguchi, H.; et al. Association between ephrin-A1 mRNA expression and poor prognosis after hepatectomy to treat hepatocellular carcinoma. Int. J. Oncol. 2014, 45, 1051–1058. [Google Scholar] [CrossRef]
- Yamamoto, H.; Tei, M.; Uemura, M.; Takemasa, I.; Uemura, Y.; Murata, K.; Fukunaga, M.; Ohue, M.; Ohnishi, T.; Ikeda, K.; et al. Ephrin-A1 mRNA is associated with poor prognosis of colorectal cancer. Int. J. Oncol. 2013, 42, 549–555. [Google Scholar] [CrossRef]
- Ieguchi, K.; Omori, T.; Komatsu, A.; Tomita, T.; Deguchi, A.; Maru, Y. Ephrin-A1 expression induced by S100A8 is mediated by the toll-like receptor 4. Biochem. Biophys. Res. Commun. 2013, 440, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Ieguchi, K.; Tomita, T.; Omori, T.; Komatsu, A.; Deguchi, A.; Masuda, J.; Duffy, S.L.; Coulthard, M.G.; Boyd, A.; Maru, Y. ADAM12-cleaved ephrin-A1 contributes to lung metastasis. Oncogene 2014, 33, 2179–2190. [Google Scholar] [CrossRef]
- Wykosky, J.; Palma, E.; Gibo, D.M.; Ringler, S.; Turner, C.P.; Debinski, W. Soluble monomeric EphrinA1 is released from tumor cells and is a functional ligand for the EphA2 receptor. Oncogene 2008, 27, 7260–7273. [Google Scholar] [CrossRef] [PubMed]
- Wewer, U.M.; Morgelin, M.; Holck, P.; Jacobsen, J.; Lydolph, M.C.; Johnsen, A.H.; Kveiborg, M.; Albrechtsen, R. ADAM12 is a four-leafed clover: The excised prodomain remains bound to the mature enzyme. J. Biol. Chem. 2006, 281, 9418–9422. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, A.; Lively, M.O.; Mintz, A.; Gibo, D.; Wykosky, J.; Debinski, W. EphrinA1 is released in three forms from cancer cells by matrix metalloproteases. Mol. Cell Biol. 2012, 32, 3253–3264. [Google Scholar] [CrossRef]
- Hirayama, R.; Yamamoto, M.; Tsukida, T.; Matsuo, K.; Obata, Y.; Sakamoto, F.; Ikeda, S. Synthesis and biological evaluation of orally active matrix metalloproteinase inhibitors. Bioorg. Med. Chem. 1997, 5, 765–778. [Google Scholar] [CrossRef]
- Hattori, M.; Osterfield, M.; Flanagan, J.G. Regulated cleavage of a contact-mediated axon repellent. Science 2000, 289, 1360–1365. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-de-Cossio, J.; Gonzalez, J.; Betancourt, L.; Besada, V.; Padron, G.; Shimonishi, Y.; Takao, T. Automated interpretation of high-energy collision-induced dissociation spectra of singly protonated peptides by ‘SeqMS’, a software aid for de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 1998, 12, 1867–1878. [Google Scholar] [CrossRef]
- Barquilla, A.; Lamberto, I.; Noberini, R.; Heynen-Genel, S.; Brill, L.M.; Pasquale, E.B. Protein kinase A can block EphA2 receptor-mediated cell repulsion by increasing EphA2 S897 phosphorylation. Mol. Biol. Cell 2016, 27, 2757–2770. [Google Scholar] [CrossRef]
- Yamazaki, T.; Masuda, J.; Omori, T.; Usui, R.; Akiyama, H.; Maru, Y. EphA1 interacts with integrin-linked kinase and regulates cell morphology and motility. J. Cell Sci. 2009, 122, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Castelli, R.; Tognolini, M.; Vacondio, F.; Incerti, M.; Pala, D.; Callegari, D.; Bertoni, S.; Giorgio, C.; Hassan-Mohamed, I.; Zanotti, I.; et al. Delta(5)-Cholenoyl-amino acids as selective and orally available antagonists of the Eph-ephrin system. Eur. J. Med. Chem. 2015, 103, 312–324. [Google Scholar] [CrossRef]
- Oh-Hashi, K.; Furuta, E.; Fujimura, K.; Hirata, Y. Application of a novel HiBiT peptide tag for monitoring ATF4 protein expression in Neuro2a cells. Biochem. Biophys. Rep. 2017, 12, 40–45. [Google Scholar] [CrossRef]
- Lozonschi, L.; Sunamura, M.; Kobari, M.; Egawa, S.; Ding, L.; Matsuno, S. Controlling tumor angiogenesis and metastasis of C26 murine colon adenocarcinoma by a new matrix metalloproteinase inhibitor, KB-R7785, in two tumor models. Cancer Res. 1999, 59, 1252–1258. [Google Scholar]
- Alford, S.; Watson-Hurthig, A.; Scott, N.; Carette, A.; Lorimer, H.; Bazowski, J.; Howard, P.L. Soluble ephrin a1 is necessary for the growth of HeLa and SK-BR3 cells. Cancer Cell Int. 2010, 10, 41. [Google Scholar] [CrossRef]
- Ieguchi, K.; Fujita, M.; Ma, Z.; Davari, P.; Taniguchi, Y.; Sekiguchi, K.; Wang, B.; Takada, Y.K.; Takada, Y. Direct binding of the EGF-like domain of neuregulin-1 to integrins ({alpha}v{beta}3 and {alpha}6{beta}4) is involved in neuregulin-1/ErbB signaling. J. Biol. Chem. 2010, 285, 31388–31398. [Google Scholar] [CrossRef] [PubMed]
- Brizzi, M.F.; Tarone, G.; Defilippi, P. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr. Opin. Cell Biol. 2012, 24, 645–651. [Google Scholar] [CrossRef]
- Cui, X.D.; Lee, M.J.; Yu, G.R.; Kim, I.H.; Yu, H.C.; Song, E.Y.; Kim, D.G. EFNA1 ligand and its receptor EphA2: Potential biomarkers for hepatocellular carcinoma. Int. J. Cancer 2010, 126, 940–949. [Google Scholar] [CrossRef]
- Tuccillo, F.M.; de Laurentiis, A.; Palmieri, C.; Fiume, G.; Bonelli, P.; Borrelli, A.; Tassone, P.; Scala, I.; Buonaguro, F.M.; Quinto, I.; et al. Aberrant glycosylation as biomarker for cancer: Focus on CD43. Biomed. Res. Int. 2014, 2014, 742831. [Google Scholar] [CrossRef] [PubMed]
- Keeley, T.S.; Yang, S.; Lau, E. The Diverse Contributions of Fucose Linkages in Cancer. Cancers 2019, 11, 1241. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ieguchi, K.; Tomita, T.; Takao, T.; Omori, T.; Mishima, T.; Shimizu, I.; Tognolini, M.; Lodola, A.; Tsunoda, T.; Kobayashi, S.; et al. Analysis of ADAM12-Mediated Ephrin-A1 Cleavage and Its Biological Functions. Int. J. Mol. Sci. 2021, 22, 2480. https://doi.org/10.3390/ijms22052480
Ieguchi K, Tomita T, Takao T, Omori T, Mishima T, Shimizu I, Tognolini M, Lodola A, Tsunoda T, Kobayashi S, et al. Analysis of ADAM12-Mediated Ephrin-A1 Cleavage and Its Biological Functions. International Journal of Molecular Sciences. 2021; 22(5):2480. https://doi.org/10.3390/ijms22052480
Chicago/Turabian StyleIeguchi, Katsuaki, Takeshi Tomita, Toshifumi Takao, Tsutomu Omori, Taishi Mishima, Isao Shimizu, Massimiliano Tognolini, Alessio Lodola, Takuya Tsunoda, Shinichi Kobayashi, and et al. 2021. "Analysis of ADAM12-Mediated Ephrin-A1 Cleavage and Its Biological Functions" International Journal of Molecular Sciences 22, no. 5: 2480. https://doi.org/10.3390/ijms22052480
APA StyleIeguchi, K., Tomita, T., Takao, T., Omori, T., Mishima, T., Shimizu, I., Tognolini, M., Lodola, A., Tsunoda, T., Kobayashi, S., Wada, S., & Maru, Y. (2021). Analysis of ADAM12-Mediated Ephrin-A1 Cleavage and Its Biological Functions. International Journal of Molecular Sciences, 22(5), 2480. https://doi.org/10.3390/ijms22052480






