Differential Morpho-Physiological and Transcriptomic Responses to Heat Stress in Two Blueberry Species
Abstract
1. Introduction
2. Results and Discussion
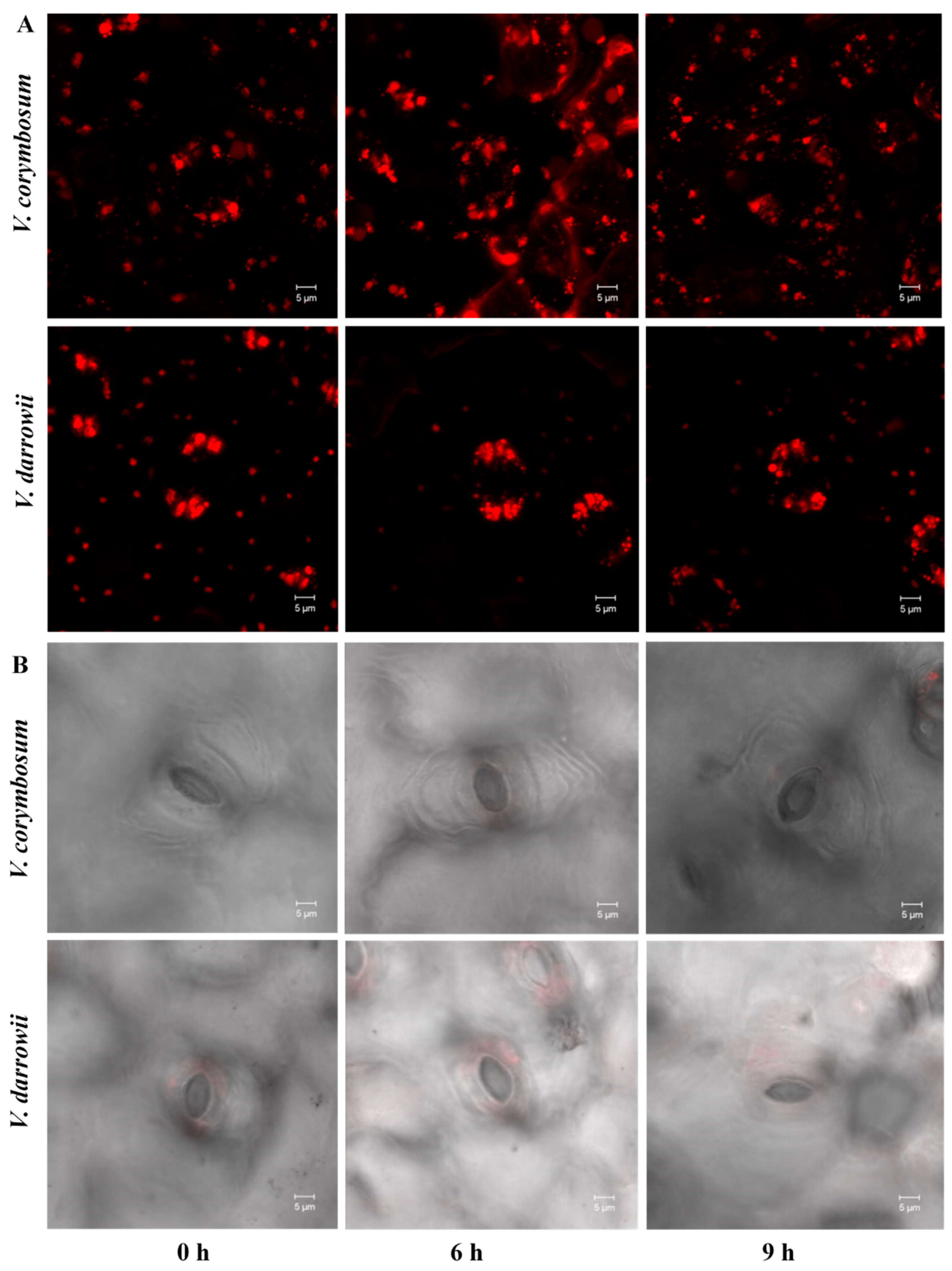
2.1. Morpho-Physiological Response to Heat Stress
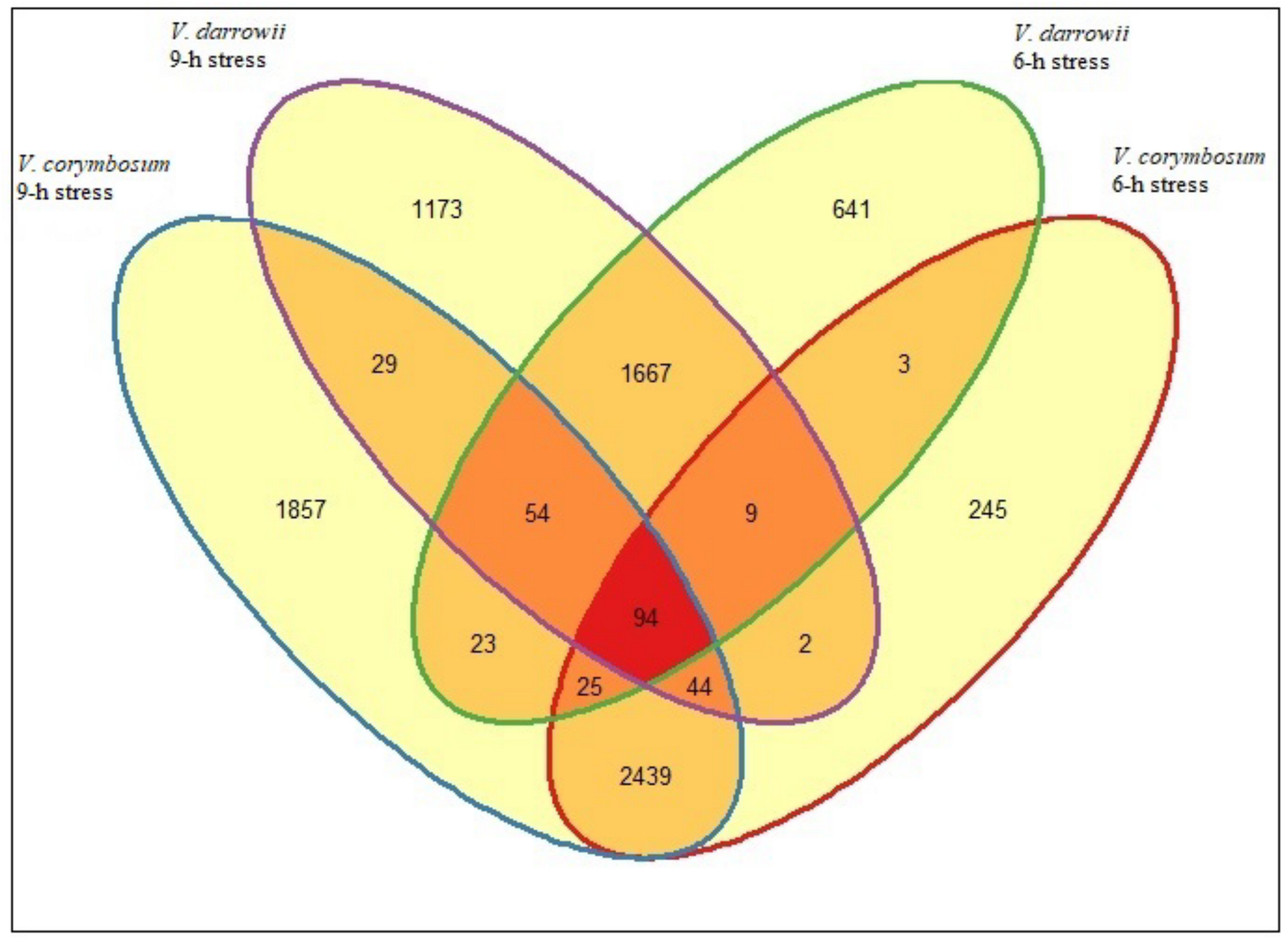
2.2. Differences in Gene Expression between V. corymbosum and V. darrowii at 6 and 9-h of Heat Stress
2.3. DEGs Commonly Enriched Pathways in Blueberry Species
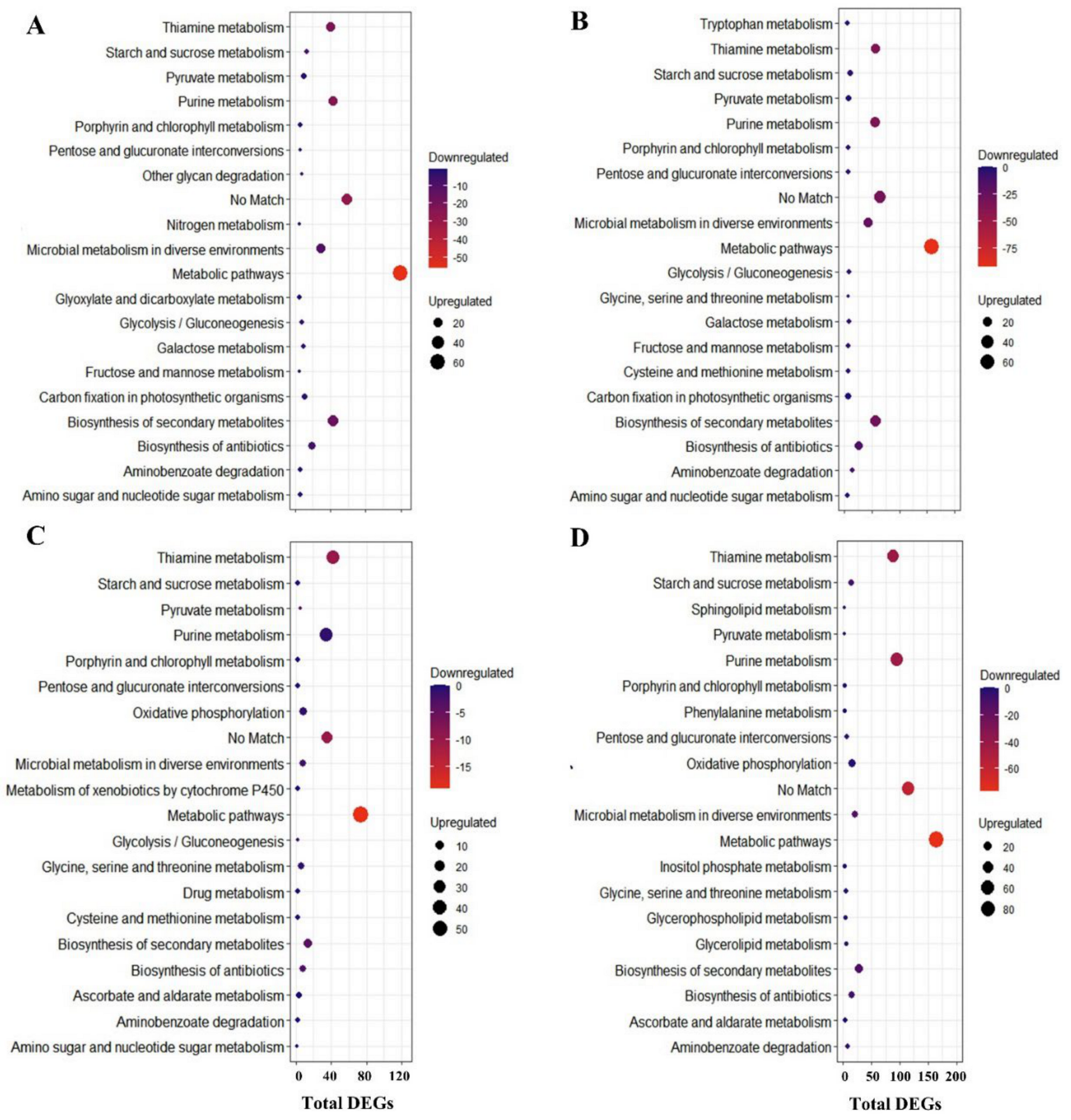
2.4. Unique DEG-Enriched Pathways in V. corymbosum
2.5. DEG Enriched Pathways in V. darrowii
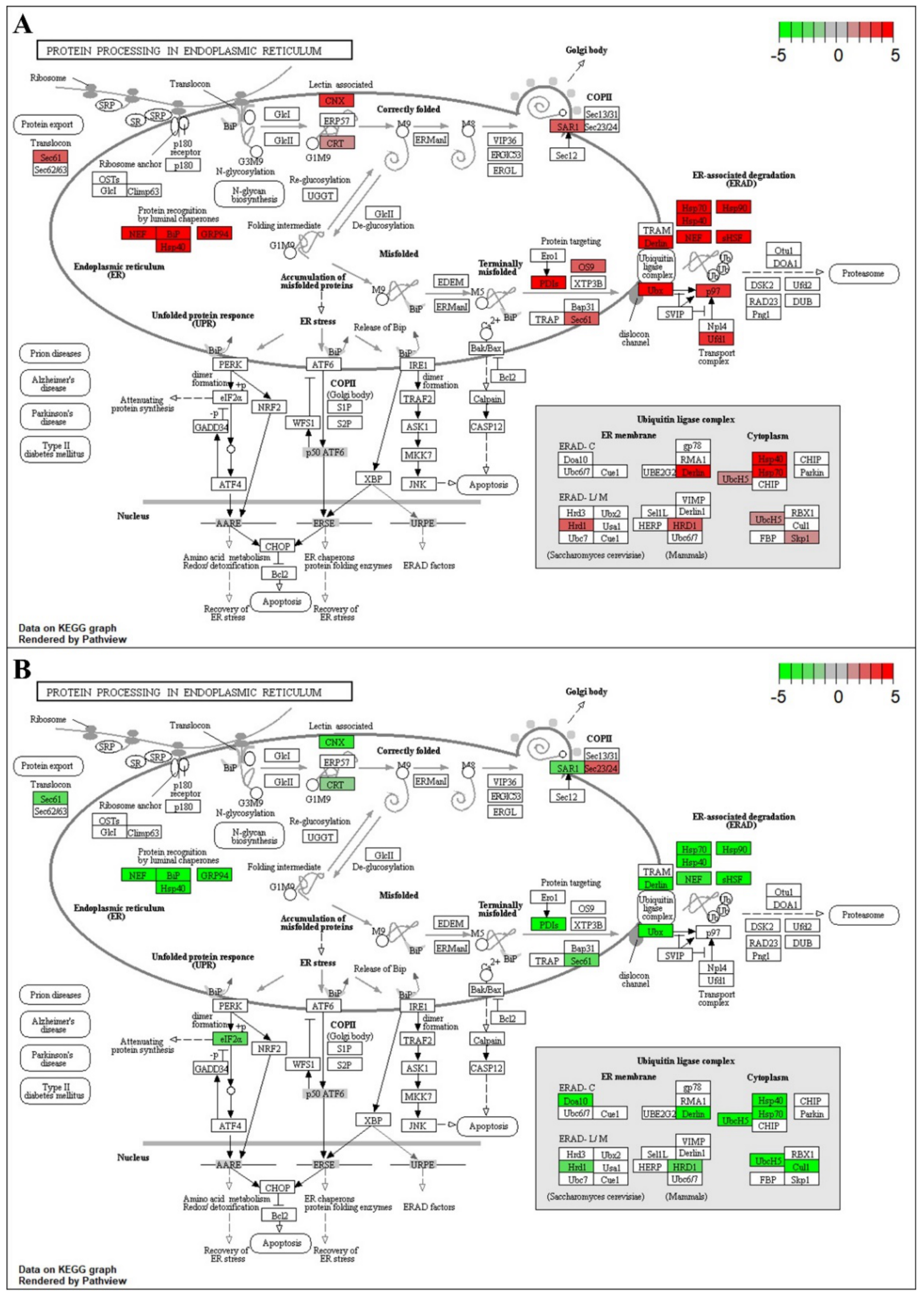
2.6. Dynamics of Key Pathways Involved in Response to Heat Stress
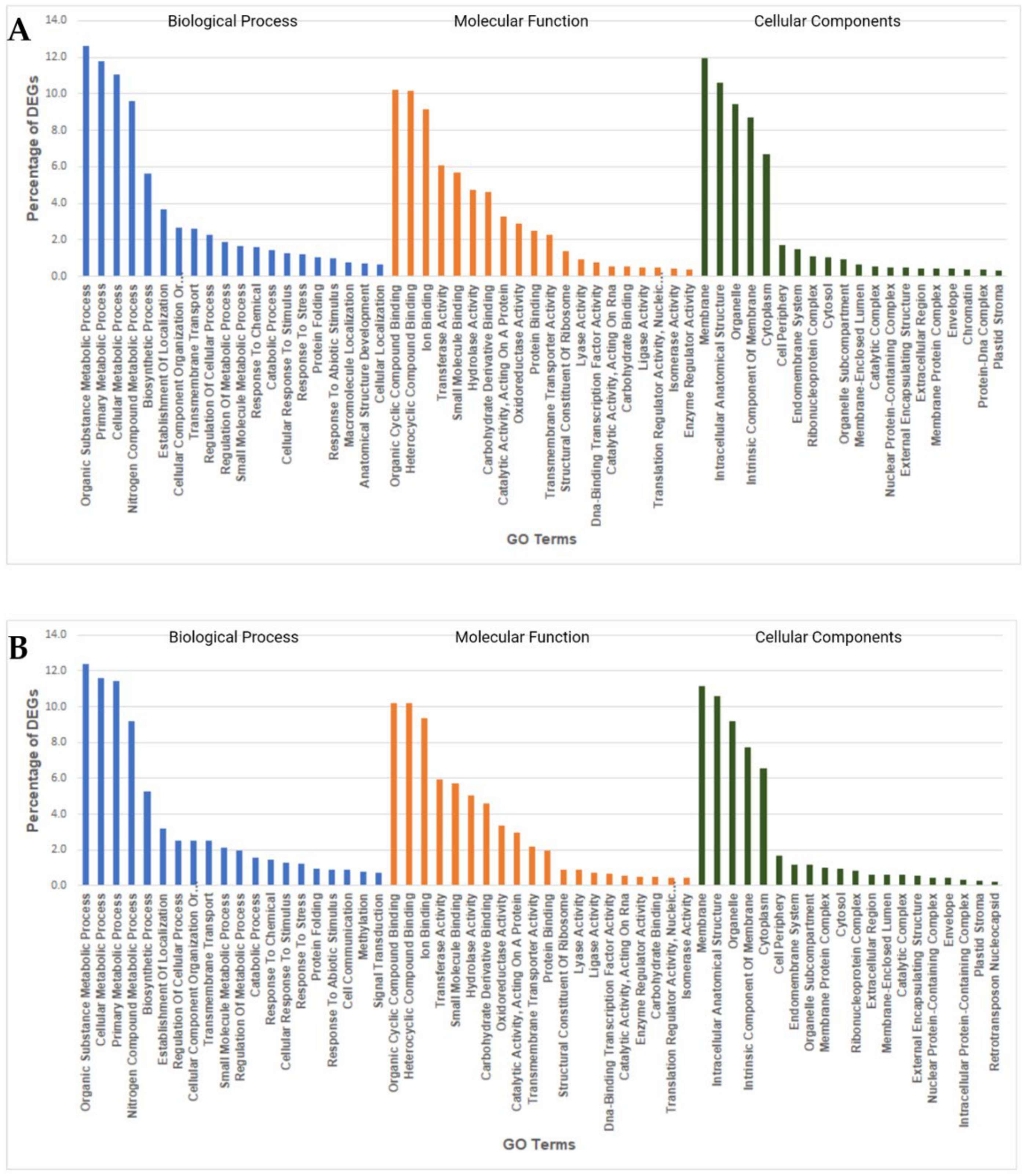
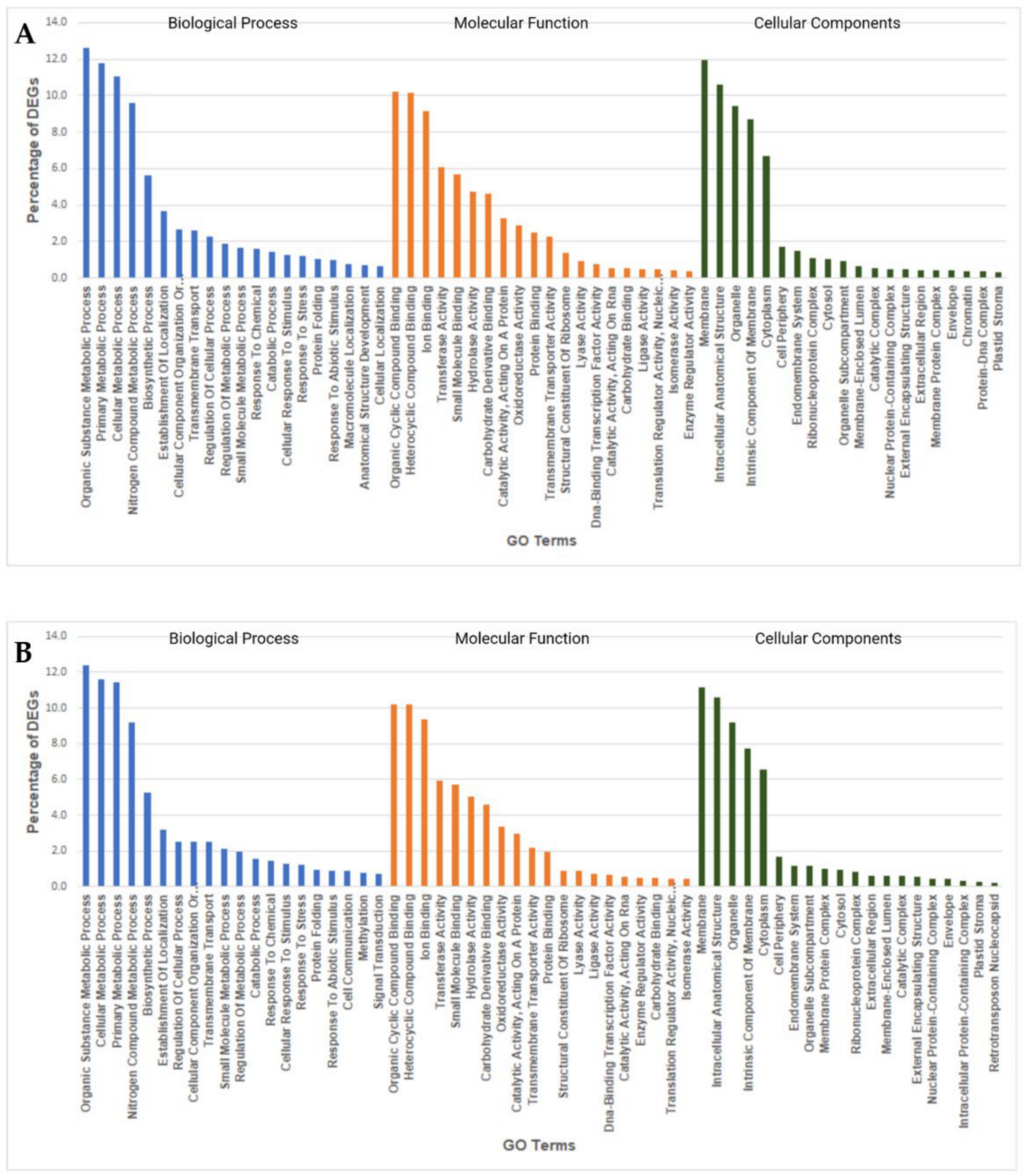
2.7. GO Enrichment Analysis of Heat Responsive DEGs
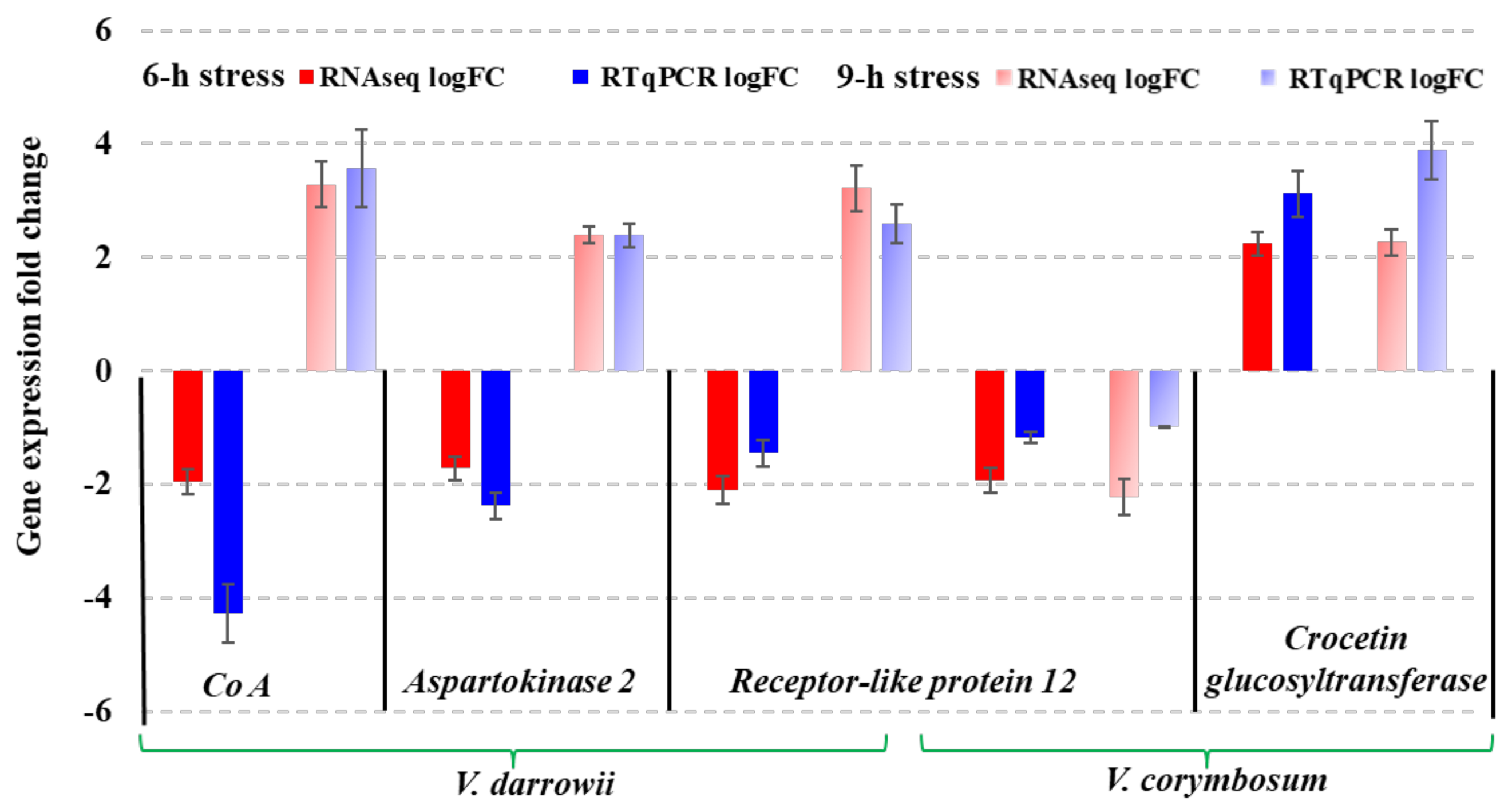
2.8. Quantitative Real-Time PCR (RT-qPCR) Validation of DEGs from RNAseq
3. Materials and Methods
3.1. Plant Materials and Phenotypic Characterization
3.2. Heat Stress Induction
3.3. Stomata and Internal Organelle Image Assay
3.4. RNA Isolation, Library Preparation, and Illumina Sequencing
3.5. De novo Assembly and Clustering
3.6. Pathway Analysis
3.7. Real-Time Quantitative PCR (RT-qPCR)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Retamales, J.B.; Hancock, J.F. Blueberries; Cabi: Cambridge, MA, USA, 2018; Volume 27. [Google Scholar]
- Pedersen, C.B.; Kyle, J.; Jenkinson, A.; Gardner, P.; McPhail, D.; Duthie, G. Effects of blueberry and cranberry juice consumption on the plasma antioxidant capacity of healthy female volunteers. Eur. J. Clin. Nutr. 2000, 54, 405–408. [Google Scholar] [CrossRef]
- Agricultural Marketing Resource Center. Blueberries. Available online: https://www.agmrc.org/commodities-products/fruits/blueberries (accessed on 5 March 2020).
- Shi, M.; Loftus, H.; McAinch, A.J.; Su, X.Q. Blueberry as a source of bioactive compounds for the treatment of obesity, type 2 diabetes and chronic inflammation. J. Funct. Foods 2017, 30, 16–29. [Google Scholar] [CrossRef]
- Davies, K.; Espley, R. Opportunities and challenges for metabolic engineering of secondary metabolite pathways for improved human health characters in fruit and vegetable crops. N. Z. J. Crop Hortic. Sci. 2013, 41, 154–177. [Google Scholar] [CrossRef]
- Norberto, S.; Silva, S.; Meireles, M.; Faria, A.; Pintado, M.; Calhau, C. Blueberry anthocyanins in health promotion: A metabolic overview. J. Funct. Foods 2013, 5, 1518–1528. [Google Scholar] [CrossRef]
- USDA ERS. Data by Commodity—Imports and Exports. 2018. Available online: https://data.ers.usda.gov/reports.aspx?ID=17851 (accessed on 4 April 2019).
- Park, K.; Cook, R. Blueberries: Is supply developing more rapidly than demand. Smart Mark 2018. Available online: https://dyson.cornell.edu/wp-content/uploads/sites/5/2019/02/smart-marketing-2018-04.pdf (accessed on 5 August 2018).
- Hortidaily News. World’s top ten blueberry producing countries. Horti. Daily 2016. Available online: https://www.hortidaily.com/article/6026165/world-s-top-ten-blueberry-producing-countries/ (accessed on 25 May 2018).
- Mainland, C.M.M.; Frederick, V. Coville and the history of north american highbush blueberry culture. Int. J. Fruit Sci. 2012, 12, 4–13. [Google Scholar] [CrossRef]
- Rowland, L.J.; Ogden, E.L.; Bassil, N.; Buck, E.J.; McCallum, S.; Graham, J.; Brown, A.; Wiedow, C.; Campbell, A.M.; Haynes, K.G. Construction of a genetic linkage map of an interspecific diploid blueberry population and identification of QTL for chilling requirement and cold hardiness. Mol. Breeding 2014, 34, 2033–2048. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Nunez, G.H.; Olmstead, J.W.; Rinehart, T.A.; Staton, M. Regulation of gene expression in roots of the pH-sensitive Vaccinium corymbosum and the pH-tolerant Vaccinium arboreum in response to near neutral pH stress using RNA-Seq. BMC Genom. 2017, 18, 580. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, R.; Sun, Y.; Xu, M.; Zhang, H.; Huang, L.; Zhu, Y.; Wang, H.; Li, G.; Liu, L. The optimal temperature for the growth of blueberry (Vaccinium corymbosum L.). Pak. J. Bot. 2017, 49, 965–979. [Google Scholar]
- Li, L.; Zhang, H.; Liu, Z.; Cui, X.; Zhang, T.; Li, Y.; Zhang, L. Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 2016, 16, 223. [Google Scholar] [CrossRef] [PubMed]
- Walworth, A.E.; Chai, B.; Song, G.Q. Transcript profile of flowering regulatory genes in VcFT-overexpressing blueberry plants. PLoS ONE 2016, 11, e0156993. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Estrada, A.D.; Blakley, I.; Reid, R.; Patel, K.; Meyer, M.D.; Andersen, S.U.; Brown, A.F.; Lila, M.A.; Loraine, A.E. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. GigaScience 2015, 4, s13742-015-0046-9. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Ogden, E.L.; Die, J.V.; Ehlenfeldt, M.K.; Polashock, J.J.; Darwish, O.; Alkharouf, N.; Rowland, L.J. Transcriptome analysis identifies genes related to the waxy coating on blueberry fruit in two northern-adapted rabbiteye breeding populations. BMC Plant Biol. 2019, 19, 460. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, Y.; Li, B.; Tan, H.; Li, D.; Li, L.; Liu, X.; Han, J.; Meng, X. Comparative transcriptome analysis of genes involved in anthocyanin synthesis in blueberry. Plant Physiol. Biochem. 2018, 127, 561–572. [Google Scholar] [CrossRef]
- Die, J.V.; Rowland, L.J. Elucidating cold acclimation pathway in blueberry by transcriptome profiling. Environ. Exp. Bot 2014, 106, 87–98. [Google Scholar] [CrossRef]
- Kulkarni, K.P.; Vorsa, N.; Natarajan, P.; Elavarthi, S.; Iorizzo, M.; Reddy, U.K.; Melmaiee, K. Admixture analysis using genotyping-by-sequencing reveals genetic relatedness and parental lineage distribution in highbush blueberry genotypes and cross derivatives. Int. J. Mol. Sci. 2021, 22, 163. [Google Scholar]
- Chen, W.; Cen, W.; Chen, L.; Di, L.; Li, Y.; Guo, W. Differential sensitivity of four highbush blueberry (Vaccinium corymbosum L.) cultivars to heat stress. Pak. J. Bot. 2012, 44, 853–860. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- James, S.A.; Bell, D.T. Leaf orientation, light interception and stomatal conductance of Eucalyptus globulus ssp. globulus leaves. Tree Physiol. 2000, 20, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Kloet, S.V. The taxonomy of Vaccinium § Cyanococcus: A summation. Can. J. Bot. 1983, 61, 256–266. [Google Scholar] [CrossRef]
- Rodríguez, V.M.; Soengas, P.; Alonso-Villaverde, V.; Sotelo, T.; Cartea, M.E.; Velasco, P. Effect of temperature stress on the early vegetative development of Brassica oleracea L. BMC Plant Biol. 2015, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Meckel, T.; Gall, L.; Semrau, S.; Homann, U.; Thiel, G. Guard cells elongate: Relationship of volume and surface area during stomatal movement. Biophys. J. 2007, 92, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, I.A.; Shalby, N.; Bai, C.; Qin, M.; Agami, R.A.; Jie, K.; Wang, B.; Zhou, G. Stomatal and photosynthetic traits are associated with investigating sodium chloride tolerance of Brassica napus L. cultivars. Plants 2020, 9, 62. [Google Scholar] [CrossRef]
- Schulze, E.D.; Lange, O.; Kappen, L.; Buschbom, U.; Evenari, M. Stomatal responses to changes in temperature at increasing water stress. Planta 1973, 110, 29–42. [Google Scholar] [CrossRef]
- Liu, J.; Sun, X.; Xu, F.; Zhang, Y.; Zhang, Q.; Miao, R.; Zhang, J.; Liang, J.; Xu, W. Suppression of OsMDHAR4 enhances heat tolerance by mediating H 2 O 2-induced stomatal closure in rice plants. Rice 2018, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Wang, J.F.; Cramer, G.; Dai, Z.W.; Duan, W.; Xu, H.G.; Wu, B.H.; Fan, P.G.; Wang, L.J.; Li, S.H. Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol. 2012, 12, 174. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von KoskullDöring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Liu, X.; Du, F.; Li, N.; Chang, Y.; Yao, D. Gene expression profile in the long-living lotus: Insights into the heat stress response mechanism. PLoS ONE 2016, 11, e0152540. [Google Scholar] [CrossRef]
- Ding, H.; Mo, S.; Qian, Y.; Yuan, G.; Wu, X.; Ge, C. Integrated proteome and transcriptome analyses revealed key factors involved in tomato (Solanum lycopersicum) under high temperature stress. Food Energy Secur. 2020, 9, e239. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Mesihovic, A.; Hu, Y.; Schleiff, E. Unfolded protein response in pollen development and heat stress tolerance. Plant Reprod. 2016, 29, 81–91. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef]
- Jin, J.; Yang, L.; Fan, D.; Liu, X.; Hao, Q. Comparative transcriptome analysis uncovers different heat stress responses in heat-resistant and heat-sensitive jujube cultivars. PLoS ONE 2020, 15, e0235763. [Google Scholar] [CrossRef]
- Wang, W.; Lin, Y.; Teng, F.; Ji, D.; Xu, Y.; Chen, C.; Xie, C. Comparative transcriptome analysis between heat-tolerant and sensitive Pyropia haitanensis strains in response to high temperature stress. Algal Res. 2018, 29, 104–112. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. Arabidopsis 3-ketoacyl-CoA thiolase-2 (KAT2), an enzyme of fatty acid β-oxidation, is involved in ABA signal transduction. Plant Cell Physiol. 2011, 52, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Aprile, A.; Havlickova, L.; Panna, R.; Marè, C.; Borrelli, G.M.; Marone, D.; Perrotta, C.; Rampino, P.; De Bellis, L.; Curn, V. Different stress responsive strategies to drought and heat in two durum wheat cultivars with contrasting water use efficiency. BMC Genom. 2013, 14, 1–18. [Google Scholar] [CrossRef]
- Vidya, S.; Kumar, H.V.; Bhatt, R.; Laxman, R.; Ravishankar, K. Transcriptional profiling and genes involved in acquired thermotolerance in banana: A non-model crop. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Zhu, Y.C.; Sun, D.X.; Yun, D.; An, G.I.; Li, W.H.; Si, W.J.; Liu, J.P.; Sun, X.W. Comparative transcriptome analysis of the effect of different heat shock periods on the unfertilized ovule in watermelon (Citrullus lanatus). J. Integr. Agric. 2020, 19, 528–540. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, B. Transcriptomic analysis reveals unique molecular factors for lipid hydrolysis, secondary cell-walls and oxidative protection associated with thermotolerance in perennial grass. BMC Genom. 2018, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Bheemanahalli, R.; Impa, S.M.; Krassovskaya, I.; Vennapusa, A.R.; Gill, K.S.; Obata, T.; Jagadish, S.V.K. Enhanced N-metabolites, ABA and IAA-conjugate in anthers instigate heat sensitivity in spring wheat. Physiol. Plant. 2020, 169, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yuan, B.; Xu, Y.; Huang, B. Differential responses of amino acids and soluble proteins to heat stress associated with genetic variations in heat tolerance for hard fescue. J. Am. Soc. Hortic. 2018, 143, 45–55. [Google Scholar] [CrossRef]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid—A potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Zhao, J.; Missihoun, T.D.; Bartels, D. The role of arabidopsis aldehyde dehydrogenase genes in response to high temperature and stress combinations. J. Exp. Bot. 2017, 68, 4295–4308. [Google Scholar] [CrossRef]
- Su, T.; Li, W.; Wang, P.; Ma, C. Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Front. Plant Sci. 2019, 10, 705. [Google Scholar] [CrossRef]
- Prasad, P.; Sreedhar, R. Identification and functional characterization of buglossoides arvensis microsomal fatty acid desaturation pathway genes involved in polyunsaturated fatty acid synthesis in seeds. J. Biotechnol. 2020, 308, 130–140. [Google Scholar]
- Shi, J.; Yan, B.; Lou, X.; Ma, H.; Ruan, S. Comparative transcriptome analysis reveals the transcriptional alterations in heat-resistant and heat-sensitive sweet maize (Zea mays L.) varieties under heat stress. BMC Plant Biol. 2017, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Yangueez, E.; Castro-Sanz, A.B.; Fernandez-Bautista, N.; Oliveros, J.C.; Castellano, M.M. Analysis of genome-wide changes in the translatome of arabidopsis seedlings subjected to heat stress. PLoS ONE 2013, 8, e71425. [Google Scholar] [CrossRef] [PubMed]
- Beine-Golovchuk, O.; Firmino, A.A.P.; Dąbrowska, A.; Schmidt, S.; Erban, A.; Walther, D.; Zuther, E.; Hincha, D.K.; Kopka, J. Plant temperature acclimation and growth rely on cytosolic ribosome biogenesis factor homologs. Plant Physiol. 2018, 176, 2251–2276. [Google Scholar] [CrossRef] [PubMed]
- Lukoszek, R.; Feist, P.; Ignatova, Z. Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-Seq. BMC Plant Biol. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Yuan, Y.; Yuan, L.; Wang, J.; Zhao, M.; Dai, Y.; Xie, S.; Zhu, S.; Hou, J.; Chen, G.; Wang, C. Upregulation of photosynthetic capacity and thylakoid membrane protein enhanced tolerance to heat stress in wucai (Brassica campestris L.). BioRxiv 2018, 493247. [Google Scholar] [CrossRef]
- Srivastava, A.; Guisse, B.; Greppin, H.; Strasser, R.J. Regulation of antenna structure and electron transport in photosystem II of Pisum sativum under elevated temperature probed by the fast polyphasic chlorophyll a fluorescence transient: OKJIP. BBABioenergetics 1997, 1320, 95–106. [Google Scholar] [CrossRef]
- Baker, N.R.; Rosenqvist, E. Applications of chlorophyll fluorescence can improve crop production strategies: An examination of future possibilities. J. Exp. Bot. 2004, 55, 1607–1621. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Deng, F.; Ren, W.J. Shading tolerance in rice is related to better light harvesting and use efficiency ad grain filling rate during grain filling period. Field Crops Res. 2015, 180, 54–62. [Google Scholar] [CrossRef]
- Teramoto, H.; Nakamori, A.; Minagawa, J.; Ono, T.A. Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiol. 2002, 130, 325–333. [Google Scholar] [CrossRef]
- Blair, E.J.; Bonnot, T.; Hummel, M.; Hay, E.; Marzolino, J.M.; Quijada, I.A.; Nagel, D.H. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in arabidopsis. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Grinevich, D.O.; Desai, J.S.; Stroup, K.P.; Duan, J.; Slabaugh, E.; Doherty, C.J. Novel transcriptional responses to heat revealed by turning up the heat at night. Plant Mol. Biol. 2019, 101, 1–19. [Google Scholar] [CrossRef]
- Gil, K.E.; Park, C.M. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef]
- Thain, S.C.; Murtas, G.; Lynn, J.R.; McGrath, R.B.; Millar, A.J. The circadian clock that controls gene expression in arabidopsis is tissue specific. Plant Physiol. 2002, 130, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Tan, B.; Yan, L.; Jiang, C.; Cheng, J.; Zheng, X.; Wang, W.; Chen, T.; Ye, X.; Li, J. Transcript profiling provides insights into molecular processes during shoot elongation in temperature-sensitive peach (Prunus persica). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Yu, E.; Fan, C.; Yang, Q.; Li, X.; Wan, B.; Dong, Y.; Wang, X.; Zhou, Y. Identification of heat responsive genes in Brassica napus siliques at the seed-filling stage through transcriptional profiling. PLoS ONE 2014, 9, e101914. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, H.; Hu, S.; Lu, X.; Yuan, C.; Zhang, C.; Wang, P.; Xiao, W.; Xiao, L.; Xue, G.P. Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA-and heat-stress-responsive nuclear genes. J. Exp. Bot. 2014, 65, 4159–4175. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, W.; Su, X.; Ge, P.; Zhou, Y.; Hao, Y.; Shu, H.; Gao, C.; Cheng, S.; Zhu, G. Early response of radish to heat stress by strand-specific transcriptome and miRNA analysis. Int. J. Mol. Sci. 2019, 20, 3321. [Google Scholar] [CrossRef] [PubMed]
- Karayekov, E.; Sellaro, R.; Legris, M.; Yanovsky, M.J.; Casal, J.J. Heat shock–induced fluctuations in clock and light signaling enhance phytochrome B–mediated arabidopsis deetiolation. Plant Cell 2013, 25, 2892–2906. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Wang, X.; Han, X.; Li, W.; Liu, T.; Chen, B.; Chen, X.; Wang-Pruski, G. Comparative transcriptome analyses revealed different heat stress responses in high-and low-GS Brassica alboglabra sprouts. BMC Genom. 2019, 20, 269. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Gupta, S.K.; Niu, S.; Li, X.Q.; Yang, Q.; Chen, G.; Zhu, W.; Haroon, M. Transcriptome analysis of heat stress response genes in potato leaves. Mol. Biol. Rep. 2020, 47, 4311–4321. [Google Scholar] [CrossRef]
- Alshameri, A.; Al-Qurainy, F.; Gaafar, A.-R.; Khan, S.; Nadeem, M.; Alansi, S. Identification of heat-responsive genes in guar [Cyamopsis tetragonoloba (L.) Taub]. Int. J. Genom. 2020, 2020, 3126592. [Google Scholar] [CrossRef]
- Ren, S.; Ma, K.; Lu, Z.; Chen, G.; Cui, J.; Tong, P.; Wang, L.; Teng, N.; Jin, B. Transcriptomic and metabolomic analysis of the heat-stress response of Populus tomentosa Carr. Forests 2019, 10, 383. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Krishnan, S.R.; Pandian, S.; Mareeswaran, N.; Aruni, W.; Pandian, S.K.; Ramesh, M. Global analysis of threonine metabolism genes unravel key players in rice to improve the abiotic stress tolerance. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Liu, L.; Lin, L. Effect of heat stress on Sargassum fusiforme leaf metabolome. J. Plant Biol. 2020, 63, 1–13. [Google Scholar] [CrossRef]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Liu, Z.; Li, S.; Li, W.; Liu, Q.; Zhang, L.; Song, X. Comparative transcriptome analysis indicates that a core transcriptional network mediates isonuclear alloplasmic male sterility in wheat (Triticum aestivum L.). BMC Plant Biol. 2020, 20, 1–19. [Google Scholar] [CrossRef]
- Higashi, Y.; Okazaki, Y.; Myouga, F.; Shinozaki, K.; Saito, K. Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci. Rep. 2015, 5, 10533. [Google Scholar] [CrossRef]
- Jayakodi, M.; Lee, S.C.; Yang, T.J. Comparative transcriptome analysis of heat stress responsiveness between two contrasting ginseng cultivars. J. Ginseng Res. 2019, 43, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Bi, Y.; Mo, J.; Zhang, R.; Qu, M.; Feng, S.; Essemine, J. Proteome and transcriptome reveal the involvement of heat shock proteins and antioxidant system in thermotolerance of Clematis florida. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Du, H.; Xu, M.; Huang, B. Metabolic responses to heat stress under elevated atmospheric CO2 concentration in a cool-season grass species. J. Am. Soc. Hortic. Sci. 2012, 137, 221–228. [Google Scholar] [CrossRef]
- Li, P.; Cao, W.; Fang, H.; Xu, S.; Yin, S.; Zhang, Y.; Lin, D.; Wang, J.; Chen, Y.; Xu, C. Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Front. Plant Sci. 2017, 8, 290. [Google Scholar] [CrossRef] [PubMed]
- Zinta, G.; AbdElgawad, H.; Peshev, D.; Weedon, J.T.; Van den Ende, W.; Nijs, I.; Janssens, I.A.; Beemster, G.T.; Asard, H. Dynamics of metabolic responses to periods of combined heat and drought in Arabidopsis thaliana under ambient and elevated atmospheric CO2. J. Exp. Bot. 2018, 69, 2159–2170. [Google Scholar] [CrossRef]
- Su, P.; Jiang, C.; Qin, H.; Hu, R.; Feng, J.; Chang, J.; Yang, G.; He, G. Identification of potential genes responsible for thermotolerance in wheat under high temperature stress. Genes 2019, 10, 174. [Google Scholar] [CrossRef]
- Zhan, X.; Qi, J.; Zhou, B.; Mao, B. Metabolomic and transcriptomic analyses reveal the regulation of pigmentation in the purple variety of Dendrobium officinale. Sci. Rep. 2020, 10, 1–15. [Google Scholar]
- Joshi, V.; Joung, J.G.; Fei, Z.; Jander, G. Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 2010, 39, 933–947. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; García-Pereira, M.J.; Gracia, M.P.; Igartua, E.; Casas, A.M.; Contreras-Moreira, B. Large differences in gene expression responses to drought and heat stress between elite barley cultivar scarlett and a spanish landrace. Front. Plant Sci. 2017, 8, 647. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.A.; Chowda-Reddy, R.; Muhle, A.A.; Tatineni, S.; Yuen, G.; Edmé, S.J.; Mitchell, R.B.; Sarath, G. Transcriptome divergence during leaf development in two contrasting switchgrass (Panicum virgatum L.) cultivars. PLoS ONE 2019, 14, e0222080. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Joshi, N.A.; Fass, J.N. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software]. 2011. Available online: https://github.com/najoshi/sickle (accessed on 15 February 2021).
- Yan, J.; Yu, L.; Xuan, J.; Lu, Y.; Lu, S.; Zhu, W. De novo transcriptome sequencing and gene expression profiling of spinach (Spinacia oleracea L.) leaves under heat stress. Sci. Rep. 2016, 6, 19473. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jaroszewski, L.; Godzik, A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 2001, 17, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Niu, B.; Fu, L.; Sun, S.; Li, W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinform. 2010, 11, 187. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; McCarthy, D.; Robinson, M.; Smyth, G.K. edgeR: Differential expression analysis of digital gene expression data User’s Guide. Bioconductor User’s Guide. 2014. Available online: http://bioconductor.org/packages/release/bioc/html/edgeR.html (accessed on 16 May 2019).
- Yu, G. ClusterProfiler: Universal enrichment tool for functional and comparative study. BioRxiv 2018, 256784. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, T.; Padmavathi, M. Application of K-means and genetic algorithms for dimension reduction by integrating SVM for diabetes diagnosis. Procedia Comput. Sci. 2015, 47, 76–83. [Google Scholar] [CrossRef]
- Eiben, A.E.; Smith, J.E. Introduction to Evolutionary Computing; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Schmittgen, T.D. Real-time quantitative PCR. Methods 2001, 25, 383–385. [Google Scholar] [CrossRef] [PubMed]


| Organism | V. corymbosum | V. darrowii |
|---|---|---|
| Total number of reads | 64,979,396 | 70,331,700 |
| Total base pairs (bp) | 9,626,737,814 | 10,494,423,421 |
| Total number of assembled transcripts | 156,183 | 183,343 |
| Total assembled bases | 162,599,434 | 208,966,135 |
| Average contig length (bp) | 1041 | 1139 |
| Total number of assembled genes | 99,093 | 109,193 |
| Percent GC (%) | 41.6 | 41.6 |
| Contig N50 (bp) | 1768 | 1967 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Callwood, J.; Melmaiee, K.; Kulkarni, K.P.; Vennapusa, A.R.; Aicha, D.; Moore, M.; Vorsa, N.; Natarajan, P.; Reddy, U.K.; Elavarthi, S. Differential Morpho-Physiological and Transcriptomic Responses to Heat Stress in Two Blueberry Species. Int. J. Mol. Sci. 2021, 22, 2481. https://doi.org/10.3390/ijms22052481
Callwood J, Melmaiee K, Kulkarni KP, Vennapusa AR, Aicha D, Moore M, Vorsa N, Natarajan P, Reddy UK, Elavarthi S. Differential Morpho-Physiological and Transcriptomic Responses to Heat Stress in Two Blueberry Species. International Journal of Molecular Sciences. 2021; 22(5):2481. https://doi.org/10.3390/ijms22052481
Chicago/Turabian StyleCallwood, Jodi, Kalpalatha Melmaiee, Krishnanand P. Kulkarni, Amaranatha R. Vennapusa, Diarra Aicha, Michael Moore, Nicholi Vorsa, Purushothaman Natarajan, Umesh K. Reddy, and Sathya Elavarthi. 2021. "Differential Morpho-Physiological and Transcriptomic Responses to Heat Stress in Two Blueberry Species" International Journal of Molecular Sciences 22, no. 5: 2481. https://doi.org/10.3390/ijms22052481
APA StyleCallwood, J., Melmaiee, K., Kulkarni, K. P., Vennapusa, A. R., Aicha, D., Moore, M., Vorsa, N., Natarajan, P., Reddy, U. K., & Elavarthi, S. (2021). Differential Morpho-Physiological and Transcriptomic Responses to Heat Stress in Two Blueberry Species. International Journal of Molecular Sciences, 22(5), 2481. https://doi.org/10.3390/ijms22052481






