The Role of TCOF1 Gene in Health and Disease: Beyond Treacher Collins Syndrome
Abstract
1. Introduction
2. TCOF1 Gene and Transcripts
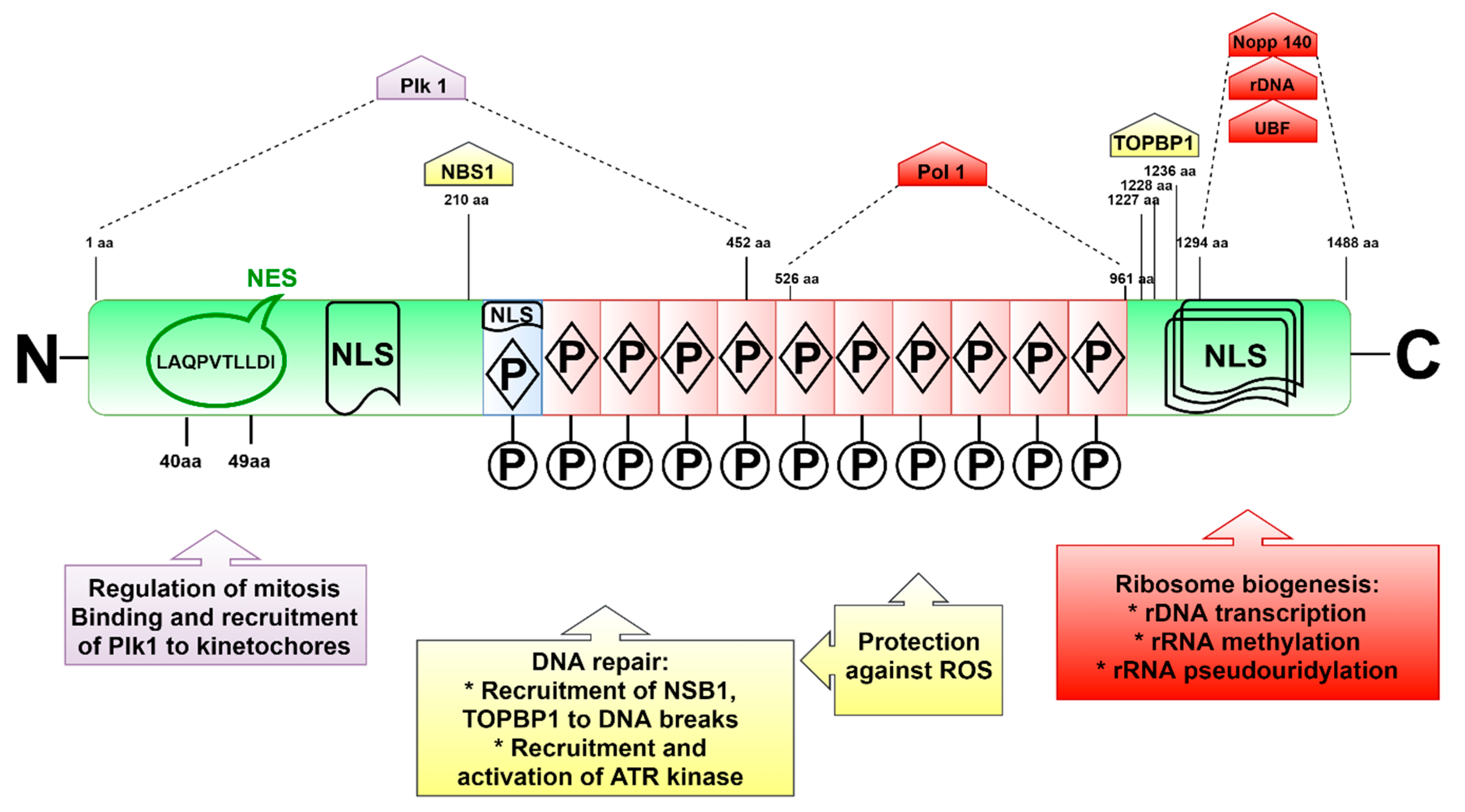
3. Structure and Localization of the Treacle Protein
4. The Role of TCOF1/Treacle in Physiological Processes
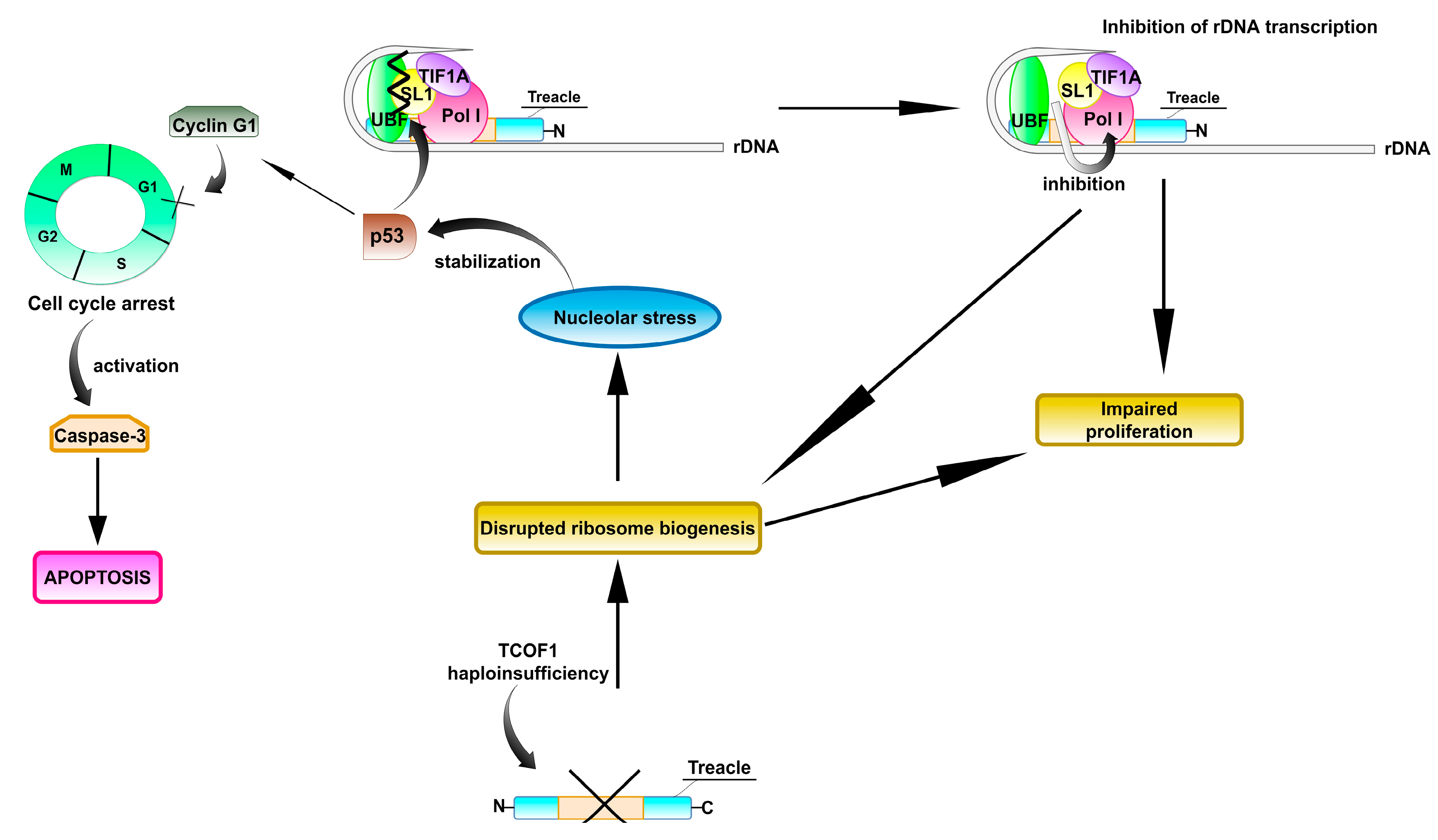
4.1. The Role of TCOF1 in Ribosome Biogenesis and Function
4.1.1. The Role of Treacle Ubiquitilation
4.2. Treacle and the Response to DNA Damage
4.3. TCOF1 and the Mitotic Regulation
4.4. TCOF1 and Telomere Integrity
5. The Role of TCOF1 in the Pathological Processes
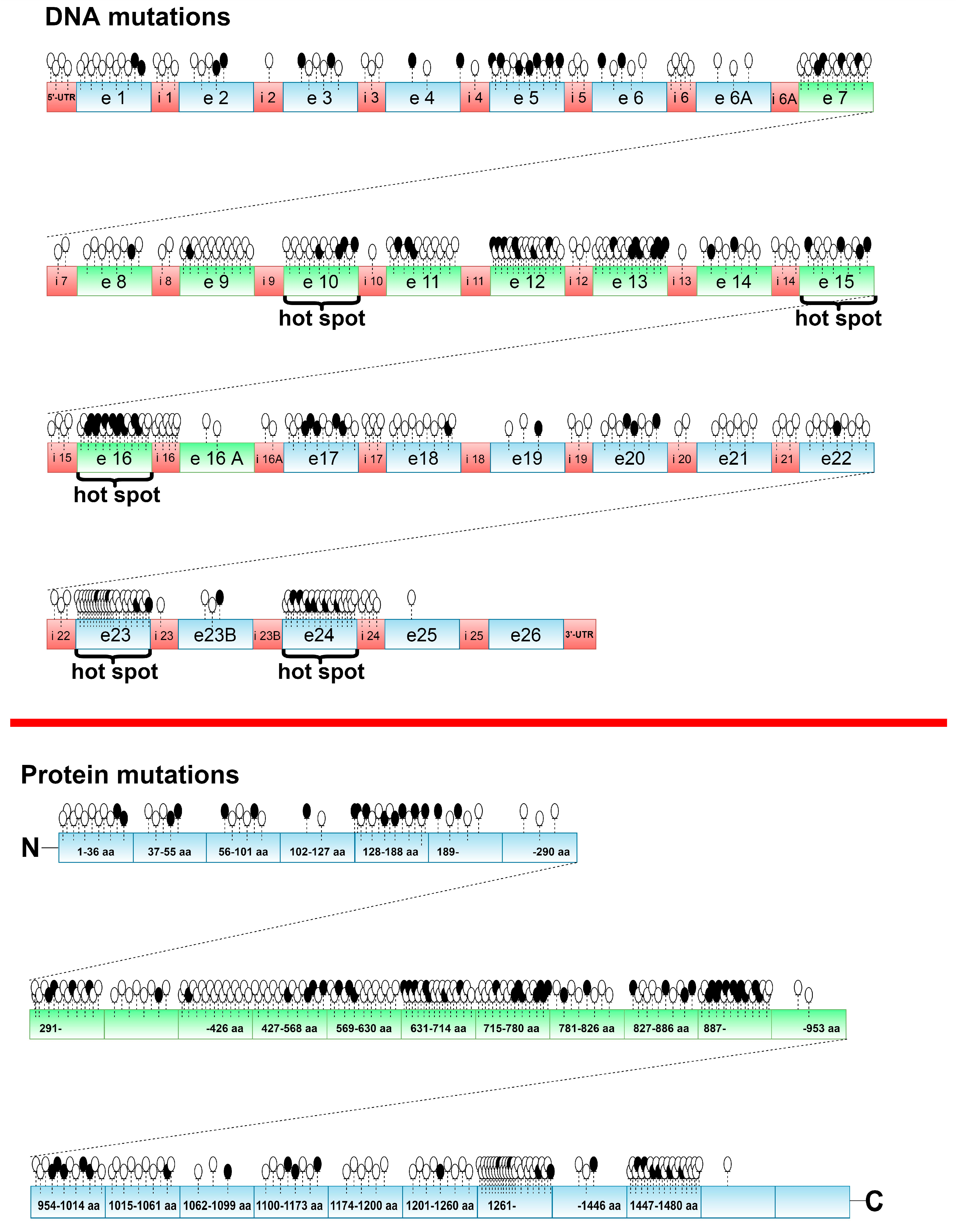
5.1. Treacher Collins Syndrome
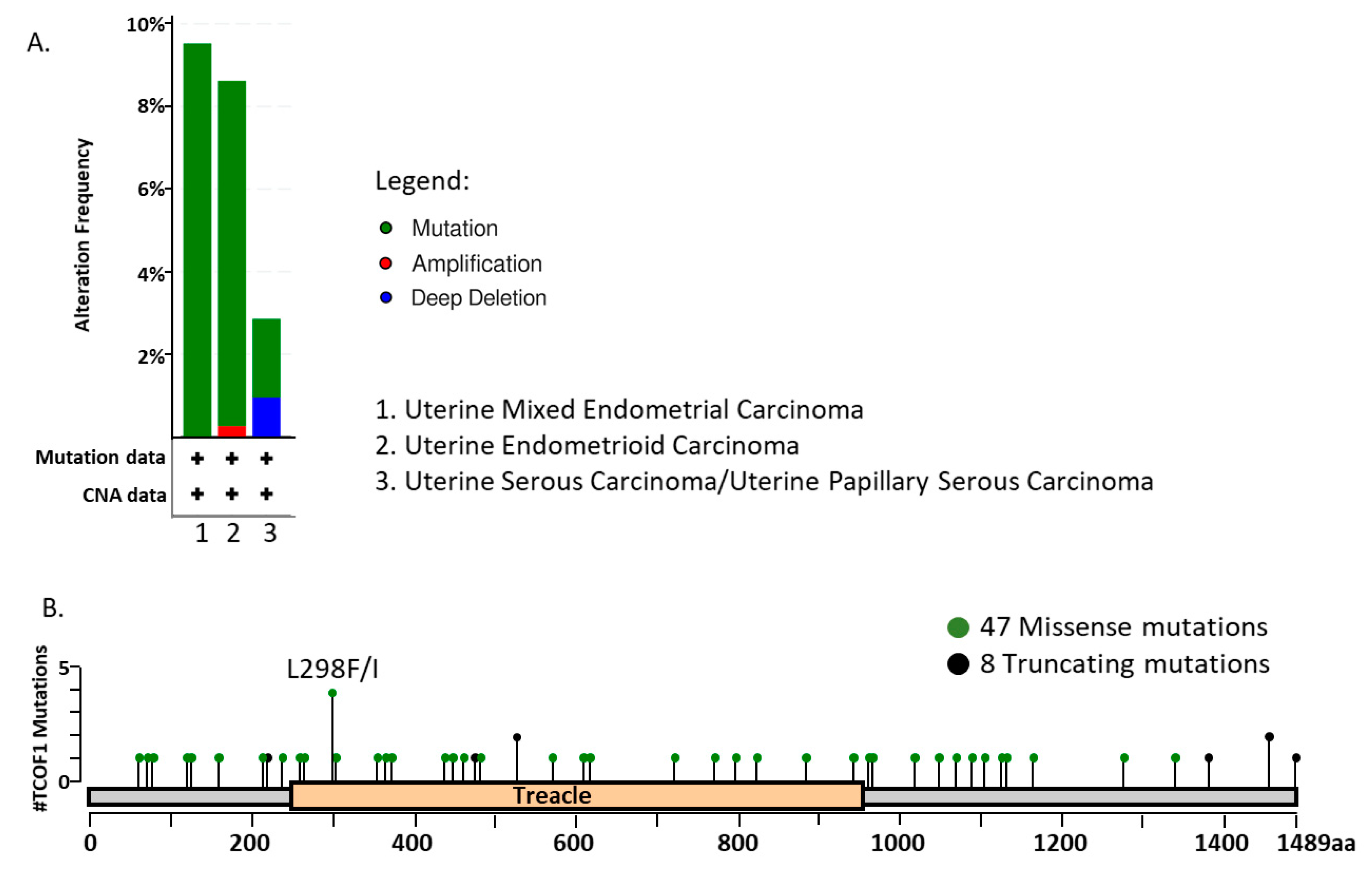
5.2. The Role of TCOF1 in Cancer
5.3. The other Pathologies Linked with TCOF1 Dysfunction
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Núñez Villacís, L.; Wong, M.S.; Ferguson, L.L.; Hein, N.; George, A.J.; Hannan, K.M. New Roles for the Nucleolus in Health and Disease. Bioessays 2018, 40, e1700233. [Google Scholar] [CrossRef]
- Turi, Z.; Lacey, M.; Mistrik, M.; Moudry, P. Impaired ribosome biogenesis: Mechanisms and relevance to cancer and aging. Aging (Albany NY) 2019, 11, 2512–2540. [Google Scholar] [CrossRef]
- Correll, C.C.; Bartek, J.; Dundr, M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells 2019, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.J.; Bridger, J.M.; Cuthbert, A.P.; Newbold, R.F.; Bickmore, W.A.; McStay, B. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 2001, 20, 2867–2874. [Google Scholar] [CrossRef]
- Hernandez-Verdun, D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011, 2, 189–194. [Google Scholar] [CrossRef]
- Pederson, T. The plurifunctional nucleolus. Nucleic Acids Res. 1998, 26, 3871–3876. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T. The nucleolus. Cold Spring Harb. Perspect Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Weeks, S.E.; Metge, B.J.; Samant, R.S. The nucleolus: A central response hub for the stressors that drive cancer progression. Cell Mol. Life Sci. 2019, 76, 4511–4524. [Google Scholar] [CrossRef] [PubMed]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis-Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef] [PubMed]
- The Treacher Collins Syndrome Collaborative Group; Dixon, J.; Edwards, S.J.; Gladwin, A.J.; Dixon, M.J.; Loftus, S.K.; Bonner, C.A.; Koprivnikar, K.; Wasmuth, J.J. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat. Genet. 1996, 12, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Wise, C.A.; Chiang, L.C.; Paznekas, W.A.; Sharma, M.; Musy, M.M.; Ashley, J.A.; Lovett, M.; Jabs, E.W. TCOF1 gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proc. Natl. Acad. Sci. USA 1997, 94, 3110–3115. [Google Scholar] [CrossRef]
- Sakai, D.; Trainor, P.A. Face off against ROS: Tcof1/Treacle safeguards neuroepithelial cells and progenitor neural crest cells from oxidative stress during craniofacial development. Dev. Growth Differ. 2016, 58, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Dixon, J.; Achilleos, A.; Dixon, M.; Trainor, P.A. Prevention of Treacher Collins syndrome craniofacial anomalies in mouse models via maternal antioxidant supplementation. Nat. Commun. 2016, 7, 10328. [Google Scholar] [CrossRef] [PubMed]
- Tse, W.K. Treacher Collins syndrome: New insights from animal models. Int. J. Biochem. Cell Biol. 2016, 81, 44–47. [Google Scholar] [CrossRef]
- Dixon, M.J.; Dixon, J.; Houseal, T.; Bhatt, M.; Ward, D.C.; Klinger, K.; Landes, G.M. Narrowing the position of the Treacher Collins syndrome locus to a small interval between three new microsatellite markers at 5q32-33.1. Am. J. Hum. Genet. 1993, 52, 907–914. [Google Scholar]
- Jabs, E.W.; Li, X.; Lovett, M.; Yamaoka, L.H.; Taylor, E.; Speer, M.C.; Coss, C.; Cadle, R.; Hall, B.; Brown, K. Genetic and physical mapping of the Treacher Collins syndrome locus with respect to loci in the chromosome 5q3 region. Genomics 1993, 18, 7–13. [Google Scholar] [CrossRef]
- Shows, K.H.; Shiang, R. Regulation of the mouse Treacher Collins syndrome homolog (Tcof1) promoter through differential repression of constitutive expression. DNA Cell Biol. 2008, 27, 589–600. [Google Scholar] [CrossRef]
- Dixon, J.; Edwards, S.J.; Anderson, I.; Brass, A.; Scambler, P.J.; Dixon, M.J. Identification of the complete coding sequence and genomic organization of the Treacher Collins syndrome gene. Genome Res. 1997, 7, 223–234. [Google Scholar] [CrossRef]
- So, R.B.; Gonzales, B.; Henning, D.; Dixon, J.; Dixon, M.J.; Valdez, B.C. Another face of the Treacher Collins syndrome (TCOF1) gene: Identification of additional exons. Gene 2004, 328, 49–57. [Google Scholar] [CrossRef]
- Larsen, D.H.; Hari, F.; Clapperton, J.A.; Gwerder, M.; Gutsche, K.; Altmeyer, M.; Jungmichel, S.; Toledo, L.I.; Fink, D.; Rask, M.B.; et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell Biol. 2014, 16, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Isaac, C.; Marsh, K.L.; Paznekas, W.A.; Dixon, J.; Dixon, M.J.; Jabs, E.W.; Meier, U.T. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol. Biol. Cell 2000, 11, 3061–3071. [Google Scholar] [CrossRef]
- Marsh, K.L.; Dixon, J.; Dixon, M.J. Mutations in the Treacher Collins syndrome gene lead to mislocalization of the nucleolar protein treacle. Hum. Mol. Genet. 1998, 7, 1795–1800. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.E. The Cajal body. Biochim. Biophys. Acta 2008, 1783, 2108–2115. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Dixon, J.; Dixon, M.J.; Trainor, P.A. Mammalian neurogenesis requires Treacle-Plk1 for precise control of spindle orientation, mitotic progression, and maintenance of neural progenitor cells. PLoS Genet. 2012, 8, e1002566. [Google Scholar] [CrossRef]
- Winokur, S.T.; Shiang, R. The Treacher Collins syndrome (TCOF1) gene product, treacle, is targeted to the nucleolus by signals in its C-terminus. Hum. Mol. Genet. 1998, 7, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Korsholm, L.M.; Gál, Z.; Lin, L.; Quevedo, O.; Ahmad, D.A.; Dulina, E.; Luo, Y.; Bartek, J.; Larsen, D.H. Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair. Nucleic Acids Res. 2019, 47, 8019–8035. [Google Scholar] [CrossRef]
- Valdez, B.C.; Henning, D.; So, R.B.; Dixon, J.; Dixon, M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA 2004, 101, 10709–10714. [Google Scholar] [CrossRef]
- Dixon, J.; Hovanes, K.; Shiang, R.; Dixon, M.J. Sequence analysis, identification of evolutionary conserved motifs and expression analysis of murine tcof1 provide further evidence for a potential function for the gene and its human homologue, TCOF1. Hum. Mol. Genet. 1997, 6, 727–737. [Google Scholar] [CrossRef]
- Dixon, J.; Jones, N.C.; Sandell, L.L.; Jayasinghe, S.M.; Crane, J.; Rey, J.P.; Dixon, M.J.; Trainor, P.A. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA 2006, 103, 13403–13408. [Google Scholar] [CrossRef]
- Barlow, A.J.; Dixon, J.; Dixon, M.; Trainor, P.A. Tcof1 acts as a modifier of Pax3 during enteric nervous system development and in the pathogenesis of colonic aganglionosis. Hum. Mol. Genet. 2013, 22, 1206–1217. [Google Scholar] [CrossRef]
- Werner, A.; Iwasaki, S.; McGourty, C.A.; Medina-Ruiz, S.; Teerikorpi, N.; Fedrigo, I.; Ingolia, N.T.; Rape, M. Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 2015, 525, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Ferreira, R.; Schneekloth, J.S.; Panov, K.I.; Hannan, K.M.; Hannan, R.D. Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 2020, 9, 266. [Google Scholar] [CrossRef]
- Xu, X.; Xiong, X.; Sun, Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci. China Life Sci. 2016, 59, 656–672. [Google Scholar] [CrossRef]
- Baßler, J.; Hurt, E. Eukaryotic Ribosome Assembly. Annu. Rev. Biochem. 2019, 88, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Bell, S.P.; Learned, R.M.; Jantzen, H.M.; Tjian, R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 1988, 241, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.I.; Yeh, N.H. Treacle recruits RNA polymerase I complex to the nucleolus that is independent of UBF. Biochem. Biophys. Res. Commun. 2009, 386, 396–401. [Google Scholar] [CrossRef]
- Gonzales, B.; Henning, D.; So, R.B.; Dixon, J.; Dixon, M.J.; Valdez, B.C. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum. Mol. Genet. 2005, 14, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Moss, T.; Mars, J.C.; Tremblay, M.G.; Sabourin-Felix, M. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019, 27, 31–40. [Google Scholar] [CrossRef]
- Monaco, P.L.; Marcel, V.; Diaz, J.J.; Catez, F. 2’-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Jerabkova, K.; Sumara, I. Cullin 3, a cellular scripter of the non-proteolytic ubiquitin code. Semin. Cell Dev. Biol. 2019, 93, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.; Baur, R.; Teerikorpi, N.; Kaya, D.U.; Rape, M. Multisite dependency of an E3 ligase controls monoubiquitylation-dependent cell fate decisions. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Giglia-Mari, G.; Zotter, A.; Vermeulen, W. DNA damage response. Cold Spring Harb. Perspect Biol. 2011, 3, a000745. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008, 283, 1–5. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. DNA double-strand break repair in a cellular context. Clin. Oncol. 2014, 26, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.G.; Pefani, D.E.; Pateras, I.S.; Trougakos, I.P. Integrating the DNA damage and protein stress responses during cancer development and treatment. J. Pathol. 2018, 246, 12–40. [Google Scholar] [CrossRef] [PubMed]
- Korsholm, L.M.; Gál, Z.; Nieto, B.; Quevedo, O.; Boukoura, S.; Lund, C.C.; Larsen, D.H. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2020, 48, 9449–9461. [Google Scholar] [CrossRef]
- Kim, J.H.; Grosbart, M.; Anand, R.; Wyman, C.; Cejka, P.; Petrini, J.H.J. The Mre11-Nbs1 Interface Is Essential for Viability and Tumor Suppression. Cell Rep. 2017, 18, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Huang, J.W.; Izhar, L.; Sowa, M.E.; Harper, J.W.; Elledge, S.J. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 18631–18636. [Google Scholar] [CrossRef] [PubMed]
- Mooser, C.; Symeonidou, I.-E.; Leimbacher, P.A.; Ribeiro, A.; Shorrocks, A.K.; Jungmichel, S.; Larsen, S.C.; Knechtle, K.; Jasrotia, A.; Zurbriggen, D.; et al. Treacle controls the nucleolar response to rDNA breaks via TOPBP1 recruitment and ATR activation. Nat. Commun. 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, A.; Lee, J.; Yoo, H.Y.; Dunphy, W.G. TopBP1 activates the ATR-ATRIP complex. Cell 2006, 124, 943–955. [Google Scholar] [CrossRef] [PubMed]
- van Sluis, M.; McStay, B. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017, 46, 81–86. [Google Scholar] [CrossRef]
- Song, B.; Liu, X.S.; Liu, X. Polo-like kinase 1 (Plk1): An Unexpected Player in DNA Replication. Cell Div. 2012, 7, 3. [Google Scholar] [CrossRef]
- Nie, X.; Xiao, D.; Ge, Y.; Xie, Y.; Zhou, H.; Zheng, T.; Li, X.; Liu, H.; Huang, H.; Zhao, Y. TRF2 recruits nucleolar protein TCOF1 to coordinate telomere transcription and replication. Cell Death Differ. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lalonde, M.; Chartrand, P. TERRA, a Multifaceted Regulator of Telomerase Activity at Telomeres. J. Mol. Biol. 2020, 432, 4232–4243. [Google Scholar] [CrossRef] [PubMed]
- Kasat, V. Franceschetti syndrome. Contemp Clin. Dent. 2011, 2, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D.; Trainor, P.A. Treacher Collins syndrome: Unmasking the role of Tcof1/treacle. Int. J. Biochem. Cell Biol. 2009, 41, 1229–1232. [Google Scholar] [CrossRef]
- Trainor, P.A.; Merrill, A.E. Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim. Biophys. Acta 2014, 1842, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Collet, C.; Verloes, A.; Lambert, L.; Herlin, C.; Blanchet, C.; Sanchez, E.; Drunat, S.; Vigneron, J.; Laplanche, J.L.; et al. Large deletions encompassing the TCOF1 and CAMK2A genes are responsible for Treacher Collins syndrome with intellectual disability. Eur. J. Hum. Genet. 2014, 22, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Lau, M.C.; Kwong, E.M.; Lai, K.P.; Li, J.W.; Ho, J.C.; Chan, T.F.; Wong, C.K.; Jiang, Y.J.; Tse, W.K. Pathogenesis of POLR1C-dependent Type 3 Treacher Collins Syndrome revealed by a zebrafish model. Biochim. Biophys. Acta 2016, 1862, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Noack Watt, K.E.; Achilleos, A.; Neben, C.L.; Merrill, A.E.; Trainor, P.A. The Roles of RNA Polymerase I and III Subunits Polr1c and Polr1d in Craniofacial Development and in Zebrafish Models of Treacher Collins Syndrome. PLoS Genet. 2016, 12, e1006187. [Google Scholar] [CrossRef] [PubMed]
- Splendore, A.; Silva, E.O.; Alonso, L.G.; Richieri-Costa, A.; Alonso, N.; Rosa, A.; Carakushanky, G.; Cavalcanti, D.P.; Brunoni, D.; Passos-Bueno, M.R. High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Hum. Mutat. 2000, 16, 315–322. [Google Scholar] [CrossRef]
- Dauwerse, J.G.; Dixon, J.; Seland, S.; Ruivenkamp, C.A.; van Haeringen, A.; Hoefsloot, L.H.; Peters, D.J.; Boers, A.C.; Daumer-Haas, C.; Maiwald, R.; et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011, 43, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Geneviève, D.; Ostertag, A.; Marlin, S.; Lacombe, D.; Martin-Coignard, D.; Coubes, C.; David, A.; Lyonnet, S.; Vilain, C.; et al. Treacher Collins syndrome: A clinical and molecular study based on a large series of patients. Genet. Med. 2016, 18, 49–56. [Google Scholar] [CrossRef]
- Conte, C.; D’Apice, M.R.; Rinaldi, F.; Gambardella, S.; Sangiuolo, F.; Novelli, G. Novel mutations of TCOF1 gene in European patients with Treacher Collins syndrome. BMC Med. Genet. 2011, 12, 125. [Google Scholar] [CrossRef]
- Edwards, S.J.; Gladwin, A.J.; Dixon, M.J. The mutational spectrum in Treacher Collins syndrome reveals a predominance of mutations that create a premature-termination codon. Am. J. Hum. Genet. 1997, 60, 515–524. [Google Scholar] [PubMed]
- Splendore, A.; Jabs, E.W.; Passos-Bueno, M.R. Screening of TCOF1 in patients from different populations: Confirmation of mutational hot spots and identification of a novel missense mutation that suggests an important functional domain in the protein treacle. J. Med. Genet. 2002, 39, 493–495. [Google Scholar] [CrossRef]
- Gladwin, A.J.; Dixon, J.; Loftus, S.K.; Edwards, S.; Wasmuth, J.J.; Hennekam, R.C.; Dixon, M.J. Treacher Collins syndrome may result from insertions, deletions or splicing mutations, which introduce a termination codon into the gene. Hum. Mol. Genet. 1996, 5, 1533–1538. [Google Scholar] [CrossRef]
- Splendore, A.; Fanganiello, R.D.; Masotti, C.; Morganti, L.S.; Passos-Bueno, M.R. TCOF1 mutation database: Novel mutation in the alternatively spliced exon 6A and update in mutation nomenclature. Hum. Mutat. 2005, 25, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Trainor, P.A.; Dixon, J.; Dixon, M.J. Treacher Collins syndrome: Etiology, pathogenesis and prevention. Eur. J. Hum. Genet. 2009, 17, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Rubbi, C.P.; Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef]
- Le Goff, S.; Boussaid, I.; Floquet, C.; Raimbault, A.; Hatin, I.; Andrieu-Soler, C.; Salma, M.; Leduc, M.; Gautier, E.F.; Guyot, B.; et al. p53 activation during ribosome biogenesis regulates normal erythroid differentiation. Blood 2021, 137, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Golomb, L.; Volarevic, S.; Oren, M. p53 and ribosome biogenesis stress: The essentials. FEBS Lett. 2014, 588, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, S.; Ivanenkov, V.V.; Teng, T.; Thomas, G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012, 26, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Pelava, A.; Schneider, C.; Watkins, N.J. The importance of ribosome production, and the 5S RNP-MDM2 pathway, in health and disease. Biochem Soc. Trans. 2016, 44, 1086–1090. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.C.; Lynn, M.L.; Gaudenz, K.; Sakai, D.; Aoto, K.; Rey, J.P.; Glynn, E.F.; Ellington, L.; Du, C.; Dixon, J.; et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008, 14, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Comai, L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell Biol. 2000, 20, 5930–5938. [Google Scholar] [CrossRef]
- De Peralta, M.S.; Mouguelar, V.S.; Sdrigotti, M.A.; Ishiy, F.A.; Fanganiello, R.D.; Passos-Bueno, M.R.; Coux, G.; Calcaterra, N.B. Cnbp ameliorates Treacher Collins Syndrome craniofacial anomalies through a pathway that involves redox-responsive genes. Cell Death Dis. 2016, 7, e2397. [Google Scholar] [CrossRef]
- Masotti, C.; Ornelas, C.C.; Splendore-Gordonos, A.; Moura, R.; Félix, T.M.; Alonso, N.; Camargo, A.A.; Passos-Bueno, M.R. Reduced transcription of TCOF1 in adult cells of Treacher Collins syndrome patients. BMC Med. Genet. 2009, 10, 136. [Google Scholar] [CrossRef]
- Dai, J.; Si, J.; Wang, M.; Huang, L.; Fang, B.; Shi, J.; Wang, X.; Shen, G. Tcof1-Related Molecular Networks in Treacher Collins Syndrome. J. Craniofac Surg. 2016, 27, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wu, N.; Han, Y.; Hou, Q.; Zhao, Y.; Pan, Y.; Xie, X.; Chen, F. DDX21 promotes gastric cancer proliferation by regulating cell cycle. Biochem. Biophys. Res. Commun. 2018, 505, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Gu, B.; Bowen, M.E.; Aryan, F.; Zalc, A.; Liang, J.; Flynn, R.A.; Swigut, T.; Chang, H.Y.; Attardi, L.D.; et al. Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 2018, 554, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Kelsen, S.G.; Clarkson, A.B.; Ji, R.; Merali, S. SILAC analysis of oxidative stress-mediated proteins in human pneumocytes: New role for treacle. Proteomics 2010, 10, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Wu, Z.; Li, S.; Tang, X.; Wang, Y.; Guo, W.; Cao, G.; Chen, K.; Zhang, M.; Guan, M.; Yang, D. Copy Number Amplification of DNA Damage Repair Pathways Potentiates Therapeutic Resistance in Cancer. Theranostics 2020, 10, 3939–3951. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Su, Y.N.; Chou, P.C.; Chiang, W.C.; Chang, M.C.; Wang, L.S.; Teng, S.C.; Wu, K.J. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J. Biol. Chem. 2005, 280, 32505–32511. [Google Scholar] [CrossRef]
- Yang, M.H.; Chang, S.Y.; Chiou, S.H.; Liu, C.J.; Chi, C.W.; Chen, P.M.; Teng, S.C.; Wu, K.J. Overexpression of NBS1 induces epithelial-mesenchymal transition and co-expression of NBS1 and Snail predicts metastasis of head and neck cancer. Oncogene 2007, 26, 1459–1467. [Google Scholar] [CrossRef]
- Rai, R.; Peng, G.; Li, K.; Lin, S.Y. DNA damage response: The players, the network and the role in tumor suppression. Cancer Genom. Proteom. 2007, 4, 99–106. [Google Scholar] [PubMed]
- Goldstein, M.; Kastan, M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.J.; Qian, W.J.; Smith, J.N.; Gritsenko, M.A.; Hu, D.; Chrisler, W.B.; Timchalk, C. Stable Acinar Progenitor Cell Model Identifies Treacle-Dependent Radioresistance. Radiat. Res. 2019, 192, 135–144. [Google Scholar] [CrossRef]
- Rocha, C.R.R.; Silva, M.M.; Quinet, A.; Cabral-Neto, J.B.; Menck, C.F.M. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics 2018, 73, e478s. [Google Scholar] [CrossRef]
- Hamdane, N.; Herdman, C.; Mars, J.C.; Stefanovsky, V.; Tremblay, M.G.; Moss, T. Depletion of the cisplatin targeted HMGB-box factor UBF selectively induces p53-independent apoptotic death in transformed cells. Oncotarget 2015, 6, 27519–27536. [Google Scholar] [CrossRef]
- Trembley, J.H.; Wang, G.; Unger, G.; Slaton, J.; Ahmed, K. Protein kinase CK2 in health and disease: CK2: A key player in cancer biology. Cell Mol. Life Sci. 2009, 66, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Buontempo, F.; McCubrey, J.A.; Orsini, E.; Ruzzene, M.; Cappellini, A.; Lonetti, A.; Evangelisti, C.; Chiarini, F.; Barata, J.T.; Martelli, A.M. Therapeutic targeting of CK2 in acute and chronic leukemias. Leukemia 2018, 32, 1–10. [Google Scholar] [CrossRef]
- Borgo, C.; Ruzzene, M. Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Cancer Res. 2019, 38, 287. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, N.M.S.; Färnegårdh, K.; Bonagas, N.; Ninou, A.H.; Groth, P.; Wiita, E.; Jönsson, M.; Hallberg, K.; Lehto, J.; Pennisi, R.; et al. Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination. Nat. Commun. 2018, 9, 3872. [Google Scholar] [CrossRef] [PubMed]
- McCool, M.A.; Bryant, C.J.; Baserga, S.J. MicroRNAs and long non-coding RNAs as novel regulators of ribosome biogenesis. Biochem. Soc. Trans. 2020, 48, 595–612. [Google Scholar] [CrossRef] [PubMed]
- Catez, F.; Dalla Venezia, N.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef]
- Mekhail, K.; Gunaratnam, L.; Bonicalzi, M.E.; Lee, S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004, 6, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Audas, T.E.; Jacob, M.D.; Lee, S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 2012, 45, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Dupin, E.; Sommer, L. Neural crest progenitors and stem cells: From early development to adulthood. Dev. Biol. 2012, 366, 83–95. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grzanka, M.; Piekiełko-Witkowska, A. The Role of TCOF1 Gene in Health and Disease: Beyond Treacher Collins Syndrome. Int. J. Mol. Sci. 2021, 22, 2482. https://doi.org/10.3390/ijms22052482
Grzanka M, Piekiełko-Witkowska A. The Role of TCOF1 Gene in Health and Disease: Beyond Treacher Collins Syndrome. International Journal of Molecular Sciences. 2021; 22(5):2482. https://doi.org/10.3390/ijms22052482
Chicago/Turabian StyleGrzanka, Małgorzata, and Agnieszka Piekiełko-Witkowska. 2021. "The Role of TCOF1 Gene in Health and Disease: Beyond Treacher Collins Syndrome" International Journal of Molecular Sciences 22, no. 5: 2482. https://doi.org/10.3390/ijms22052482
APA StyleGrzanka, M., & Piekiełko-Witkowska, A. (2021). The Role of TCOF1 Gene in Health and Disease: Beyond Treacher Collins Syndrome. International Journal of Molecular Sciences, 22(5), 2482. https://doi.org/10.3390/ijms22052482





