Overexpression of AtBBD1, Arabidopsis Bifunctional Nuclease, Confers Drought Tolerance by Enhancing the Expression of Regulatory Genes in ABA-Mediated Drought Stress Signaling
Abstract
1. Introduction
2. Results and Discussion
2.1. AtBBD1 Localizes to the Nucleus and Cytoplasm, and Is Strongly Expressed in Trichomes and Stomatal Guard Cells of Leaves
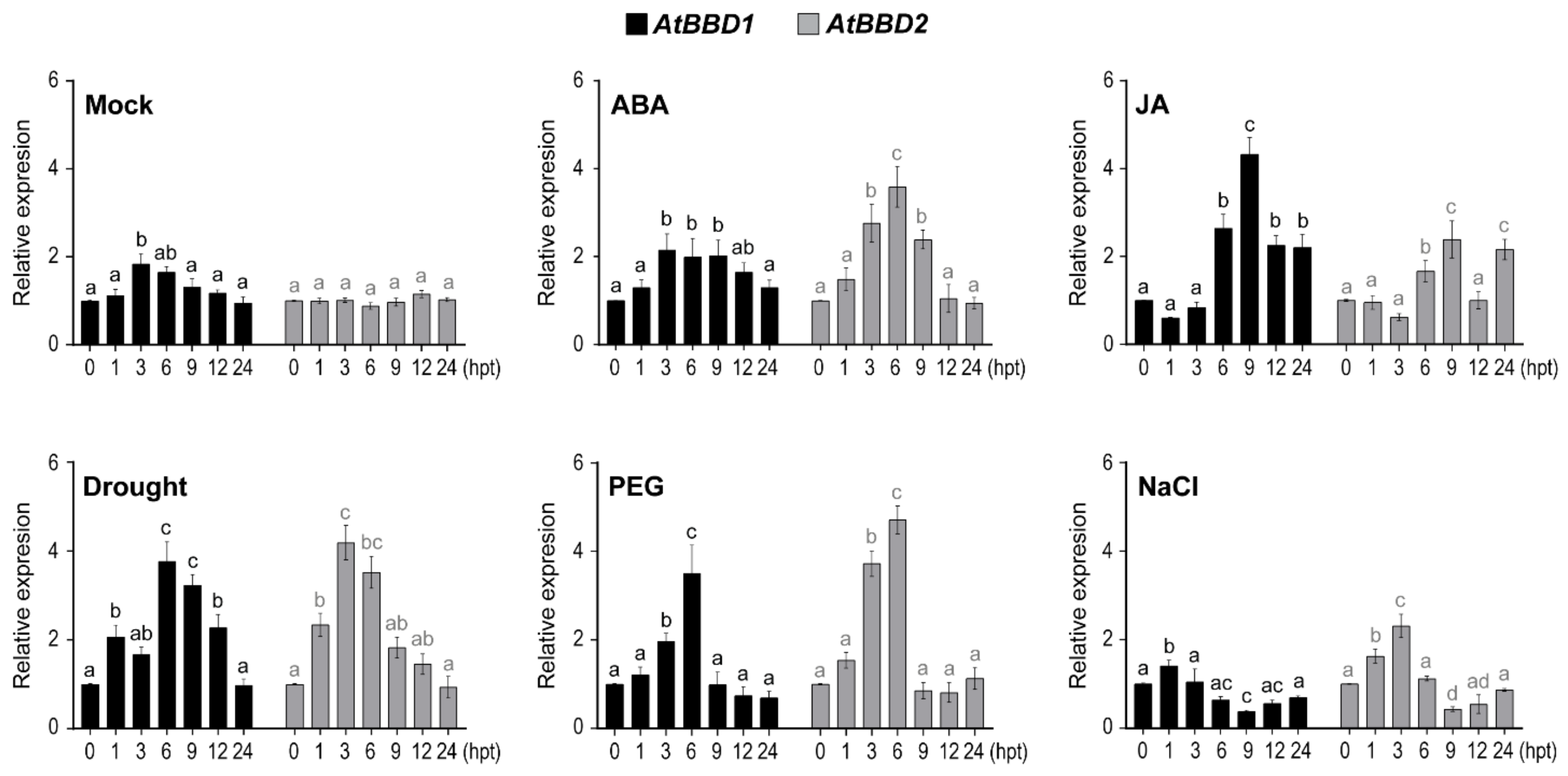
2.2. AtBBD1 and AtBBD2 Are Strongly Induced by ABA and Drought, and AtBBD1, but Not AtBBD2, Is also Strongly Responsive to JA
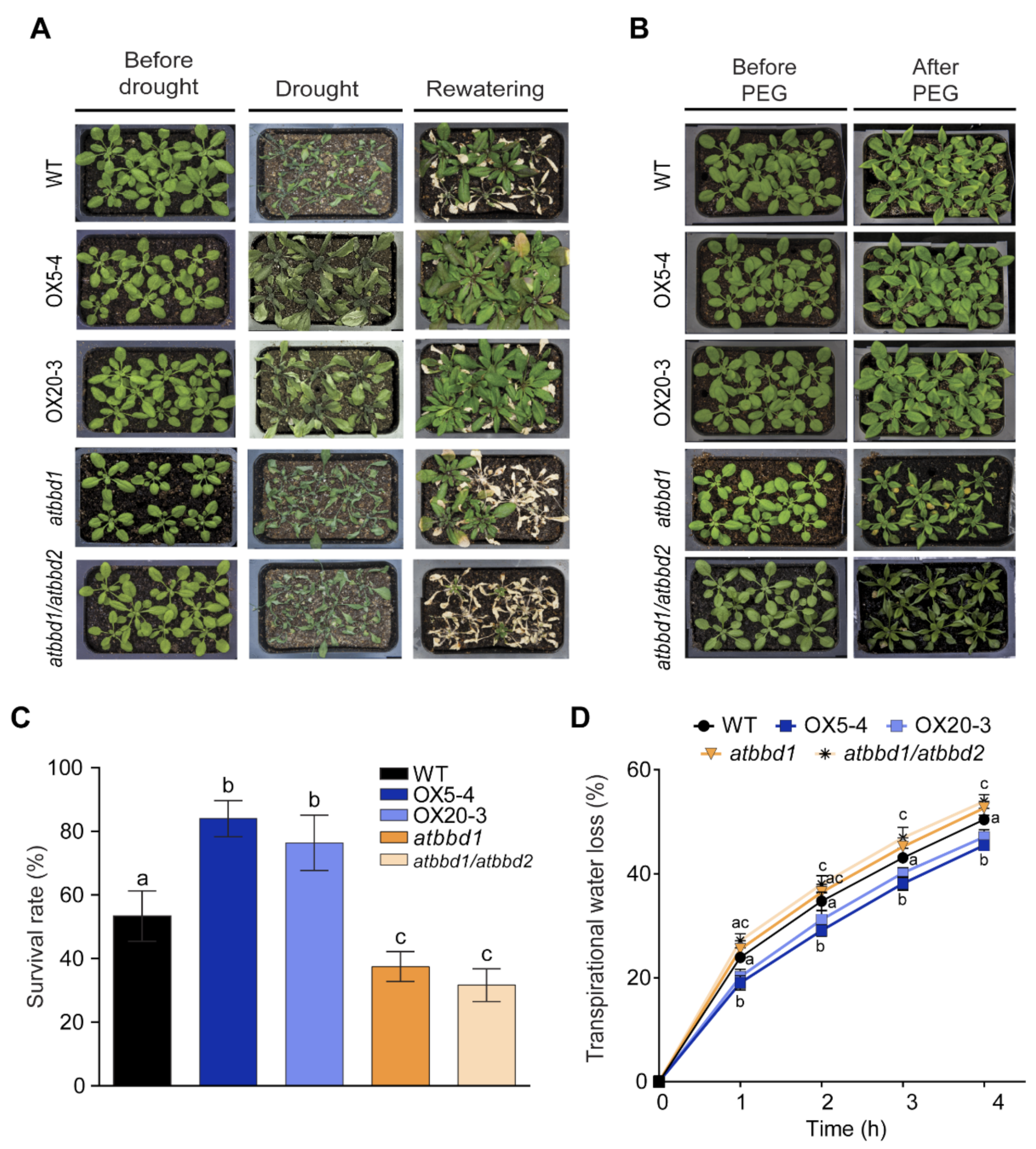
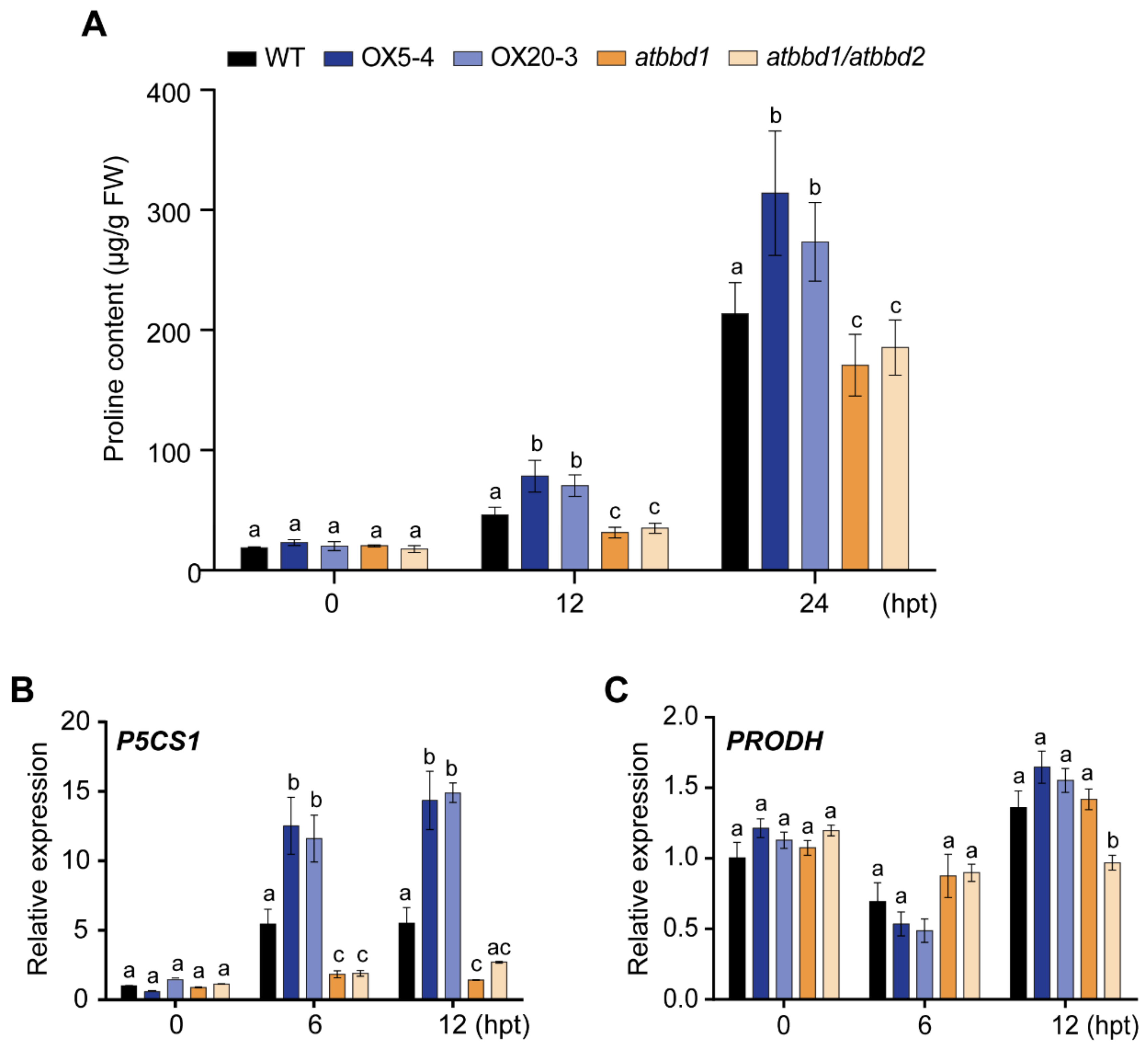
2.3. Overexpression of AtBBD1 Reduces Water Loss and Increases Proline Content, Conferring Drought Tolerance
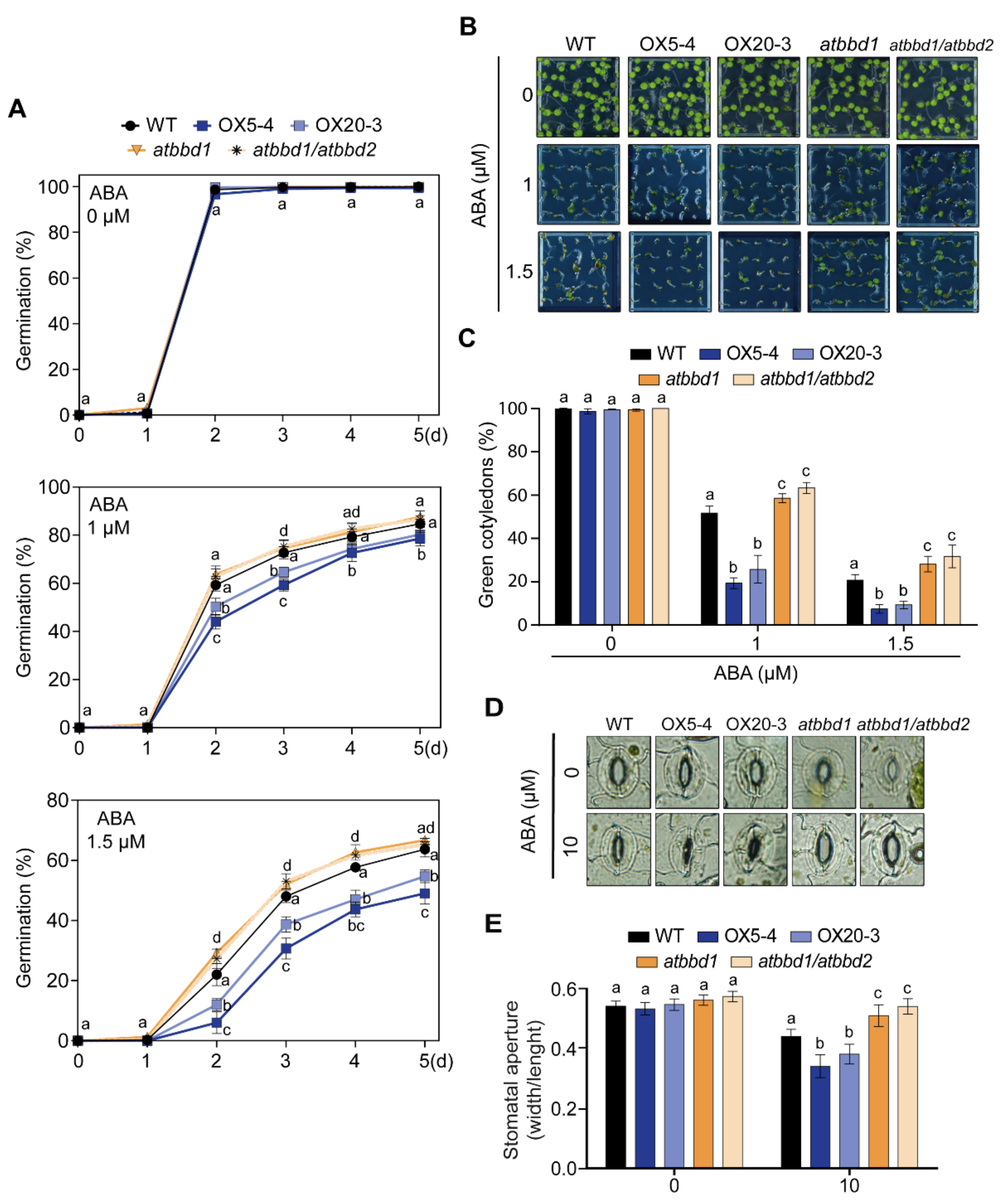
2.4. AtBBD1 Enhances the ABA-Mediated Stomatal Closure
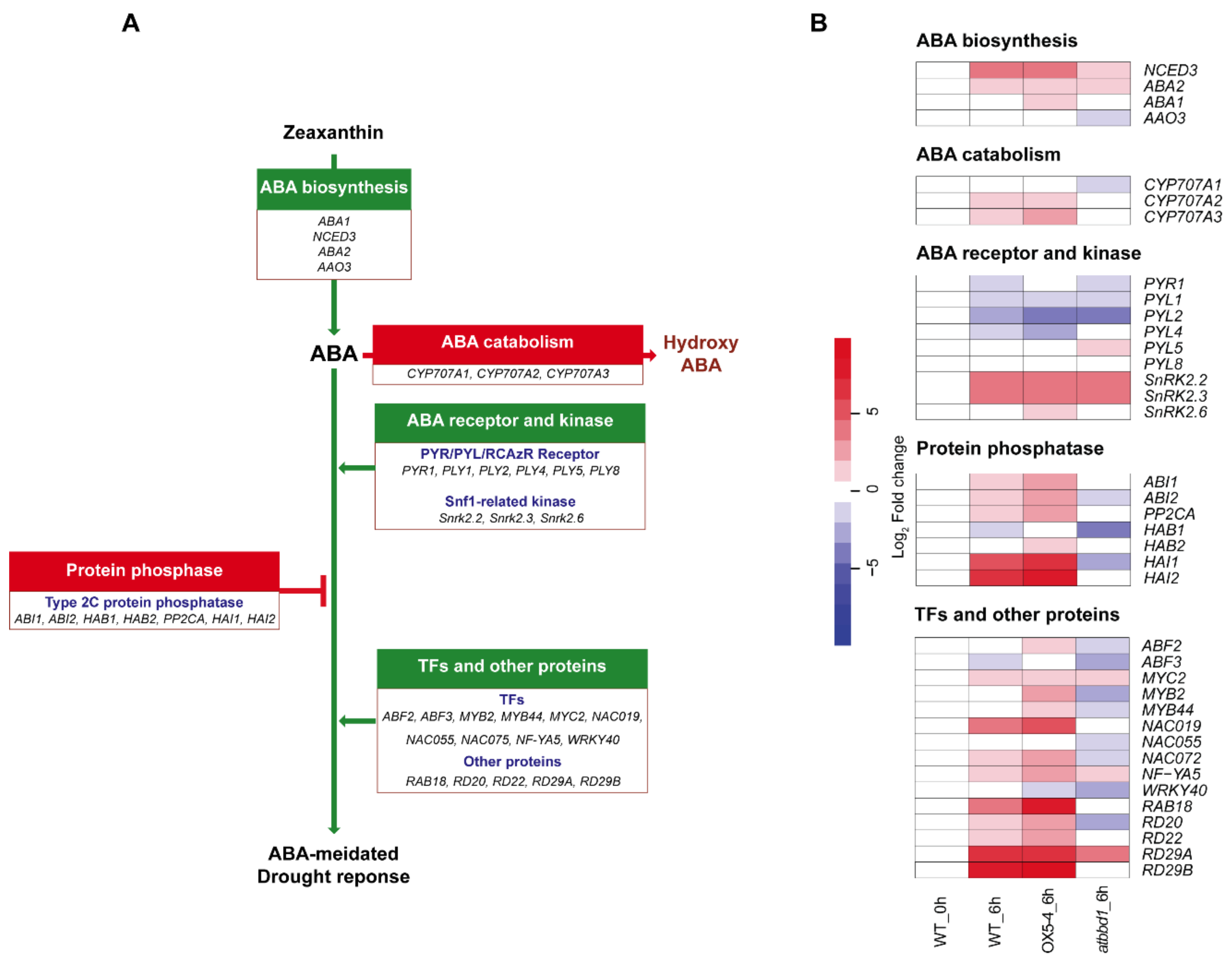
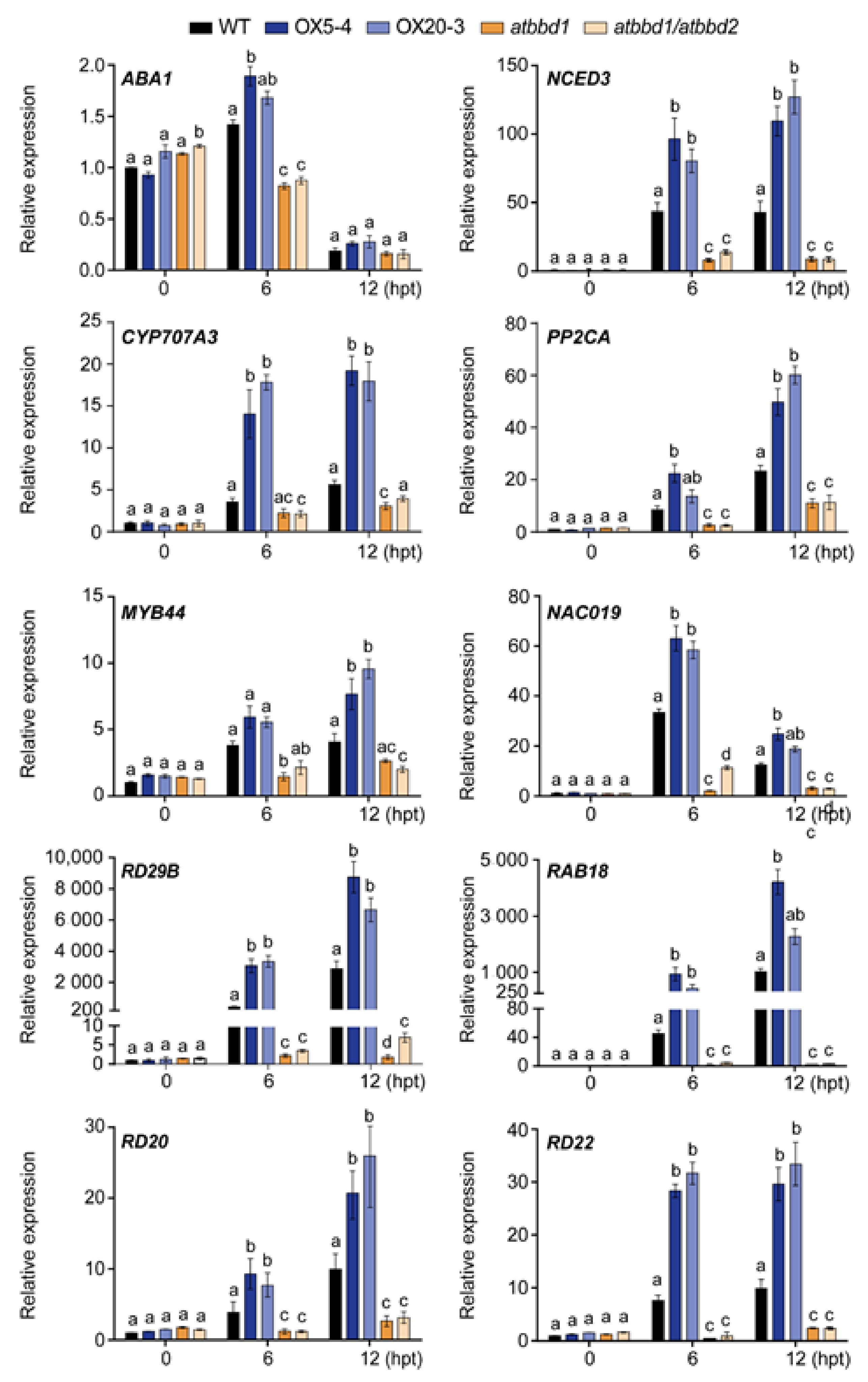
2.5. Overexpression of AtBBD1 Enhances the Expression of Key Regulatory Genes in ABA-Dependent Pathway
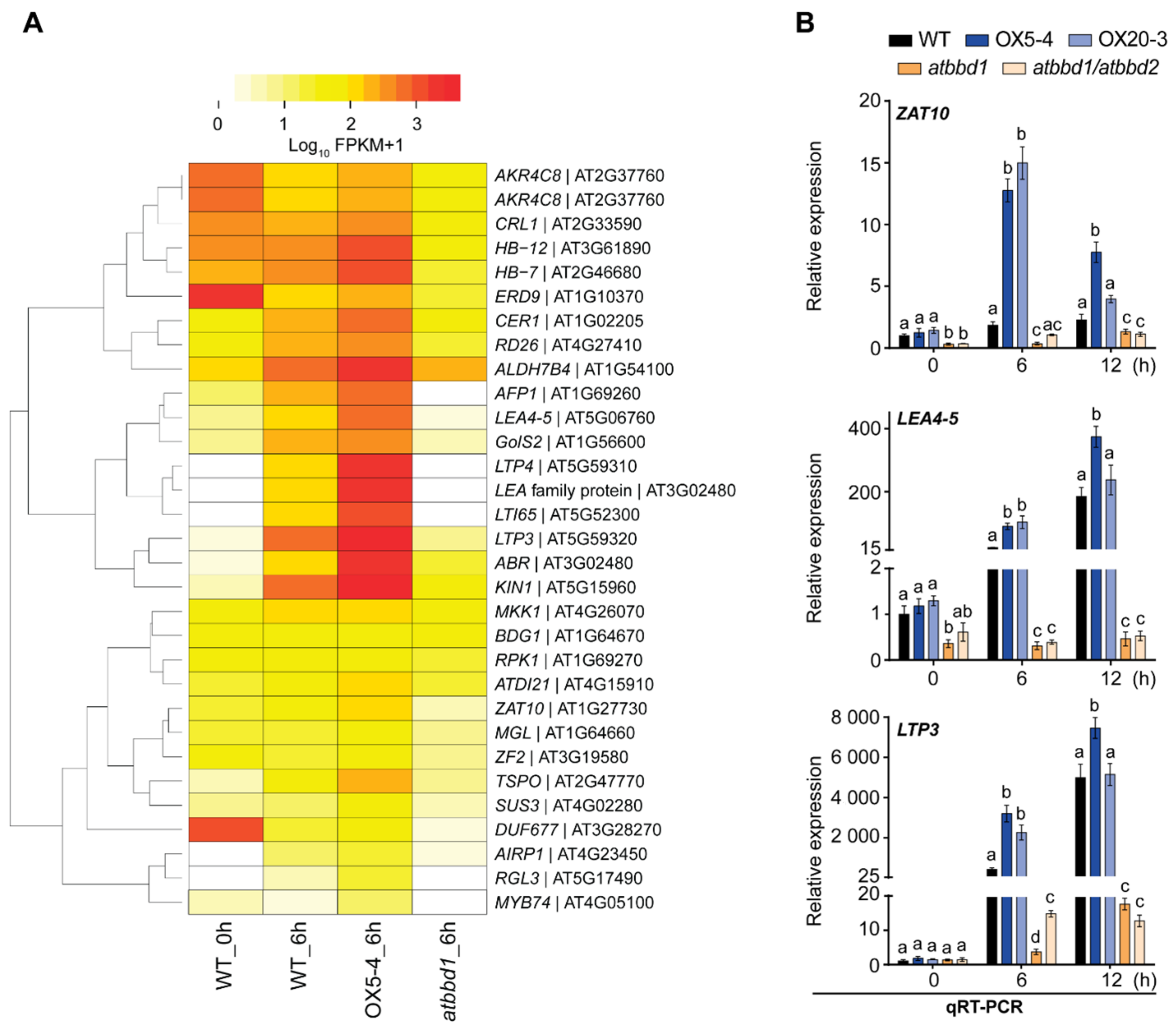
2.6. Overexpression of AtBBD1 Increases the Expression of Drought Stress-Responsive Genes
3. Materials and Methods
3.1. Plant Materials
3.2. Construction of Transgenic Plants
3.3. Subcellular Localization Analysis
3.4. GUS Histochemical Staining Analysis
3.5. RNA Isolation and qPCR Analysis
3.6. Drought Treatment and Water Loss
3.7. Quantification of Proline Contents
3.8. Seed Germination Assays
3.9. Measurement of Stomatal Aperture
3.10. RNA Sequencing
3.11. Data Access
3.12. Transcriptome Data Analysis
3.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oliver, M.J.; Farrant, J.M.; Hilhorst, H.W.M.; Mundree, S.; Williams, B.; Bewley, J.D. Desiccation Tolerance: Avoiding Cellular Damage During Drying and Rehydration. Annu. Rev. Plant Biol. 2020, 71, 435–460. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Mori, I.C.; Munemasa, S. Diverse Stomatal Signaling and the Signal Integration Mechanism. Annu. Rev. Plant Biol. 2015, 66, 369–392. [Google Scholar] [CrossRef]
- Lee, S.C.; Lim, C.W.; Lan, W.; He, K.; Luan, S. ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 2013, 6, 528–538. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Jiang, H.; Li, H.; Bu, Q.; Li, C. The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating ABA response and jasmonate response. Plant Signal. Behav. 2009, 4, 464–466. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef]
- Scholz, S.S.; Vadassery, J.; Heyer, M.; Reichelt, M.; Bender, K.W.; Snedden, W.A.; Boland, W.; Mithöfer, A. Mutation of the Arabidopsis Calmodulin-Like Protein CML37 Deregulates the Jasmonate Pathway and Enhances Susceptibility to Herbivory. Mol. Plant 2014, 7, 1712–1726. [Google Scholar] [CrossRef]
- Scholz, S.S.; Reichelt, M.; Vadassery, J.; Mithöfer, A. Calmodulin-like protein CML37 is a positive regulator of ABA during drought stress in Arabidopsis. Plant Signal. Behav. 2015, 10, e1011951. [Google Scholar] [CrossRef]
- Lambert, R.; Quiles, F.A.; Galvez-Valdivieso, G.; Piedras, P. Nucleases activities during French bean leaf aging and dark-induced senescence. J. Plant Physiol. 2017, 218, 235–242. [Google Scholar] [CrossRef]
- Lombardi, L.; Ceccarelli, N.; Picciarelli, P.; Lorenzi, R. DNA degradation during programmed cell death in Phaseolus coccineus suspensor. Plant Physiol. Biochem. 2007, 45, 221–227. [Google Scholar] [CrossRef]
- Chen, H.M.; Pang, Y.; Zeng, J.; Ding, Q.; Yin, S.Y.; Liu, C.; Lu, M.Z.; Cui, K.M.; He, X.Q. The Ca2+ -dependent DNases are involved in secondary xylem development in Eucommia ulmoides. J. Integr. Plant Biol. 2012, 54, 456–470. [Google Scholar] [CrossRef]
- Sui, W.; Guo, K.; Li, L.; Liu, S.; Takano, T.; Zhang, X. Arabidopsis Ca(2+)-dependent nuclease AtCaN2 plays a negative role in plant responses to salt stress. Plant Sci. 2019, 281, 213–222. [Google Scholar] [CrossRef] [PubMed]
- You, M.K.; Shin, H.Y.; Kim, Y.J.; Ok, S.H.; Cho, S.K.; Jeung, J.U.; Yoo, S.D.; Kim, J.K.; Shin, J.S. Novel bifunctional nucleases, OmBBD and AtBBD1, are involved in abscisic acid-mediated callose deposition in Arabidopsis. Plant Physiol. 2010, 152, 1015–1029. [Google Scholar] [CrossRef]
- Jiang, A.L.; Cheng, Y.; Li, J.; Zhang, W. A zinc-dependent nuclear endonuclease is responsible for DNA laddering during salt-induced programmed cell death in root tip cells of rice. J. Plant Physiol. 2008, 165, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Yupsanis, T.; Kefalas, P.S.; Eleftheriou, P.; Kotinis, K. RNase and DNase activities in the alfalfa and lentil grown in iso-osmotic solutions of NaCl and mannitol. J. Plant Physiol. 2001, 158, 921–927. [Google Scholar] [CrossRef]
- Li, C.; Li, C.; Wang, B.; Zhang, R.; Fu, K.; Gale, W.J.; Li, C. Programmed cell death in wheat (Triticum aestivum L.) endosperm cells is affected by drought stress. Protoplasma 2018, 255, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Huque, A.K.M.M.; So, W.M.; You, M.K.; Shin, J.S. Phylogenetic Analysis and In Vitro Bifunctional Nuclease Assay of Arabidopsis BBD1 and BBD2. Molecules 2020, 25, 2169. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Cui, M.H.; Noh, M.S.; Jung, K.W.; Shin, J.S. The FBA motif-containing protein AFBA1 acts as a novel positive regulator of ABA response in Arabidopsis. Plant Cell Physiol. 2017, 58, 574–586. [Google Scholar] [CrossRef]
- Li, C.-L.; Wang, M.; Wu, X.-M.; Chen, D.-H.; Lv, H.-J.; Shen, J.-L.; Qiao, Z.; Zhang, W. THI1, a Thiamine Thiazole Synthase, Interacts with Ca2+-Dependent Protein Kinase CPK33 and Modulates the S-Type Anion Channels and Stomatal Closure in Arabidopsis. Plant Physiol. 2016, 170, 1090–1104. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32(7), 959–970. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Yoshiba, Y.; Nanjo, T.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 1999, 261, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Satoh, R.; Kiyosue, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol. 1998, 118, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 Differentially Modulates Diverse Jasmonate-Dependent Functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Takahashi, Y. FRET kinase sensor development reveals SnRK2/OST1 activation by ABA but not by MeJA and high CO(2) during stomatal closure. eLife 2020, 9, e56351. [Google Scholar] [CrossRef]
- Marin, E.; Nussaume, L.; Quesada, A.; Gonneau, M.; Sotta, B.; Hugueney, P.; Frey, A.; Marion-Poll, A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996, 15, 2331–2342. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Osakabe, Y.; Qin, F.; Simpson, S.D.; Maruyama, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007, 49, 46–63. [Google Scholar] [CrossRef]
- Baek, W.; Lim, C.W.; Lee, S.C. A DEAD-box RNA helicase, RH8, is critical for regulation of ABA signalling and the drought stress response via inhibition of PP2CA activity. Plant Cell Environ. 2018, 41, 1593–1604. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8’-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef] [PubMed]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Mizuno, S.; Tanaka, H.; Maruyama, K.; Osakabe, K.; Todaka, D.; Fujita, Y.; Kobayashi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 2010, 285, 9190–9201. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Alshareef, S.; Butt, H.; Lozano-Juste, J.; Li, L.; Galal, A.A.; Moustafa, A.; Momin, A.A.; Tashkandi, M.; Richardson, D.N.; et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 2017, 89, 291–309. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Verdaguer, B.; de Kochko, A.; Fux, C.I.; Beachy, R.N.; Fauquet, C. Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol. Biol. 1998, 37, 1055–1067. [Google Scholar] [CrossRef]
- Xie, M.; He, Y.; Gan, S. Bidirectionalization of polar promoters in plants. Nat. Biotechnol. 2001, 19, 677–679. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.B.; Kim, Y.A.; Kim, S.J.; Lee, S.H.; Kim, D.H.; Cheong, G.W.; Hwang, I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 2001, 13, 1511–1526. [Google Scholar] [CrossRef]
- So, W.M.; Huque, A.K.M.M.; Shin, H.-Y.; Kim, S.Y.; Shin, J.S.; Cui, M.; Shin, J.S. AtMYB109 negatively regulates stomatal closure under osmotic stress in Arabidopsis thaliana. J. Plant Physiol. 2020, 255, 153292. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huque, A.K.M.M.; So, W.; Noh, M.; You, M.K.; Shin, J.S. Overexpression of AtBBD1, Arabidopsis Bifunctional Nuclease, Confers Drought Tolerance by Enhancing the Expression of Regulatory Genes in ABA-Mediated Drought Stress Signaling. Int. J. Mol. Sci. 2021, 22, 2936. https://doi.org/10.3390/ijms22062936
Huque AKMM, So W, Noh M, You MK, Shin JS. Overexpression of AtBBD1, Arabidopsis Bifunctional Nuclease, Confers Drought Tolerance by Enhancing the Expression of Regulatory Genes in ABA-Mediated Drought Stress Signaling. International Journal of Molecular Sciences. 2021; 22(6):2936. https://doi.org/10.3390/ijms22062936
Chicago/Turabian StyleHuque, A. K. M. Mahmudul, Wonmi So, Minsoo Noh, Min Kyoung You, and Jeong Sheop Shin. 2021. "Overexpression of AtBBD1, Arabidopsis Bifunctional Nuclease, Confers Drought Tolerance by Enhancing the Expression of Regulatory Genes in ABA-Mediated Drought Stress Signaling" International Journal of Molecular Sciences 22, no. 6: 2936. https://doi.org/10.3390/ijms22062936
APA StyleHuque, A. K. M. M., So, W., Noh, M., You, M. K., & Shin, J. S. (2021). Overexpression of AtBBD1, Arabidopsis Bifunctional Nuclease, Confers Drought Tolerance by Enhancing the Expression of Regulatory Genes in ABA-Mediated Drought Stress Signaling. International Journal of Molecular Sciences, 22(6), 2936. https://doi.org/10.3390/ijms22062936







