Genetic Deletion of Trace-Amine Associated Receptor 9 (TAAR9) in Rats Leads to Decreased Blood Cholesterol Levels
Abstract
:1. Introduction
2. Results
2.1. Generation of TAAR9-KO Rats
2.2. Validation of TAAR9-KO Rats
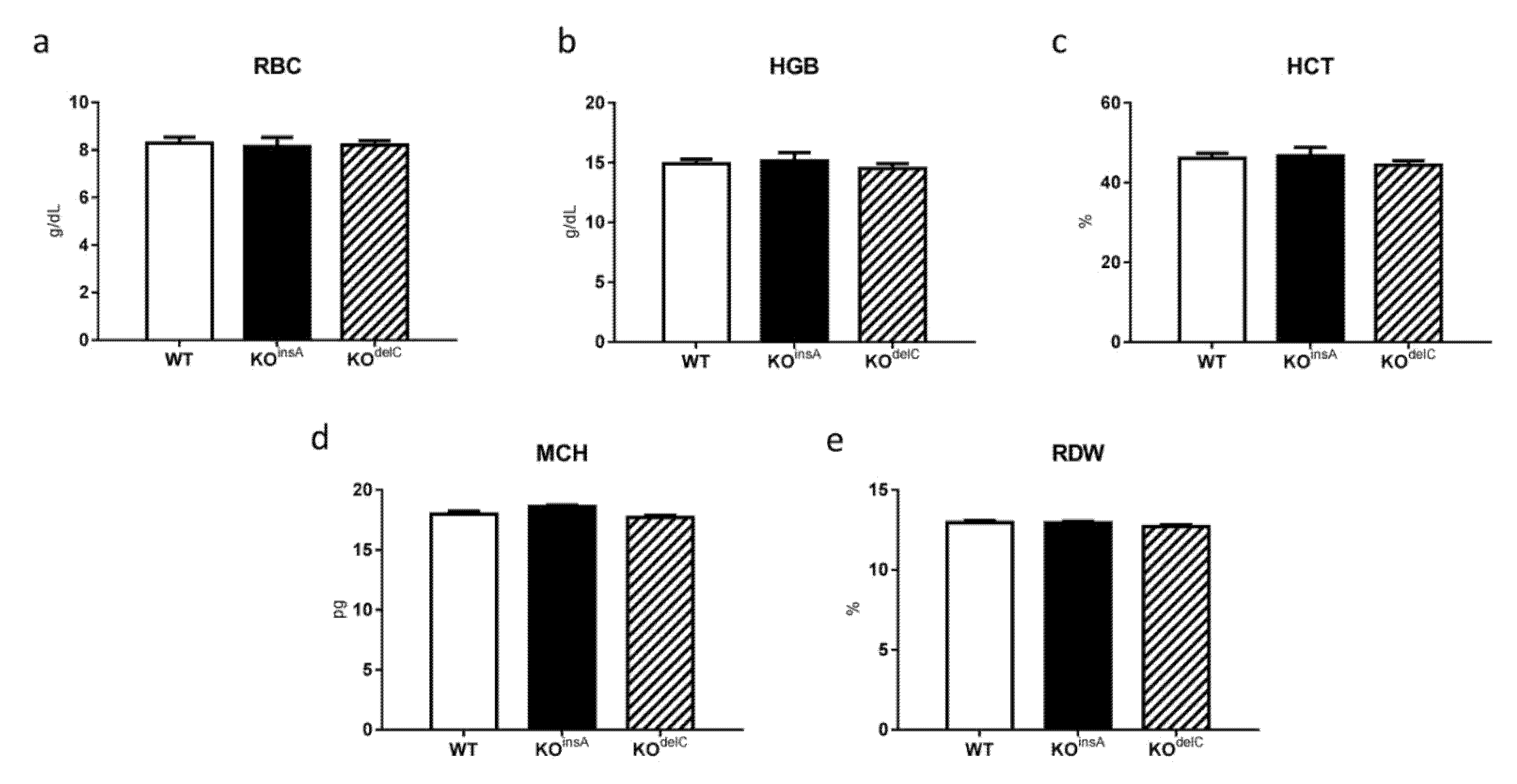
2.3. Hematological Analysis of TAAR9-KO Rats
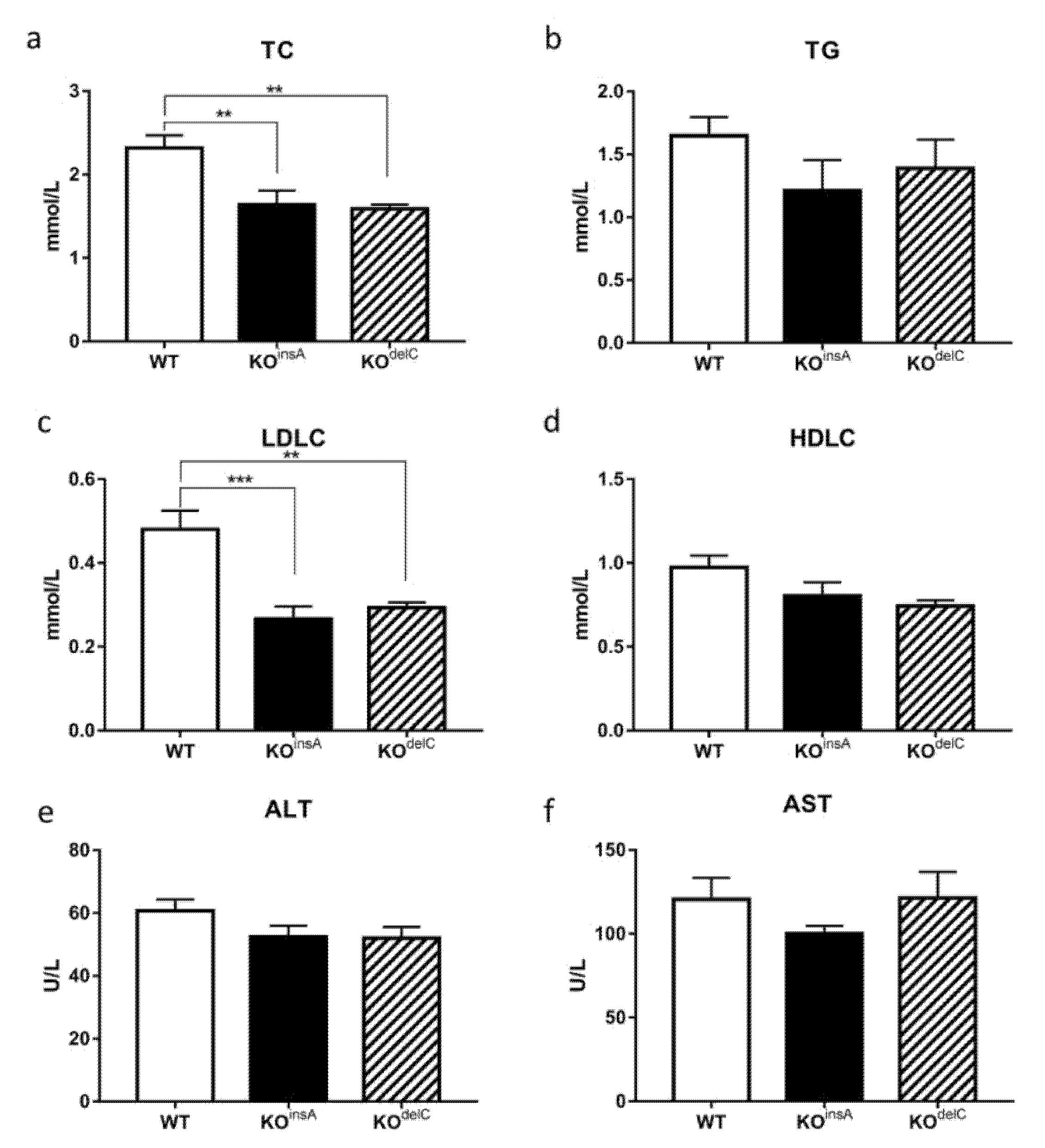
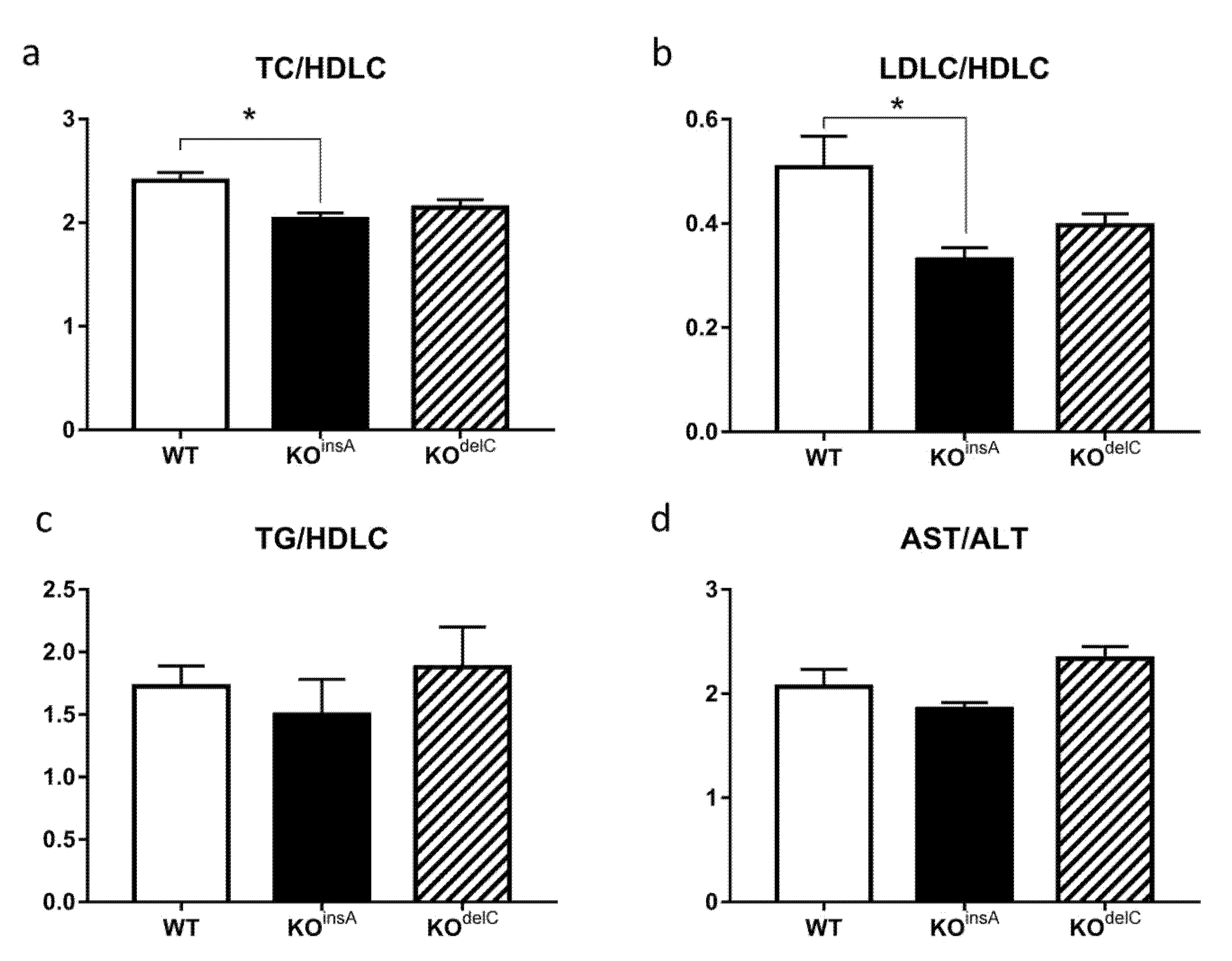
2.4. Biochemical Analysis of TAAR9-KO Rats
3. Discussion
4. Materials and Methods
4.1. Generation of TAAR9-Knockout Rats
4.2. Genotyping
4.3. Gene Expression Analysis by qPCR
4.4. Animals
4.5. Sample Collection and Storage
4.6. Measurement of Hematological Parameters
4.7. Osmotic Fragility Test (OFT)
4.8. Measurement of Biochemical Parameters
4.9. Interassay Repeatability
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borowsky, B.; Adham, N.; Jones, K.A.; Raddatz, R.; Artymyshyn, R.; Ogozalek, K.L.; Durkin, M.M.; Lakhlani, P.P.; Bonini, J.A.; Pathirana, S.; et al. Trace amines: Identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 8966–8971. [Google Scholar] [CrossRef] [Green Version]
- Bunzow, J.R.; Sonders, M.S.; Arttamangkul, S.; Harrison, L.M.; Zhang, G.; Quigley, D.I.; Darland, T.; Suchland, K.L.; Pasumamula, S.; Kennedy, J.L.; et al. Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol. Pharmacol. 2001, 60, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, R.R.; Hoener, M.C.; Berry, M.D. Trace amines and their receptors. Pharmacol. Rev. 2018, 70, 549–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, M.D.; Gainetdinov, R.R.; Hoener, M.C.; Shahid, M. Pharmacology of human trace amine-associated receptors: Therapeutic opportunities and challenges. Pharmacol. Ther. 2017, 180, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Koblan, K.S.; Kent, J.; Hopkins, S.C.; Krystal, J.H.; Cheng, H.; Goldman, R.; Loebel, A. A Non-D2-Receptor-Binding Drug for the Treatment of Schizophrenia. N. Engl. J. Med. 2020, 382, 1497–1506. [Google Scholar] [CrossRef]
- Pacifico, R.; Dewan, A.; Cawley, D.; Guo, C.; Bozza, T. An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Rep. 2012, 2, 76–88. [Google Scholar] [CrossRef] [Green Version]
- Liberles, S.D.; Buck, L.B. A second class of chemosensory receptors in the olfactory epithelium. Nature 2006, 442, 645–650. [Google Scholar] [CrossRef]
- Espinoza, S.; Sukhanov, I.; Efimova, E.V.; Kozlova, A.; Antonova, K.A.; Illiano, P.; Leo, D.; Merkulyeva, N.; Kalinina, D.; Musienko, P.; et al. Trace Amine-Associated Receptor 5 Provides Olfactory Input Into Limbic Brain Areas and Modulates Emotional Behaviors and Serotonin Transmission. Front. Mol. Neurosci. 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Efimova, E.V.; Kozlova, A.A.; Razenkova, V.; Katolikova, N.V.; Antonova, K.A.; Sotnikova, T.D.; Merkulyeva, N.S.; Veshchitskii, A.S.; Kalinina, D.S.; Korzhevskii, D.E.; et al. Increased dopamine transmission and adult neurogenesis in trace amine-associated receptor 5 (TAAR5) knockout mice. Neuropharmacology 2021, 182, 108373. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, D.M.; Lemon, J.K.; Fluegge, D.; Pashkovski, S.L.; Korzan, W.J.; Datta, S.R.; Spehr, M.; Fendt, M.; Liberles, S.D. Detection and avoidance of a carnivore odor by prey. Proc. Natl. Acad. Sci. USA 2011, 108, 11235–11240. [Google Scholar] [CrossRef] [Green Version]
- Ferrero, D.M.; Wacker, D.; Roque, M.A.; Baldwin, M.W.; Stevens, R.C.; Liberles, S.D. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem. Biol. 2012, 7, 1184–1189. [Google Scholar] [CrossRef] [Green Version]
- Ohta, H.; Takebe, Y.; Murakami, Y.; Takahama, Y.; Morimura, S. Tyramine and β-phenylethylamine, from fermented food products, as agonists for the human trace amine-associated receptor 1 (hTAAR1) in the stomach. Biosci. Biotechnol. Biochem. 2017, 81, 1002–1006. [Google Scholar] [CrossRef] [Green Version]
- Vanti, W.B.; Muglia, P.; Nguyen, T.; Cheng, R.; Kennedy, J.L.; George, S.R.; O’Dowd, B.F. Discovery of a null mutation in a human trace amine receptor gene. Genomics 2003, 82, 531–536. [Google Scholar] [CrossRef]
- Ito, J.; Ito, M.; Nambu, H.; Fujikawa, T.; Tanaka, K.; Iwaasa, H.; Tokita, S. Anatomical and histological profiling of orphan G-protein-coupled receptor expression in gastrointestinal tract of C57BL/6J mice. Cell Tissue Res. 2009, 338, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Gozal, E.A.; O’Neill, B.E.; Sawchuk, M.A.; Zhu, H.; Halder, M.; Chou, C.-C.; Hochman, S. Anatomical and functional evidence for trace amines as unique modulators of locomotor function in the mammalian spinal cord. Front. Neural Circuits 2014, 8, 134. [Google Scholar] [CrossRef] [Green Version]
- Regard, J.B.; Sato, I.T.; Coughlin, S.R. Anatomical Profiling of G Protein-Coupled Receptor Expression. Cell 2008, 135, 561–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babusyte, A.; Kotthoff, M.; Fiedler, J.; Krautwurst, D. Biogenic amines activate blood leukocytes via trace amine-associated receptors TAAR1 and TAAR2. J. Leukoc. Biol. 2013, 93, 387–394. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, G.; Terrazzino, S.; Fortin, D.; Farruggio, A.; Rinaldi, L.; Leon, A. HPLC electrochemical detection of trace amines in human plasma and platelets and expression of mRNA transcripts of trace amine receptors in circulating leukocytes. Neurosci. Lett. 2003, 346, 89–92. [Google Scholar] [CrossRef]
- Harmeier, A.; Meyer, C.A.; Staempfli, A.; Casagrande, F.; Petrinovic, M.M.; Zhang, Y.P.; Künnecke, B.; Iglesias, A.; Höner, O.P.; Hoener, M.C. How female mice attract males: A urinary volatile amine activates a trace amine-associated receptor that induces male sexual interest. Front. Pharmacol. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Zhukov, I.S.; Kubarskaya, L.G.; Tissen, I.Y.; Kozlova, A.A.; Dagayev, S.G.; Kashuro, V.A.; Vlasova, O.L.; Sinitca, E.L.; Karpova, I.V.; Gainetdinov, R.R. Minimal Age-Related Alterations in Behavioral and Hematological Parameters in Trace Amine‑Associated Receptor 1 (TAAR1) Knockout Mice. Cell. Mol. Neurobiol. 2020, 1, 273–282. [Google Scholar] [CrossRef]
- Leonova, E.I.; Gainetdinov, R.R. CRISPR/Cas9 technology in translational biomedicine. Cell. Physiol. Biochem. 2020, 54, 354–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popp, M.W.; Maquat, L.E. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell 2016, 165, 1319–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vettore, L.; Zanella, A.; Molaro, L.; De Matteis, C.; Pavesi, M.; Mariani, M. A New Test for the Laboratory Diagnosis of Spherocytosis. Acta Haematol. 1984, 72, 258–263. [Google Scholar] [CrossRef]
- Huisjes, R.; Bogdanova, A.; van Solinge, W.W.; Schiffelers, R.M.; Kaestner, L.; van Wijk, R. Squeezing for life—Properties of red blood cell deformability. Front. Physiol. 2018, 9, 656. [Google Scholar] [CrossRef] [PubMed]
- Millán, J.; Pintó, X.; Muñoz, A.; Zúñiga, M.; Rubiés-Prat, J.; Pallardo, L.F.; Masana, L.; Mangas, A.; González-Santos, A.H.-M.P.; Ascaso, J.F.; et al. Lipoprotein ratios: Physiological significance and clinical usefulness in cardiovascular prevention. Vasc. Health Risk Manag. 2009, 5, 757–765. [Google Scholar] [PubMed]
- Ozer, J.; Ratner, M.; Shaw, M.; Bailey, W.; Schomaker, S. The current state of serum biomarkers of hepatotoxicity. Toxicology 2008, 245, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Giannini, E.G.; Testa, R.; Savarino, V. Liver enzyme alteration: A guide for clinicians. CMAJ 2005, 172, 367–379. [Google Scholar] [CrossRef] [Green Version]
- Gelissen, I.C.; Brown, A.J. An overview of cholesterol homeostasis. Methods Mol. Biol. 2017, 1583, 1–6. [Google Scholar] [CrossRef]
- Abrams, J.J.; Grundy, S.M.; Ginsberg, H. Metabolism of plasma triglycerides in hypothyroidism and hyperthyroidism in man. J. Lipid Res. 1981, 22, 307–322. [Google Scholar] [CrossRef]
- Faselis, C.; Imprialos, K.; Grassos, H.; Pittaras, A.; Kallistratos, M.; Manolis, A. Is very low LDL-C harmful? Curr. Pharm. Des. 2018, 24, 3658–3664. [Google Scholar] [CrossRef] [PubMed]
- Kapourchali, F.R.; Surendiran, G.; Goulet, A.; Moghadasian, M.H. The Role of Dietary Cholesterol in Lipoprotein Metabolism and Related Metabolic Abnormalities: A Mini-review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2408–2415. [Google Scholar] [CrossRef] [PubMed]
- Bernecker, C.; Köfeler, H.; Pabst, G.; Trötzmüller, M.; Kolb, D.; Strohmayer, K.; Trajanoski, S.; Holzapfel, G.A.; Schlenke, P.; Dorn, I. Cholesterol Deficiency Causes Impaired Osmotic Stability of Cultured Red Blood Cells. Front. Physiol. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Da Luz, P.L.; Favarato, D.; Faria-Neto, J.R.; Lemos, P.; Chagas, A.C.P. High ratio of triglycerides to HDL-cholesterol predicts extensive coronary disease. Clinics 2008, 63, 427–432. [Google Scholar] [CrossRef] [Green Version]
- McGill, M.R. The past and present of serum aminotransferases and the future of liver injury biomarkers. EXCLI J. 2016, 15, 817–828. [Google Scholar] [CrossRef]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of trimethylamine n-oxide (TMAO) in disease: Potential biomarker or new therapeutic target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef] [Green Version]
- Wallrabenstein, I.; Kuklan, J.; Weber, L.; Zborala, S.; Werner, M.; Altmüller, J.; Becker, C.; Schmidt, A.; Hatt, H.; Hummel, T.; et al. Human Trace Amine-Associated Receptor TAAR5 Can Be Activated by Trimethylamine. PLoS ONE 2013, 8, e54950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fennema, D.; Phillips, I.R.; Shephard, E.A. Trimethylamine and trimethylamine N-oxide, a Flavin-Containing Monooxygenase 3 (FMO3)-mediated host-microbiome metabolic axis implicated in health and disease. Drug Metab. Dispos. 2016, 44, 1839–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016, 8, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Zucchi, R.; Accorroni, A.; Chiellini, G. Update on 3-iodothyronamine and its neurological and metabolic actions. Front. Physiol. 2014, 5, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxfield, F.R.; Tabas, I. Role of cholesterol and lipid organization in disease. Nature 2005, 438, 612–621. [Google Scholar] [CrossRef]
- Herrington, W.; Lacey, B.; Sherliker, P.; Armitage, J.; Lewington, S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ. Res. 2016, 118, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, E.; Krivokharchenko, A.; Ganten, D.; Bader, M. Efficiency of transgenic rat production is independent of transgene-construct and overnight embryo culture. Theriogenology 2004, 61, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Lasnier, E.; Mario, N.; Boque, M.C.; You, S.N.; Vaubourdolle, M. Evaluation of the clinical chemistry analyser olympus AU400. Clin. Chem. Lab. Med. 2000, 38, 1043–1049. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murtazina, R.Z.; Zhukov, I.S.; Korenkova, O.M.; Popova, E.A.; Kuvarzin, S.R.; Efimova, E.V.; Kubarskaya, L.G.; Batotsyrenova, E.G.; Zolotoverkhaya, E.A.; Vaganova, A.N.; et al. Genetic Deletion of Trace-Amine Associated Receptor 9 (TAAR9) in Rats Leads to Decreased Blood Cholesterol Levels. Int. J. Mol. Sci. 2021, 22, 2942. https://doi.org/10.3390/ijms22062942
Murtazina RZ, Zhukov IS, Korenkova OM, Popova EA, Kuvarzin SR, Efimova EV, Kubarskaya LG, Batotsyrenova EG, Zolotoverkhaya EA, Vaganova AN, et al. Genetic Deletion of Trace-Amine Associated Receptor 9 (TAAR9) in Rats Leads to Decreased Blood Cholesterol Levels. International Journal of Molecular Sciences. 2021; 22(6):2942. https://doi.org/10.3390/ijms22062942
Chicago/Turabian StyleMurtazina, Ramilya Z., Ilya S. Zhukov, Olga M. Korenkova, Elena A. Popova, Savelii R. Kuvarzin, Evgeniya V. Efimova, Larisa G. Kubarskaya, Ekaterina G. Batotsyrenova, Ekaterina A. Zolotoverkhaya, Anastasia N. Vaganova, and et al. 2021. "Genetic Deletion of Trace-Amine Associated Receptor 9 (TAAR9) in Rats Leads to Decreased Blood Cholesterol Levels" International Journal of Molecular Sciences 22, no. 6: 2942. https://doi.org/10.3390/ijms22062942







