Mitochondrial Fuel Dependence on Glutamine Drives Chemo-Resistance in the Cancer Stem Cells of Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
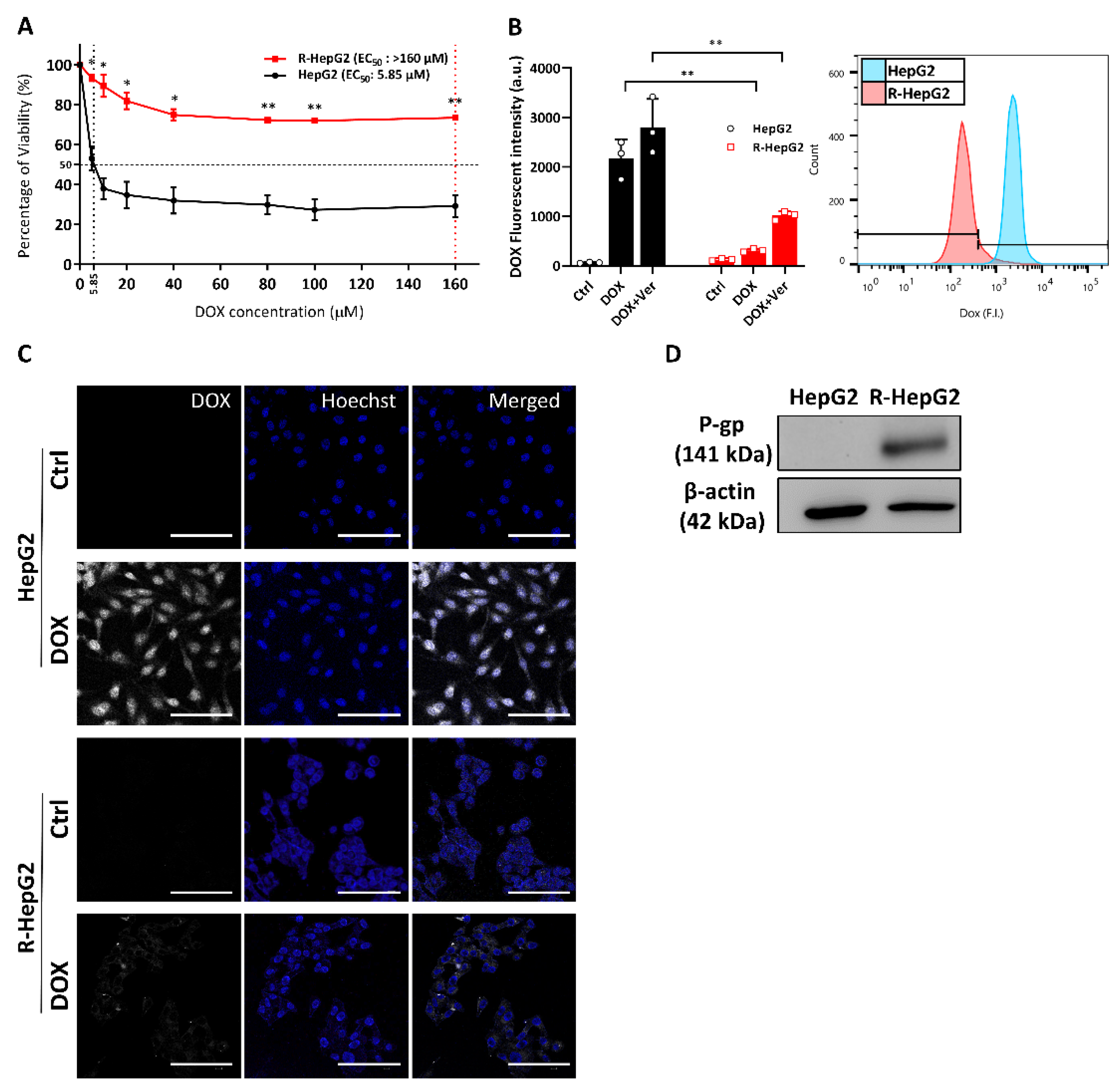
2.1. Enhanced Chemo-Resistance Is Due to the Drug Efflux by P-Glycoprotein in the R-HepG2
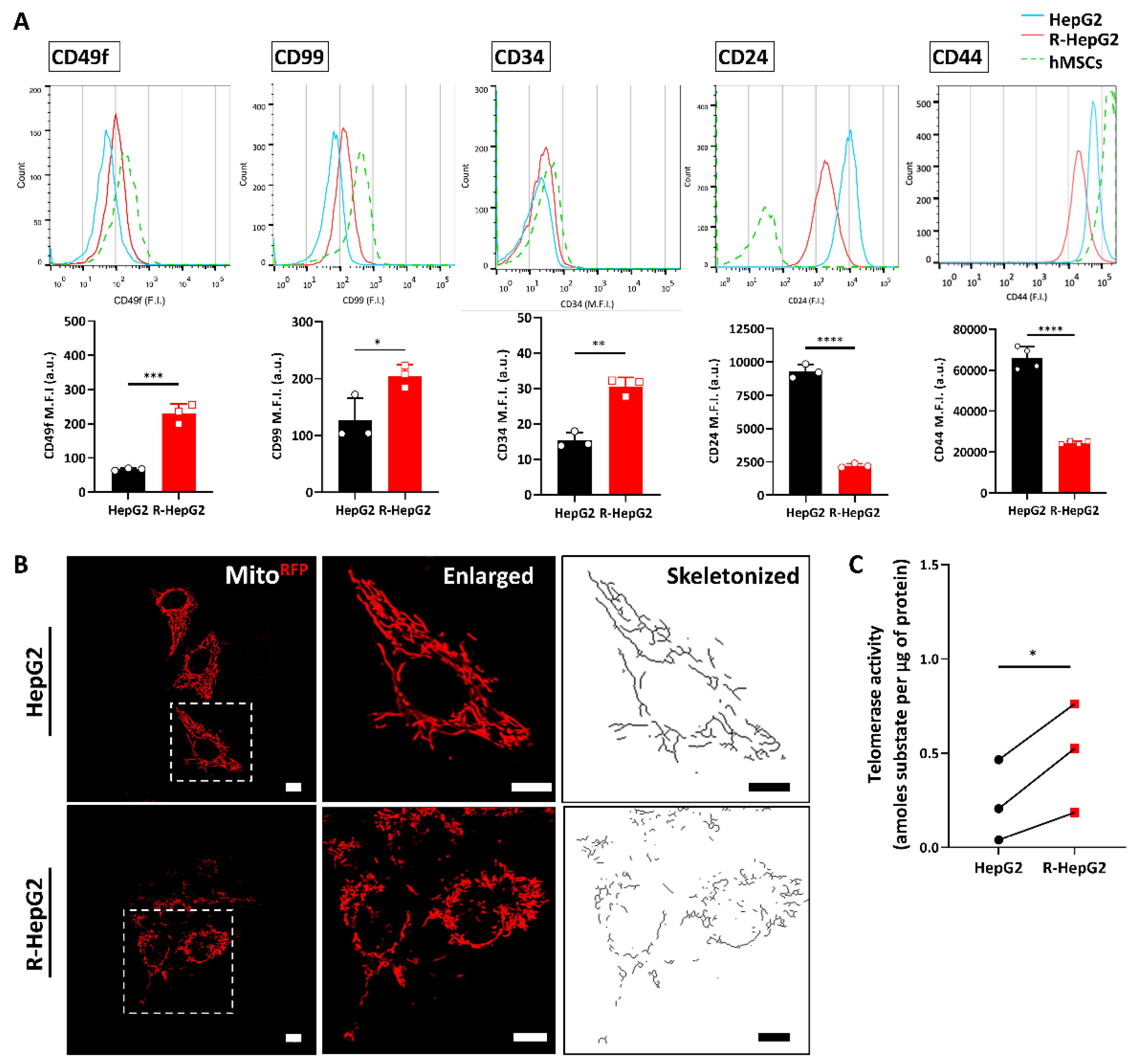
2.2. Chemo-Resistant R-HepG2 Is Enriched with CSC Markers
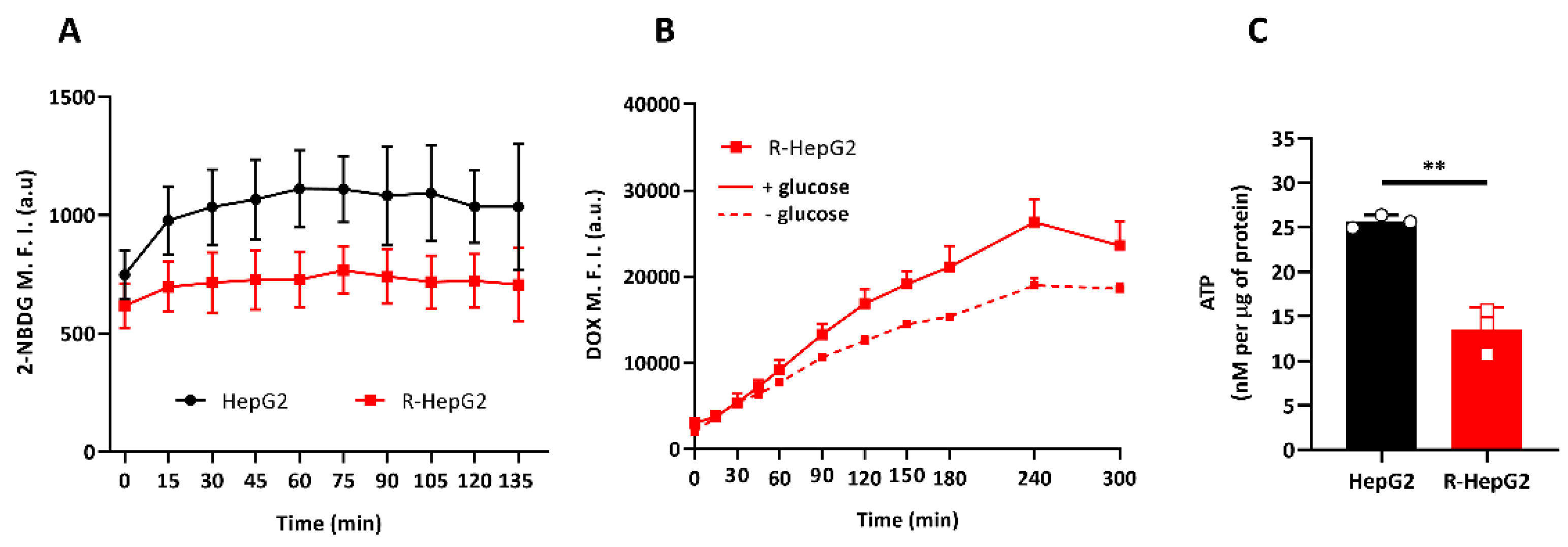
2.3. R-HepG2 Has Low Glucose Dependence and Low Endogenous ATP Content
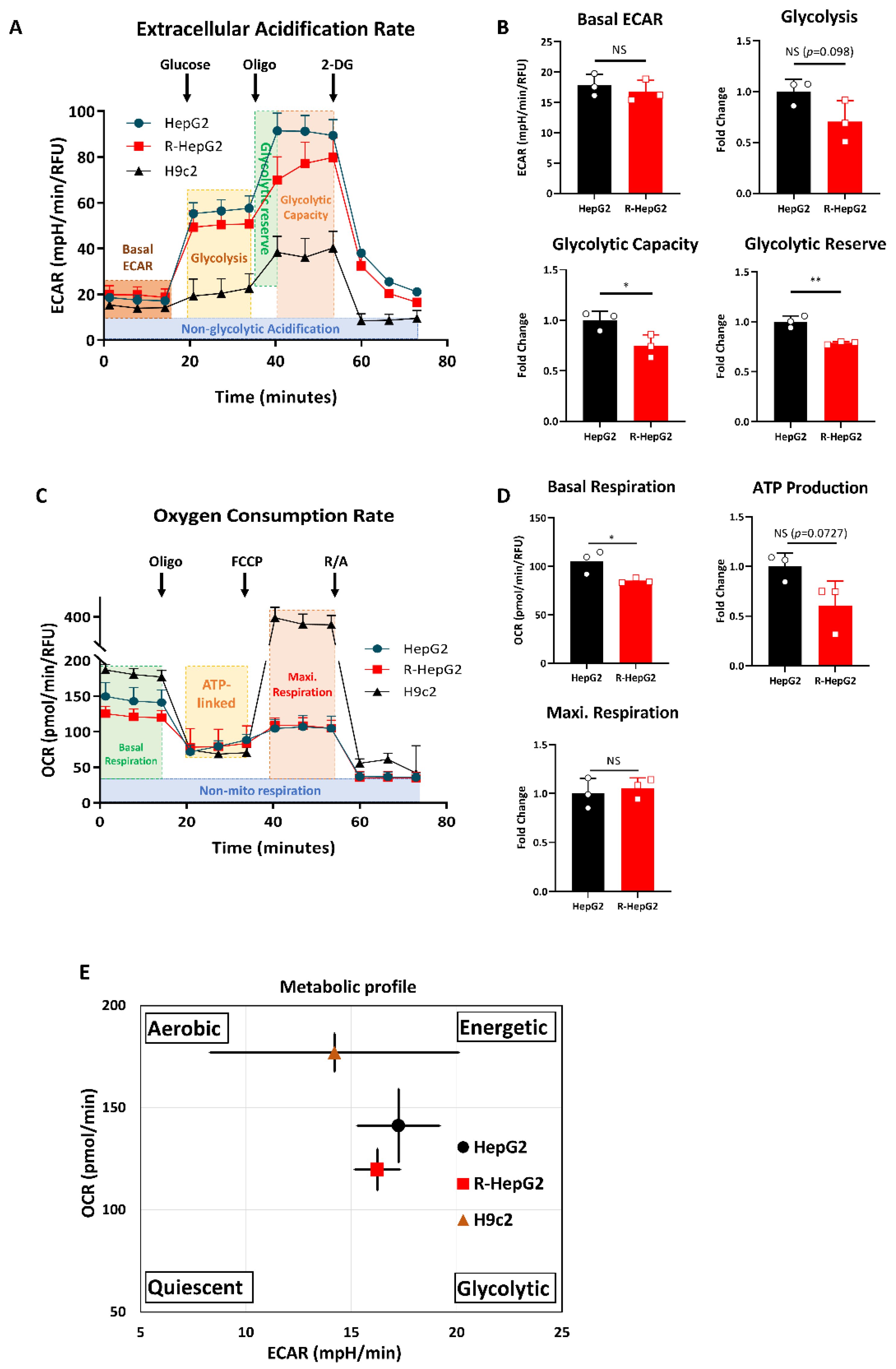
2.4. R-HepG2 Displays a Distinctive Metabolic Programme to Its Parental HepG2
2.5. Altered Metabolic Phenotype Drives Chemo-Resistance by Regulating Drug Efflux in the R-HepG2
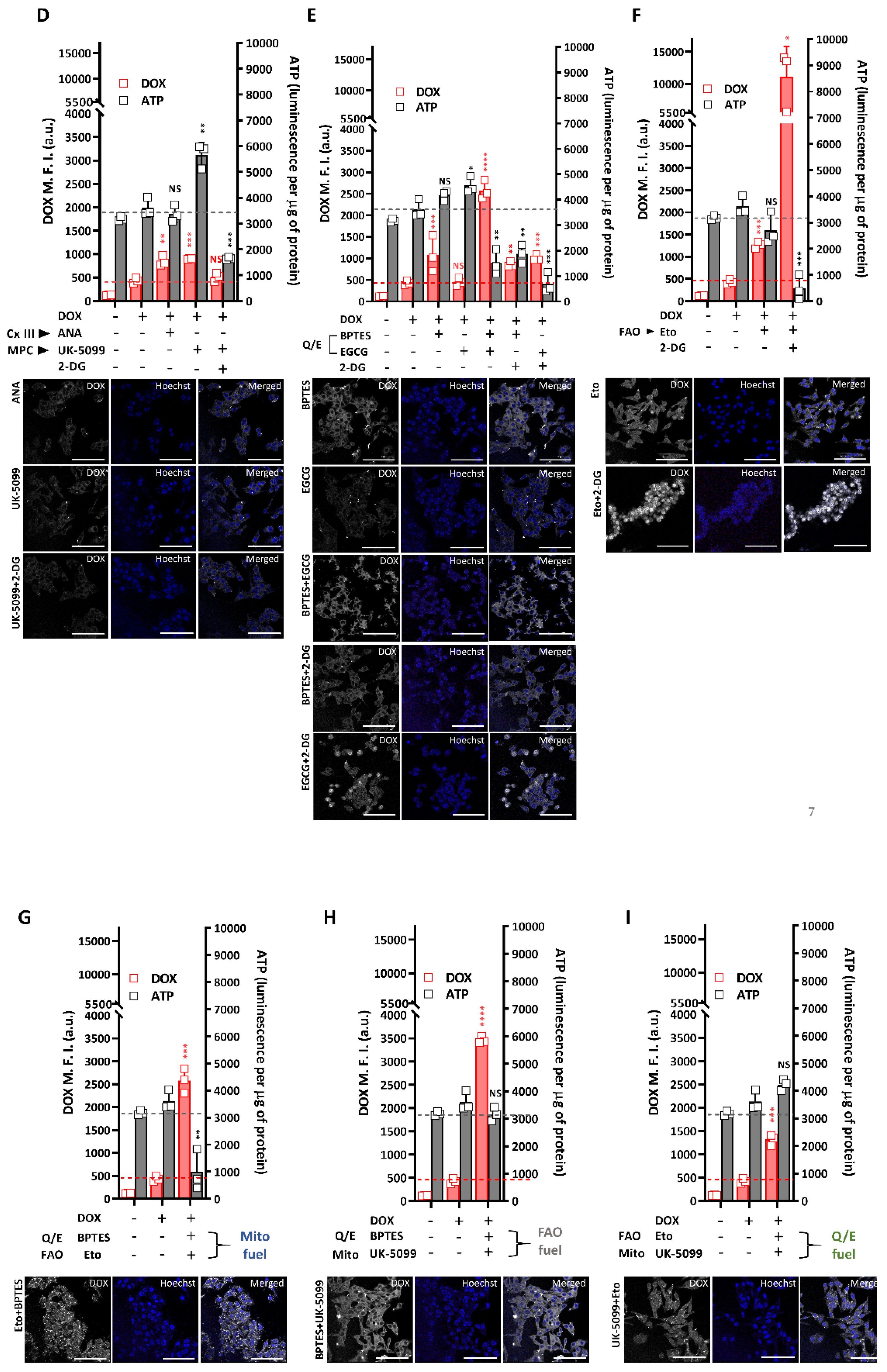
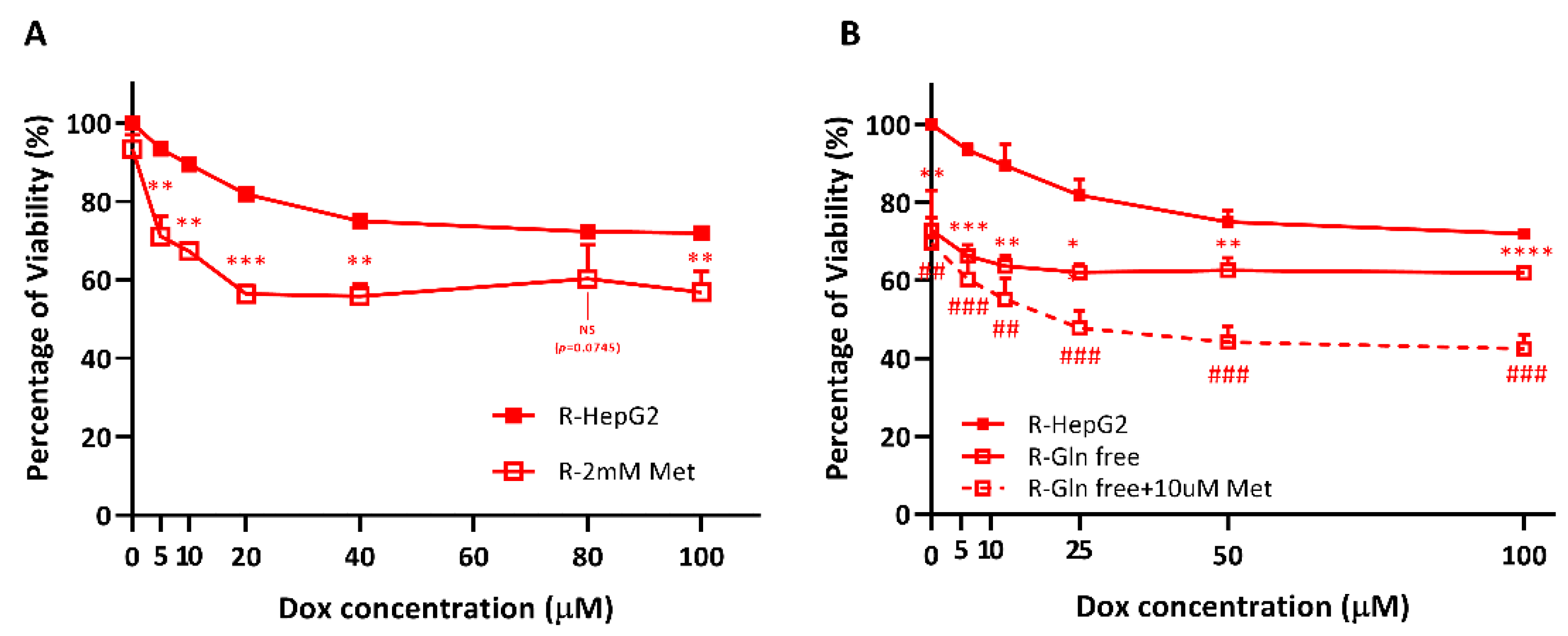
2.6. R-HepG2 Is Senstized to DOX Treatment When the Mitochondrial OXPHOS Is Inhibited and Deprived of Glutamine Fuel
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture and Transfection
4.3. Cell Viability Assay
4.4. Drug Efflux, CSC Surface Marker Immunocytochemistry and Glucose Uptake by Flow Cytometry
4.5. Telomerase Activity Assay
4.6. Metabolic Profiling by Glycolysis-Stress and Mito-Stress Assays
4.7. Endogenous ATP Measurement
4.8. Mitochondrial Fuel Dependence Assay
4.9. Mitochondrial Morphology
4.10. SDS-PAGE and Western Blotting
4.11. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| αKG | α-ketoglutarate |
| 2-DG | 2-deoxy-D-glucose |
| 2-NBDG | 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose |
| ATP | adenosine triphosphate |
| BPTES | bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide |
| DOX | doxorubicin |
| ECAR | Extracellular acidification rate |
| EGCG | Epigallocatechin gallate |
| Eto | Etomoxir |
| FCCP | carbonyl cyanide p-trifluoro-methoxy-phenyl-hydrazone |
| mSLP | mitochondrial substrate level phosphorylation |
| OCR | Oxygen consumption rate |
| P-gp | P-glycoprotein |
| UK-5099 | 2-Cyano-3-(1-phenyl-1H-indol-3-yl)-2-propenoic acid |
| Ver | Verapamil |
| OXPHOS | Oxidative phosphorylation |
References
- Alfarouk, K.O.; Stock, C.M.; Taylor, S.; Walsh, M.; Muddathir, A.K.; Verduzco, D.; Bashir, A.H.H.; Mohammed, O.Y.; Elhassan, G.O.; Harguindey, S.; et al. Resistance to cancer chemotherapy: Failure in drug response from ADME to P-gp. Cancer Cell Int. 2015, 15, 71–83. [Google Scholar] [CrossRef]
- Abdullah, L.N.; Chow, E.K. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Cojoc, M.; Mäbert, K.; Muders, M.H.; Dubrovska, A. A role for cancer stem cells in therapy resistance: Cellular and molecular mechanisms. Semin. Cancer Biol. 2015, 31, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Koren, E.; Fuchs, Y. The bad seed: Cancer stem cells in tumor development and resistance. Drug Resist. Updat. 2016, 28, 1–12. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Sotgia, F.; Lisanti, M.P. Cancer stem cells (CSCs): Metabolic strategies for their identification and eradication. Biochem. J. 2018, 475, 1611–1634. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef]
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312. [Google Scholar] [CrossRef]
- Sancho, P.; Burgos-Ramos, E.; Tavera, A.; Bou Kheir, T.; Jagust, P.; Schoenhals, M.; Barneda, D.; Sellers, K.; Campos-Olivas, R.; Graña, O.; et al. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015, 22, 590–605. [Google Scholar] [CrossRef]
- Shen, Y.A.; Wang, C.Y.; Hsieh, Y.T.; Chen, Y.J.; Wei, Y.H. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle 2015, 14, 86–98. [Google Scholar] [CrossRef]
- Peiris-Pagès, M.; Martinez-Outschoorn, U.E.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer stem cell metabolism. Breast Cancer Res. 2016, 18, 55–64. [Google Scholar] [CrossRef]
- Peixoto, J.; Lima, J. Metabolic traits of cancer stem cells. Dis. Model. Mech. 2018, 11, 1–13. [Google Scholar] [CrossRef]
- Kim, E.J.; Jin, X.; Kim, O.R.; Ham, S.W.; Park, S.H.; Kim, H. Glioma stem cells and their non-stem differentiated glioma cells exhibit differences in mitochondrial structure and function. Oncol. Rep. 2018, 39, 411–416. [Google Scholar] [CrossRef]
- Chen, H.; Chan, D.C. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017, 26, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Srinivasan, S.; Ruthel, G.; Kashina, A.K.; Carstens, R.P.; Mendoza, A.; Khanna, C.; van Winkle, T.; Avadhani, N.G. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene 2014, 33, 5238–5250. [Google Scholar] [CrossRef]
- Lee, K.M.; Giltnane, J.M.; Balko, J.M.; Schwarz, L.J.; Guerrero-Zotano, A.L.; Hutchinson, K.E.; Nixon, M.J.; Estrada, M.V.; Sánchez, V.; Sanders, M.E.; et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017, 26, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Oizel, K.; Chauvin, C.; Oliver, L.; Gratas, C.; Geraldo, F.; Jarry, U.; Scotet, E.; Rabe, M.; Alves-Guerra, M.C.; Teusan, R.; et al. Efficient mitochondrial glutamine targeting prevails over glioblastoma metabolic plasticity. Clin. Cancer Res. 2017, 23, 6292–6304. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.Y.; Ong, R.C.; Suen, Y.K.; Ooi, V.; Wong, H.N.; Mak, T.C.; Fung, K.P.; Yu, B.; Kong, S.K. Polyphyllin D is a potent apoptosis inducer in drug-resistant HepG2 cells. Cancer Lett. 2005, 217, 203–211. [Google Scholar] [CrossRef]
- Ju, Z.; Rudolph, K.L. Telomeres and telomerase in cancer stem cells. Eur. J. Cancer 2006, 42, 1197–1203. [Google Scholar] [CrossRef]
- Bosc, C.; Selak, M.A.; Sarry, J.E. Resistance is futile: Targeting mitochondrial energetics and metabolism to overcome drug resistance in cancer treatment. Cell Metab. 2017, 26, 705–707. [Google Scholar] [CrossRef]
- Van der Windt, G.J.W.; Chang, C.H.; Pearce, E.L. Measuring bioenergetics in T cells using a seahorse extracellular flux analyzer. Curr. Protoc. Immunol. 2016, 113, 3–16. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 7–14. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Arismendi-Morillo, G.; Mukherjee, P.; Chinopoulos, C. On the origin of ATP synthesis in cancer. iScience 2020, 23, 101761. [Google Scholar] [CrossRef] [PubMed]
- Herzig, S.; Raemy, E.; Montessuit, S.; Veuthey, J.L.; Zamboni, N.; Westermann, B.; Kunji, E.R.; Martinou, J.C. Identification and functional expression of the mitochondrial pyruvate carrier. Science 2012, 337, 93–96. [Google Scholar] [CrossRef]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef]
- Andrzejewski, S.; Gravel, S.P.; Pollak, M.; St-Pierre, J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2014, 28, 2–12. [Google Scholar] [CrossRef]
- Vial, G.; Detaille, D.; Guigas, B. Role of mitochondria in the mechanism(s) of action of metformin. Front. Endocrinol. Lausanne 2019, 10, 294–301. [Google Scholar] [CrossRef]
- Lathia, J.D.; Gallagher, J.; Heddleston, J.M.; Wang, J.; Eyler, C.E.; Macswords, J.; Wu, Q.; Vasanji, A.; McLendon, R.E.; Hjelmeland, A.B.; et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell 2010, 6, 421–432. [Google Scholar] [CrossRef]
- Chung, S.S.; Eng, W.S.; Hu, W.; Khalaj, M.; Garrett-Bakelman, F.E.; Tavakkoli, M.; Levine, R.L.; Carroll, M.; Klimek, V.M.; Melnick, A.M.; et al. CD99 is a therapeutic target on disease stem cells in myeloid malignancies. Sci. Transl. Med. 2017, 9, eaaj2025. [Google Scholar] [CrossRef]
- Saito, Y.; Kitamura, H.; Hijikata, A.; Tomizawa-Murasawa, M.; Tanaka, S.; Takagi, S.; Uchida, N.; Suzuki, N.; Sone, A.; Najima, Y.; et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci. Transl. Med. 2010, 2, 17ra9. [Google Scholar] [CrossRef]
- Yu, K.R.; Yang, S.R.; Jung, J.W.; Kim, H.; Ko, K.; Han, D.W.; Park, S.B.; Choi, S.W.; Kang, S.K.; Schöler, H.; et al. CD49f enhances multipotency and maintains stemness through the direct regulation of OCT4 and SOX2. Stem Cells 2012, 30, 876–887. [Google Scholar] [CrossRef]
- Desai, R.; East, D.A.; Hardy, L.; Faccendam, D.; Rigon, M.; Crosby, J.; Alvarez, M.S.; Singh, A.; Mainenti, M.; Hussey, L.K.; et al. Mitochondria form contact sites with the nucleus to couple prosurvival retrograde response. Sci. Adv. 2020, 6, eabc9955. [Google Scholar] [CrossRef]
- Xia, M.; Zhang, Y.; Jin, K.; Lu, Z.; Zeng, Z.; Xiong, W. Communication between mitochondria and other organelles: A brand-new perspective on mitochondria in cancer. Cell Biosci. 2019, 9, 27. [Google Scholar] [CrossRef]
- Amin, M.L. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights 2013, 7, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Ong, R.C.; Lei, J.; Lee, R.K.; Cheung, J.Y.; Fung, K.P.; Lin, C.; Ho, H.P.; Yu, B.; Li, M.; Kong, S.K. Polyphyllin D induces mitochondrial fragmentation and acts directly on the mitochondria to induce apoptosis in drug-resistant HepG2 cells. Cancer Lett. 2008, 261, 158–164. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, F.; Wen, H.; Shi, W.; Huang, Q.; Huang, Y.; Xie, J.; Li, P.; Chen, J.; Qin, L.; et al. Tumor-and mitochondria-targeted nanoparticles eradicate drug resistant lung cancer through mitochondrial pathway of apoptosis. J. Nanobiotechnol. 2020, 18, 8–28. [Google Scholar] [CrossRef]
- Yang, R.; Huang, B.; Zhu, Y.; Li, Y.; Liu, F.; Shi, J. Cell type-dependent bimodal p53 activation engenders a dynamic mechanism of chemoresistance. Sci. Adv. 2018, 4, eaat5077. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.H.; Pelicano, H.; Zhou, Y.; Carew, J.S.; Feng, L.; Bhalla, K.N.; Keating, M.J.; Huang, P. Inhibition of glycolysis in cancer cells: A novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005, 65, 613–621. [Google Scholar]
- Chen, Q.; Kirk, K.; Shurubor, Y.I.; Zhao, D.; Arreguin, A.J.; Shahi, I.; Valsecchi, F.; Primiano, G.; Calder, E.L.; Carelli, V. Rewiring of glutamine metabolism is a bioenergetic adaptation of human cells with mitochondrial DNA mutations. Cell Metab. 2018, 27, 1007–1025.e5. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.; Khiste, S.; Freinkman, E.; Dhawan, A.; Majumder, B.; Mondal, J.; Pinkerton, A.B.; Eton, E.; Medhi, R.; Chandrasekar, V.; et al. Targeting tumor phenotypic plasticity and metabolic remodeling in adaptive cross-drug tolerance. Sci. Signal 2019, eaas8779. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Vidal, C.; Dey, S.; Zhang, L. Mitochondria targeting as an effective strategy for cancer therapy. Int. J. Mol. Sci. 2020, 21, 3363. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, A.C.K.; Lau, P.M.; Kwan, Y.W.; Kong, S.K. Mitochondrial Fuel Dependence on Glutamine Drives Chemo-Resistance in the Cancer Stem Cells of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 3315. https://doi.org/10.3390/ijms22073315
Lee ACK, Lau PM, Kwan YW, Kong SK. Mitochondrial Fuel Dependence on Glutamine Drives Chemo-Resistance in the Cancer Stem Cells of Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2021; 22(7):3315. https://doi.org/10.3390/ijms22073315
Chicago/Turabian StyleLee, Alan Chun Kit, Pui Man Lau, Yiu Wa Kwan, and Siu Kai Kong. 2021. "Mitochondrial Fuel Dependence on Glutamine Drives Chemo-Resistance in the Cancer Stem Cells of Hepatocellular Carcinoma" International Journal of Molecular Sciences 22, no. 7: 3315. https://doi.org/10.3390/ijms22073315
APA StyleLee, A. C. K., Lau, P. M., Kwan, Y. W., & Kong, S. K. (2021). Mitochondrial Fuel Dependence on Glutamine Drives Chemo-Resistance in the Cancer Stem Cells of Hepatocellular Carcinoma. International Journal of Molecular Sciences, 22(7), 3315. https://doi.org/10.3390/ijms22073315





