A Tale of Two Transcriptomic Responses in Agricultural Pests via Host Defenses and Viral Replication
Abstract
:1. Introduction
2. Results
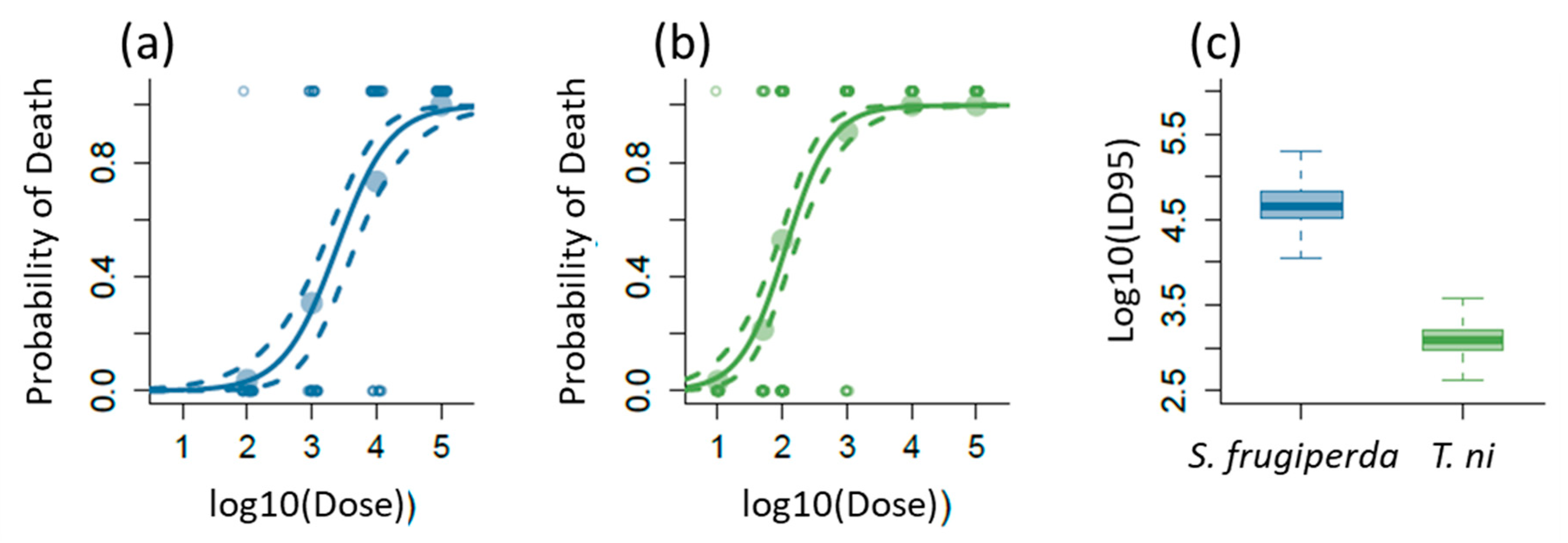
2.1. Lethal Effects of AcMNPV on S. frugiperda and T. ni
2.2. De Novo Assembly and Annotation of S. frugiperda and T. ni 4th Instar Reference Transcriptomes
2.3. Host Transcriptomic Responses to the AcMNPV Infection
2.4. Transcripts Associated with Chitin Metabolism and Epithelial Membrane Formation and Stability Were Suppressed in AcMNPV-Infected Hosts
2.5. Transcripts Associated with Hemocyte-Induced Defenses and Immune Responses Were Suppressed during Systemic Infection
2.6. Lipid Metabolism and Oxidative Stress Responses Emerge as the Most Prominent Functional Processes Induced by Both Hosts in Response to Infection
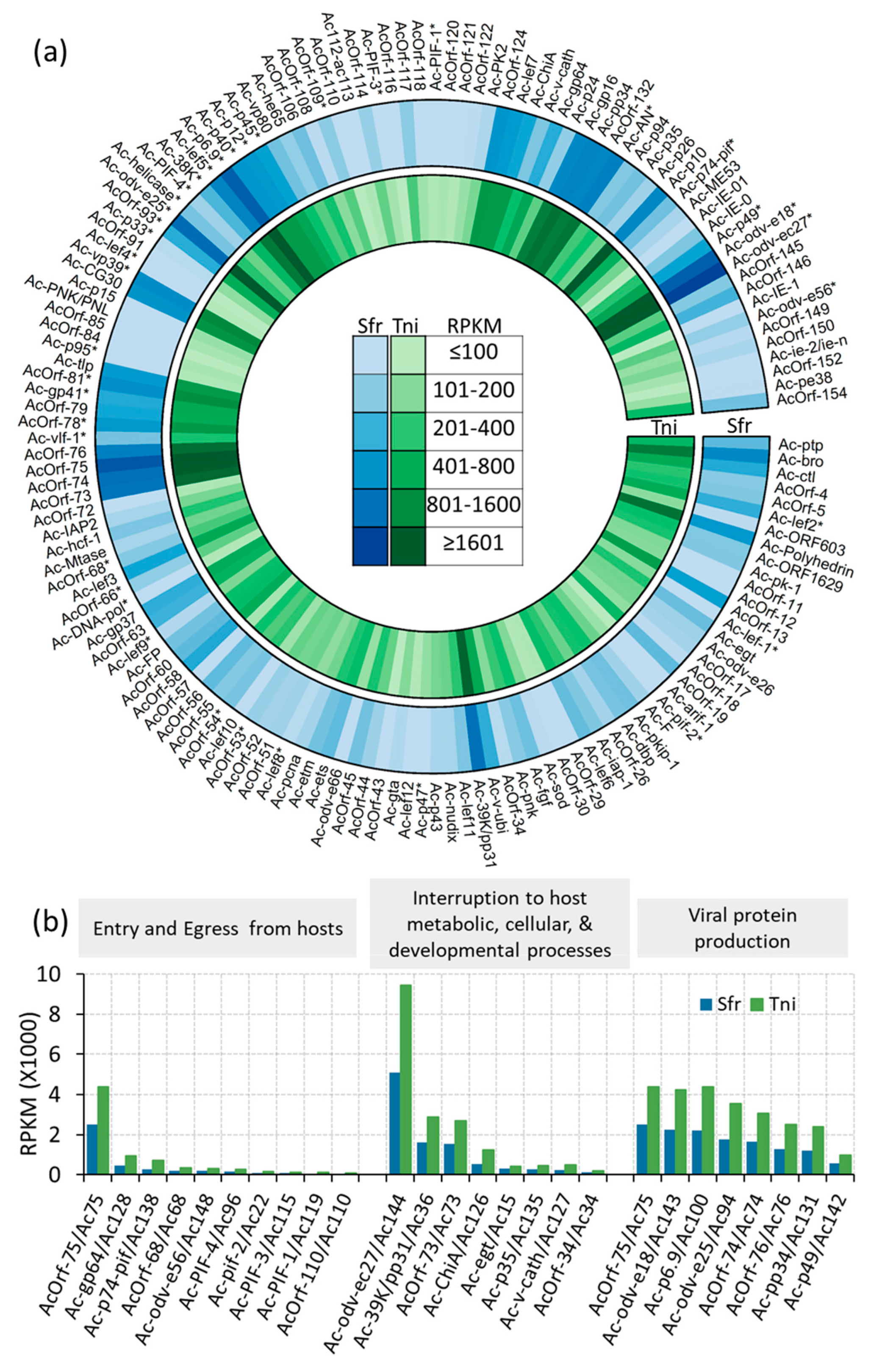
2.7. AcMNPV Genome Response to the Insect Hosts
2.8. Viral-Host Co-Transcriptional Interactions
3. Discussion
3.1. Host Transcriptomic Signatures Suggest Impaired Membrane Integrity, Enabling Viral Proliferation
3.2. Hemocyte-Mediated Innate Immunity Is Suppressed during Systemic Infection
3.3. Transcriptomic Signatures Indicative of Imbalances in Energy and Redox Homeostasis with Substantial Consequences to the Development of the Entire Organism during Disease Progression
3.4. Signaling Processes Associated with AcMNPV Infection
3.5. The Role of AcMNPV Genes Found in the Host Hemolymph
3.6. Host Transcriptional Responses to the Budded Virus during the Systemic Infection Stage Differs from the Midgut Responses to the Occlusion-Derived Virus during the Primary Infection Stage
3.7. Key Host Genes Affected by the AcPNMV Infection Are Targets of Commercially Available Pesticides Used against Lepidopteran Pests
4. Material and Methods
4.1. Insect and Virus Source Material
4.2. Determination of LD95 of AcMNPV for S. frugiperda and T. ni
4.3. Insect Treatment with AcMNPV Virus
4.4. Extraction of Hemolymph and Preparation of RNA-Seq Libraries
4.5. Sequencing, Assembly, and Annotation of the Reference Transcriptome
4.6. RNA-Seq Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuxa, J.R. Prevalence of viral infections in populations of fall armyworm, Spodoptera frugiperda, in Southeastern Louisiana. Environ. Entomol. 1982, 11, 239–242. [Google Scholar] [CrossRef]
- Miller, L. The Baculoviruses; Miller, L.K., Ed.; Kluwer Academic: Dordrecht, The Netherlands, 1997. [Google Scholar]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; Lopez-Ferber, M.; Possee, R.D. The compelete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef]
- Maghodia, A.B.; Jarvis, D.L.; Geisler, C. Complete genome sequence of the Autographa californica multiple nucleopolyhedrovirus strain E2. Genome Announc. 2014, 2, 6–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.C. Baculovirus vectors for gene therapy. Adv. Virus Res. 2006, 68, 287–320. [Google Scholar] [CrossRef] [PubMed]
- Blissard, G.W.; Theilmann, D.A. Baculovirus entry and egress from insect cells. Annu. Rev. Virol. 2018, 5, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Elderd, B.D. Developing models of disease transmission: Insights from the ecology of baculovirus-driven systems. PLoS Pathog. 2013, 9, e1003372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cory, J.S.; Myers, J.H. The ecology and evolution of insect baculoviruses. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 239–272. [Google Scholar] [CrossRef] [Green Version]
- Haas-stapleton, E.J.; Washburn, J.O.; Volkman, L.E. Pathogenesis of Autographa californica M nucleopolyhedrovirus in fifth instar Spodoptera frugiperda. J. Gen. Virol. 2003, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- FAO and CABI Community-based fall armyworm (Spodoptera frugiperda) monitoring, early warning and management. In Training of Trainers Manual, 1st ed.; FAO: Rome, Italy, 2019; ISBN 9789251312315.
- Mota-Sanchez, D.; Wise, J.C. The Arthropod Pesticide Resistance Database; Michigan State University: East Lansing, MI, USA, 2020; Available online: www.pesticideresistance.org (accessed on 31 July 2020).
- Wang, P.; Zhao, J.Z.; Rodrigo-Simón, A.; Kain, W.; Janmaat, A.F.; Shelton, A.M.; Ferré, J.; Myers, J. Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in a greenhouse population of the cabbage looper, Trichoplusia ni. Appl. Environ. Microbiol. 2007, 73, 1199–1207. [Google Scholar] [CrossRef] [Green Version]
- Lindgren, P.D.; Greene, G.L. Suppression and mAnagement of Cabbage Looper Populations; USDA Technical Bulletin: Washington, DC, USA, 1984.
- Sparks, A.N. Review of the biology of the fall armyworm (Lepidoptera, Noctuidae). Fla. Entomol. 1979, 62, 82–87. [Google Scholar] [CrossRef]
- Richter, A.R.; Fuxa, J.R.; Abdel-Fattah, M. Effect of host plant on the susceptibility of Spodoptera frugiperda (Lepidoptera: Noctuidae) to a nuclear polyhedrosis virus. Environ. Entomol. 1987, 16, 1004–1006. [Google Scholar] [CrossRef]
- Pitre, H.N.; Hogg, D.B. Development of the fall armyworm (Lepidoptera, Noctuidae) on cotton, soybean and corn. J. Georg. Entomol. Soc. 1983, 18, 182–187. [Google Scholar]
- Shorey, H.H.; Andres, L.A.; Hale, R.L. The biology of Trichoplusia ni (Lepidoptera: Noctuidae). I. life history and behavior. Ann. Entomol. Soc. Am. 1962, 55, 591–597. [Google Scholar] [CrossRef]
- Salem, T.Z.; Zhang, F.; Xie, Y.; Thiem, S.M. Comprehensive analysis of host gene expression in Autographa californica nucleopolyhedrovirus-infected Spodoptera frugiperda cells. Virology 2011, 412, 167–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Cao, L.; Miao, Y.; Wu, S.; Xu, S.; Wang, R.; Du, J.; Liang, A.; Fu, Y. Transcriptome analysis of Spodoptera frugiperda 9 (Sf9) cells infected with baculovirus, AcMNPV or AcMNPV-BmK IT. Biotechnol. Lett. 2017, 39, 1129–1139. [Google Scholar] [CrossRef]
- Chen, Y.-R.; Zhong, S.; Fei, Z.; Hashimoto, Y.; Xiang, J.Z.; Zhang, S.; Blissard, G.W. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J. Virol. 2013, 87, 6391–6405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-R.; Zhong, S.; Fei, Z.; Gao, S.; Zhang, S.; Li, Z.; Wang, P.; Blissard, G.W. Transcriptome responses of the host Trichoplusia ni to infection by the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Virol. 2014, 88, 13781–13797. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, A.; Bao, K.; Chen, W.; Wang, P.; Fei, Z.; Blissard, G.W. Transcriptional responses of the Trichoplusia ni midgut to oral infection by the baculovirus Autographa californica Multiple Nucleopolyhedrovirus. J. Virol. 2019, 93, e00353-19. [Google Scholar] [CrossRef] [Green Version]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [CrossRef] [Green Version]
- Pearce, S.L.; Clarke, D.F.; East, P.D.; Elfekih, S.; Gordon, K.H.J.; Jermiin, L.S.; McGaughran, A.; Oakeshott, J.G.; Papanikolaou, A.; Perera, O.P.; et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017, 15, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Cheng, T.; Wu, J.; Wu, Y.; Chilukuri, R.V.; Huang, L.; Yamamoto, K.; Feng, L.; Li, W.; Chen, Z.; Guo, H.; et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 2017, 1, 1747–1756. [Google Scholar] [CrossRef]
- Chen, W.; Yang, X.; Tetreau, G.; Song, X.; Coutu, C.; Hegedus, D.; Blissard, G.; Fei, Z.; Wang, P. A high-quality chromosome-level genome assembly of a generalist herbivore, Trichoplusia ni. Mol. Ecol. Resour. 2019, 19, 485–496. [Google Scholar] [CrossRef]
- Zeng, X.D.; Nan, F.; Liang, C.Y.; Song, J.H.; Wang, Q.; Vlak, J.M.; Chen, X.W. Functional analysis of the Autographa californica nucleopolyhedrovirus IAP1 and IAP2. Sci. China Ser. C Life. Sci. 2009, 52, 761–770. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y.; et al. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Kakumani, P.K.; Malhotra, P.; Mukherjee, S.K.; Bhatnagar, R.K. A draft genome assembly of the army worm, Spodoptera frugiperda. Genomics 2014, 104, 134–143. [Google Scholar] [CrossRef]
- Legeai, F.; Gimenez, S.; Duvic, B.; Escoubas, J.-M.; Gosselin Grenet, A.-S.; Blanc, F.; Cousserans, F.; Séninet, I.; Bretaudeau, A.; Mutuel, D.; et al. Establishment and analysis of a reference transcriptome for Spodoptera frugiperda. BMC Genom. 2014, 15, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.R.; Morandin, C.; Noureddine, M.; Pant, S. Conserved roles of Osiris genes in insect development, polymorphism and protection. J. Evol. Biol. 2018, 31, 516–529. [Google Scholar] [CrossRef] [Green Version]
- Nagaraj, R.; Adler, P.N. Dusky-like functions as a Rab11 effector for the deposition of cuticle during Drosophila bristle development. Development 2012, 139, 906–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, P.N.; Sobala, L.F.; Thom, D.; Nagaraj, R. dusky-like is required to maintain the integrity and planar cell polarity of hairs during the development of the Drosophila wing. Dev. Biol. 2013, 379, 76–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roch, F.; Alonso, C.R.; Akam, M. Drosophila miniature and dusky encode ZP proteins required for cytoskeletal reorganisation during wing morphogenesis. J. Cell Sci. 2003, 116, 1199–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonning, A.; Hemphälä, J.; Tång, E.; Nannmark, U.; Samakovlis, C.; Uv, A. A transient luminal chitinous matrix is required to model epithelial tube diameter in the Drosophila trachea. Dev. Cell 2005, 9, 423–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Jayaram, S.A.; Hemphala¨, J.; Senti, K.-A.; Tsarouhas, V.; Jin, H.; Samakovlis, C. Septate-junction-dependent luminal deposition of chitin eeacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 2006, 16, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Luschnig, S.; Ba, T.; Armbruster, K.; Krasnow, M.A. serpentine and vermiform encode matrix proteins with chitin binding and Deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 2006, 16, 186–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petkau, G.; Wingen, C.; Jussen, L.C.A.; Radtke, T.; Behr, M. Obstructor-A is required for epithelial extracellular matrix dynamics, exoskeleton function, and tubulogenesis. J. Biol. Chem. 2012, 287, 21396–21405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Oh, D.; Dassanayake, M. GOMCL: A toolkit to cluster, evaluate, and extract non-redundant associations of Gene Ontology-based functions. BMC Bioinform. 2020, 21, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.R.; Timpl, R. Laminin and other basement membrane components. Ann. Rev. Cell Biol. 1987, 3, 57–85. [Google Scholar] [CrossRef]
- Chi, H.-C.; Hui, C.-F. Primary structure of the Drosophila Laminin B2 chain and comparison with Human, Mouse, and Drosophila Laminin B1 and B2 chains. J. Biol. Chem. 1989, 264, 1543–1550. [Google Scholar] [CrossRef]
- Martinek, N.; Shahab, J.; Saathoff, M.; Ringuette, M. Haemocyte-derived SPARC is required for collagen-IV-dependent stability of basal laminae in Drosophila embryos. J. Cell Sci. 2011, 124, 1671–1680. [Google Scholar] [CrossRef] [Green Version]
- Shahab, J.; Baratta, C.; Scuric, B.; Godt, D.; Venken, K.J.T.; Ringuette, M.J. Loss of SPARC dysregulates basal lamina assembly to disrupt larval fat body homeostasis in Drosophila melanogaster. Dev. Dyn. 2015, 244, 540–552. [Google Scholar] [CrossRef]
- Yamakawa, M.; Tanaka, H. Immune proteins and their gene expression in the silkworm, Bombyx mori. Dev. Comp. Immunol. 1999, 23, 281–289. [Google Scholar] [CrossRef]
- Arai, I.; Ohta, M.; Suzuki, A.; Tanaka, S.; Yoshizawa, Y.; Sato, R. Immunohistochemical analysis of the role of hemocytin in nodule formation in the larvae of the silkworm, Bombyx mori. J. Insect Sci. 2013, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Bao, J.; Mo, B.; Liu, L.; Li, T.; Pan, G.; Chen, J.; Zhou, Z. Hemocytin facilitates host immune responses against Nosema bombycis. Dev. Comp. Immunol. 2019, 103, 103495. [Google Scholar] [CrossRef] [PubMed]
- Ponnuvel, K.M.; Yamakawa, M. Immune responses against bacterial infection in Bombyx mori and regulation of host gene expression. Curr. Sci. 2002, 83, 447–454. [Google Scholar]
- Lesch, C.; Goto, A.; Lindgren, M.; Bidla, G.; Dushay, M.S.; Theopold, U. A role for Hemolectin in coagulation and immunity in Drosophila melanogaster. Dev. Comp. Immunol. 2007, 31, 1255–1263. [Google Scholar] [CrossRef]
- Jagdale, S.; Bansode, S.; Joshi, R. Insect Proteases: Structural-Functional Outlook. In Proteases in Physiology and Pathology; Springer Nature: Singapore, 2017; ISBN 9789811025129. [Google Scholar]
- Houot, B.; Bousquet, F.; Ferveur, J.F. The consequences of regulation of desat1 expression for pheromone emission and detection in Drosophila melanogaster. Genetics 2010, 185, 1297–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione Transferases. Annu Rev. Pharm. Toxicol 2005, 45, 51–88. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol. Biol. 2010, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.; Shikano, I.; Felton, G.W.; Liu, T.-X.; Hoover, K. Host permissiveness to baculovirus influences time-dependent immune responses and fitness costs. Insect Sci. 2021, 28, 103–114. [Google Scholar] [CrossRef]
- Katsuma, S.; Kawaoka, S.; Mita, K.; Shimada, T. Genome-wide survey for baculoviral host homologs using the Bombyx genome sequence. Insect Biochem. Mol. Biol. 2008, 38, 1080–1086. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Chateigner, A.; Ernenwein, L.; Barbe, V.; Annie, B.; Herniou2, E.A.; Cordaux, R. Population genomics supports baculoviruses as vectors of horizontal transfer of insect transposons. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, T.M.; Huijskens, I.; Willis, L.G.; Theilmann, D.A. The Autographa californica Multiple Nucleopolyhedrovirus ie0 - ie1 gene complex is essential for wild-type virus replication, but either IE0 or IE1 can support virus growth. J. Virol. 2005, 79, 4619–4629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Sebastián, S.; López-Vidal, J.; Escribano, J.M. Significant productivity improvement of the baculovirus expression vector system by engineering a novel expression cassette. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, G.E.; Henner, D.J. Multiple early transcripts and splicing of the Autographa californica nuclear polyhedrosis virus IE-1 gene. J. Virol. 1988, 62, 3193–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacs, G.R.; Guarino, L.A.; Summers, M.D. Novel regulatory properties of the IE1 and IE0 transactivators encoded by the baculovirus Autographa californica multicapsid nuclear polyhedrosis virus. J. Virol. 1991, 65, 5281–5288. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.Y.; Roh, J.Y.; Wang, Y.; Zhen, Z.; Tao, X.Y.; Lee, J.H.; Liu, Q.; Kim, J.S.; Shin, S.W.; Je, Y.H. Analysis of genes expression of Spodoptera exigua Larvae upon AcMNPV infection. PLoS ONE 2012, 7, e42462. [Google Scholar] [CrossRef] [Green Version]
- Guarino, L.A.; Dong, W.E.N.; Xu, B.I.N.; Broussard, D.R.; Davis, R.W.; Jarvis, D.L. Baculovirus phosphoprotein pp3l is associated with virogenic stroma. J. Virol. 1992, 66, 7113–7120. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.J.; Qiu, L.H.; An, S.H.; Yao, Q.; Park, E.Y.; Chen, K.P.; Zhang, C.X. Open reading frame 60 of the Bombyx mori nucleopolyhedrovirus plays a role in budded virus production. Virus Res. 2010, 151, 185–191. [Google Scholar] [CrossRef]
- Ishimwe, E.; Hodgson, J.J.; Clem, R.J.; Passarelli, A.L. Reaching the melting point: Degradative enzymes and protease inhibitors involved in baculovirus infection and dissemination. Virology 2015, 479–480, 637–649. [Google Scholar] [CrossRef] [Green Version]
- Braunagel, S.C.; He, H.; Ramamurthy, P.; Summers, M.D. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology 1996, 114, 100–114. [Google Scholar] [CrossRef] [Green Version]
- Heaton, N.S.; Randall, G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011, 19, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, T.; Welch, M.D. Baculovirus Actin-based motility drives nuclear envelope disruption and nuclear egress report. Curr. Biol. 2018, 28, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, J.; Zhang, Y.; Hu, Y.; Hu, X.; Zhou, Y.; Zhao, H.; Pei, R.; Wu, C.; Chen, J.; Zhao, H.; et al. Autographa californica multiple nucleopolyhedrovirus Ac34 protein retains cellular Actin-Related protein 2/3 complex in the nucleus by subversion of CRM1- dependent nuclear export. PLoS Pathog. 2016, 12, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Tang, Z.; Yuan, M.; Wu, W.; Yang, K. The 91-205 amino acid region of AcMNPV ORF34 (Ac34), which comprises a potential C3H zinc finger, is required for its nuclear localization and optimal virus multiplication. Virus Res. 2017, 228, 79–89. [Google Scholar] [CrossRef]
- Cai, Y.; Long, Z.; Qiu, J.; Yuan, M.; Li, G.; Yang, K. An ac34 deletion mutant of Autographa californica Nucleopolyhedrovirus exhibits delayed late gene expression and a lack of virulence in vivo. J. Virol. 2012, 86, 10432–10443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, K.K.; Meadows, S.M.; Cripps, R.M. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002, 110, 39–50. [Google Scholar] [CrossRef]
- Clem, R.J.; Miller, L.K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell Biol. 1994, 14, 5212–5222. [Google Scholar] [CrossRef] [Green Version]
- Lu, A.; Miller, L.K. Species-specific effects of the hcf-1 gene on baculovirus virulence. J. Virol. 1996, 70, 5123–5130. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, A.; Hamajima, R.; Tomizaki, M.; Kondo, T.; Nanba, Y.; Yamada, H.; Ikeda, M. HCF-1 encoded by baculovirus AcMNPV is required for productive nucleopolyhedrovirus infection of. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Clem, R.J.; Miller, L.K. Apoptosis reduces both the in vitro replication and the in vivo infectivity of a baculovirus. J. Virol. 1993, 67, 3730–3738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, M.; Yamada, H.; Ito, H.; Kobayashi, M. Baculovirus IAP1 induces caspase-dependent apoptosis in insect cells. J. Gen. Virol 2011, 92, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- McLachlin, J.R.; Escobar, J.C.; Harrelson, J.A.; Clem, R.J.; Miller, L.K. Deletions in the Ac-iap1 gene of the baculovirus AcMNPV occur spontaneously during serial passage and confer a cell line-specific replication advantage. Virus Res. 2001, 81, 77–91. [Google Scholar] [CrossRef]
- Reilly, D.R.O.; Miller, L.K. A Baculovirus blocks insect molting by producing Ecdysteroid UDP-Glucosyl Transferase. Science 1989, 245, 1110–1112. [Google Scholar] [CrossRef]
- Rao, R.; Fiandra, L.; Giordana, B.; De Eguileor, M.; Congiu, T.; Burlini, N.; Arciello, S.; Corrado, G.; Pennacchio, F. AcMNPV ChiA protein disrupts the peritrophic membrane and alters midgut physiology of Bombyx mori larvae. Insect Biochem. Mol. Biol. 2004, 34, 1205–1213. [Google Scholar] [CrossRef]
- Erlandson, M.A.; Toprak, U.; Hegedus, D.D. Role of the peritrophic matrix in insect-pathogen interactions. J. Insect Physiol. 2019, 117, 103894. [Google Scholar] [CrossRef]
- He, L.; Li, N.; Chen, Y.; Liu, S. Regulation of Chitinase in Spodoptera exigua (Hübner) (Lepidoptera:Noctuidae) during infection by Heliothis virescens. Front. Physiol. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Romoser, W.S.; Turell, M.J.; Lerdthusnee, K.; Neira, M.; Dohm, D.; Ludwig, G.; Wasieloski, L. Pathogenesis of Rift Valley fever virus in mosquitoes—Tracheal conduits & the basal lamina as an extra-cellular barrier. In Infectious Diseases from Nature: Mechanisms of Viral Emergence and Persistence; Peters, C.J., Calisher, C.H., Eds.; Springer: Vienna, Austria, 2005; pp. 89–100. [Google Scholar]
- Passarelli, A.L. Barriers to success: How baculoviruses establish efficient systemic infections. Virology 2012, 411, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Engelhard, E.K.; Kam-Morgan, L.N.; Washburn, J.O.; Volkman, L.E. The insect tracheal system: A conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc. Natl. Acad. Sci. USA 1994, 91, 3224–3227. [Google Scholar] [CrossRef] [Green Version]
- Hawtin, R.E.; Arnold, K.; Ayres, M.D.; Zanotto, P.M.d.A.; Howard, S.C.; Gooday, G.W.; Chappell, L.H.; Kitts, P.A.; King, L.A.; Possee, R.D. Identification and preliminary characterization of a Chitinase gene in the Autographa californica nuclear polyhedrosis-virus genome. Virology 1995, 212, 673–685. [Google Scholar] [CrossRef] [Green Version]
- Daimon, T.; Katsuma, S.; Kang, W.K.; Shimada, T. Comparative studies of Bombyx mori nucleopolyhedrovirus chitinase and its host ortholog, BmChi-h. Biochem. Biophys. Res. Commun. 2006, 345, 825–833. [Google Scholar] [CrossRef]
- Hawtin, R.E.; Zarkowska, T.; Arnold, K.; Thomas, C.J.; Gooday, G.W.; King, L.A.; Kuzio, J.A.; Possee, R.D. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 1997, 238, 243–253. [Google Scholar] [CrossRef] [Green Version]
- Campbell, A.G.; Fessler, L.I.; Salo, T.; Fesslerq, J.H. Papilin: A Drosophila proteoglycan-like sulfated glycoprotein from basement membranes. J. Biol. Chem. 1987, 262, 17605–17612. [Google Scholar] [CrossRef]
- Hortsch, M.; Marikar, Y.; Fishman, S.; Soneral, S.N.; Dong, R.; Jacobs, J.R. The expression of MDP-1, a component of Drosophila embryonic basement membranes, is modulated by apoptotic cell death. Int, J. Dev. Biol. 1998, 42, 33–42. [Google Scholar]
- Kramerova, I.A.; Kawaguchi, N.; Fessler, L.I.; Nelson, R.E.; Chen, Y.; Kramerov, A.A.; Kusche-gullberg, M.; Kramer, J.M.; Ackley, B.D.; Sieron, A.L.; et al. Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development 2000, 127, 5475–5485. [Google Scholar]
- Beck, K.; Hunter, I.; Engel, J. Structure and function of laminin: Anatomy of a multidomain glycoprotein. FASEB J. 1990, 4, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Keeley, D.P.; Hastie, E.; Jayadev, R.; Kelley, L.C.; Chi, Q.; Payne, S.G.; Jeger, J.L.; Hoffman, B.D.; Sherwood, D.R. Comprehensive endogenous tagging of basement membrane components reveals dynamic movement within the matrix scaffolding. Dev. Cell 2020, 54, 60–74.e7. [Google Scholar] [CrossRef]
- Means, J.C.; Passarelli, A.L. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc. Natl. Acad. Sci. USA 2010, 107, 9825–9830. [Google Scholar] [CrossRef] [Green Version]
- Monsma, S.A.; Oomens, A.G.P.; Blissard, G.W. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 1996, 70, 4607–4616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Sun, Y.; Li, Y.; Liu, X.; Yue, Q.; Li, Z. Inhibition of cellular fatty acid synthase impairs replication of budded virions of Autographa californica multiple nucleopolyhedrovirus in Spodoptera frugiperda cells. Virus Res. 2018, 252, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Munger, J.; Bennett, B.D.; Parikh, A.; Feng, X.; Rabitz, H.A.; Shenk, T.; Rabinowitz, J.D. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 2010, 26, 1179–1186. [Google Scholar] [CrossRef] [Green Version]
- Breitenbach, J.E.; Shelby, K.S.; Popham, H.J.R. Baculovirus induced transcripts in hemocytes from the larvae of Heliothis virescens. Viruses 2011, 3, 2047–2064. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, F.; Ferveur, J.F. desat1 A Swiss army knife for pheromonal communication and reproduction? Fly (Austin) 2012, 6, 102–107. [Google Scholar] [CrossRef] [Green Version]
- Köhler, K.; Brunner, E.; Xue, L.G.; Boucke, K.; Greber, U.F.; Mohanty, S.; Barth, J.M.I.; Wenk, M.R.; Hafen, E. A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila. Autophagy 2009, 5, 980–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [Green Version]
- Roxstrom-Lindquist, K.; Terenius, O.; Faye, I. Parasite-specific immune response in adult Drosophila melanogaster: A genomic study. EMBO Rep. 2004, 5, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Tittiger, C.; Wicker-Thomas, C.; Le Goff, G.; Young, S.; Wajnberg, E.; Fricaux, T.; Taquet, N.; Blomquist, G.J.; Feyereisen, R. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 14858–14863. [Google Scholar] [CrossRef] [Green Version]
- Cerenius, L.; Kawabata, S.I.; Lee, B.L.; Nonaka, M.; Söderhäll, K. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem. Sci. 2010, 35, 575–583. [Google Scholar] [CrossRef]
- Lavine, M.D.; Strand, M.R. Insect hemocytes and their role in immunity. Insect Immunol. 2008, 32, 25–47. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, G.H.; Dong, Z.M.; Duan, J.; Xu, P.Z.; Cheng, T.C.; Xiang, Z.H.; Xia, Q.Y. Genome-wide identification and expression analysis of serine proteases and homologs in the silkworm Bombyx mori. BMC Genom. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Washburn, J.O.; Haas-Stapleton, E.J.; Tan, F.F.; Beckage, N.E.; Volkman, L.E. Co-infection of Manduca sexta larvae with polydnavirus from Cotesia congregata increases susceptibility to fatal infection by Autographa californica M Nucleopolyhedrovirus. J. Insect Physiol. 2000, 46, 179–190. [Google Scholar] [CrossRef]
- Lavington, E.; Cogni, R.; Kuczynski, C.; Koury, S.; Behrman, E.L.; O’brien, K.R.; Schmidt, P.S.; Eanes, W.F. A small system-high-resolution study of metabolic adaptation in the central metabolic pathway to temperate climates in Drosophila melanogaster. Mol. Biol. Evol. 2014, 31, 2032–2041. [Google Scholar] [CrossRef] [Green Version]
- Wise, E.M.; Ball, E.G. Malic enzyme and lipogenesis. Proc. Natl. Acad. Sci. USA 1964, 52, 1255–1263. [Google Scholar] [CrossRef] [Green Version]
- Merritt, T.J.S.; Duvernell, D.; Eanes, W.F. Natural and synthetic alleles provide complementary insights into the nature of selection acting on the Men polymorphism of Drosophila melanogaster. Genetics 2005, 171, 1707–1718. [Google Scholar] [CrossRef] [Green Version]
- Geer, B.W.; Krochko, D.; Williamson, J.H. Ontogeny, cell distribution, and the physiological role of NADP-malic enzyme in Drosophila melanogaster. Biochem. Genet. 1979, 17, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Yoon, C.S.; Yi, J.; Cho, J.R.; Kim, H.S. Cellular immune responses and FAD-glucose dehydrogenase activity of Mamestra brassicae (Lepidoptera: Noctuidae) challenged with three species of entomopathogenic fungi. Physiol. Entomol. 2005, 30, 287–292. [Google Scholar] [CrossRef]
- Trienens, M.; Rohlfs, M. A potential collective defense of Drosophila larvae against the invasion of a harmful fungus. Front. Ecol. Evol. 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Gomez-Diaz, C.; Bargeton, B.; Abuin, L.; Bukar, N.; Reina, J.H.; Bartoi, T.; Graf, M.; Ong, H.; Ulbrich, M.H.; Masson, J.F.; et al. A CD36 ectodomain mediates insect pheromone detection via a putative tunnelling mechanism. Nat. Commun. 2016, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Vosshall, L.B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 2007, 450, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Mast, J.D.; De Moraes, C.M.; Alborn, H.T.; Lavis, L.D.; Stern, D.L. Evolved differences in larval social behavior mediated by novel pheromones. Elife 2014, 3, e04205. [Google Scholar] [CrossRef]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.L.; Bulet, P.; Imler, J.L. Pherokine-2 and -3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 2003, 270, 3398–3407. [Google Scholar] [CrossRef]
- Labeur, C.; Dallerac, R.; Wicker-Thomas, C. Involvement of desat1 gene in the control of Drosophila melanogaster pheromone biosynthesis. Genetica 2002, 114, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, F.; Nojima, T.; Houot, B.; Chauvel, I.; Chaudy, S.; Dupas, S.; Yamamoto, D.; Ferveur, J.-F. Expression of a desaturase gene, desat1, in neural and nonneural tissues separately affects perception and emission of sex pheromones in Drosophila. Proc. Natl. Acad. Sci. USA 2012, 109, 249–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, S.; Ramirez, L.; Dimopoulos, G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartholomay, L.C.; Cho, W.L.; Rocheleau, T.A.; Boyle, J.P.; Beck, E.T.; Fuchs, J.F.; Liss, P.; Rusch, M.; Butler, K.M.; Wu, R.C.C.; et al. Description of the transcriptomes of immune response-activated hemocytes from the mosquito vectors Aedes aegypti and Armigeres subalbatus. Infect. Immun 2004, 72, 4114–4126. [Google Scholar] [CrossRef] [Green Version]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling. Curr. Biol. 2014, 24, 453–462. [Google Scholar] [CrossRef] [Green Version]
- Rohrmann, G.F. Chapter 12, The AcMNPV Genome: Gene Content, Conservation, and Function. In Baculovirus Molecular Biology, 3rd ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2013; pp. 1–84. [Google Scholar]
- O’Reilly, D.R.; Miller, L.K. Regulation of expression of a baculovirus ecdysteroid UDPglucosyltransferase gene. J. Virol. 1990, 64, 1321–1328. [Google Scholar] [CrossRef] [Green Version]
- Hoover, K.; Grove, M.; Gardner, M.; Hughes, D.P.; Mcneil, J.; Slavicek, J. A Gene for an extended phenotype. Science 2011, 333, 1401. [Google Scholar] [CrossRef] [Green Version]
- Ros, V.I.D.; Van Houte, S.; Hemerik, L.; Van Oers, M.M. Baculovirus-induced tree-top disease: How extended is the role of egt as a gene for the extended phenotype? Mol. Ecol. 2015, 24, 249–258. [Google Scholar] [CrossRef]
- Van Houte, S.; Ros, V.I.D.; Van Oers, M.M. Hyperactivity and tree-top disease induced by the baculovirus AcMNPV in Spodoptera exigua larvae are governed by independent mechanisms. Naturwissenschaften 2014, 101, 347–350. [Google Scholar] [CrossRef]
- Herrero, S.; Ansems, M.; Van Oers, M.M.; Vlak, J.M.; Bakker, P.L.; de Maagd, R.A. REPAT, a new family of proteins induced by bacterial toxins and baculovirus infection in Spodoptera exigua. Insect Biochem. Mol. Biol. 2007, 37, 1109–1118. [Google Scholar] [CrossRef]
- Merzendorfer, H. Chitin synthesis inhibitors: Old molecules and new developments. Insect Sci. 2013, 20, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, turnover, and functions of chitin in insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Douris, V.; Steinbach, D.; Panteleri, R.; Livadaras, I.; Pickett, J.A.; Van Leeuwen, T.; Nauen, R.; Vontas, J. Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 14692–14697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moussian, B.; Schwarz, H.; Bartoszewski, S.; Nu, C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster. J. Morphol. 2005, 264, 117–130. [Google Scholar] [CrossRef]
- Cook, D.R.; Leonard, B.R.; Gore, J. Field and laboratory performance of novel insecticides against armyworms (Lepidoptera: Noctuidae). Fla. Entomol. 2006, 87, 433–439. [Google Scholar] [CrossRef]
- IRAC South Africa. Integrated Pest Management (IPM) & Insect Resistance Management (IRM) for Fall Armyworm in South African Maize; IRAC: Pretoria, South Africa, 2018; pp. 1–21. [Google Scholar]
- Powell, G.F.; Ward, D.A.; Prescott, M.C.; Spiller, D.G.; White, M.R.H.; Turner, P.C.; Earley, F.G.P.; Phillips, J.; Rees, H.H. The molecular action of the novel insecticide, Pyridalyl. Insect Biochem. Mol. Biol. 2011, 41, 459–469. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: DsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, X.; Dai, Y.; Wei, X.; Ni, M.; Zhang, L.; Zhu, Z. Expressing double-stranded RNAs of insect hormone-related genes enhances baculovirus insecticidal activity. Int. J. Mol. Sci. 2019, 20, 419. [Google Scholar] [CrossRef] [Green Version]
- Bonning, B.C.; Hirst, M.; Possee, R.D.; Hammock, B.D. Further development of a recombinant baculovirus insecticide expressing the enzyme juvenile hormone esterase from Heliothis virescens. Insect Biochem. Mol. Biol. 1992, 22, 453–458. [Google Scholar] [CrossRef]
- Chen, E.; Kolosov, D.; O’Donnell, M.J.; Erlandson, M.A.; McNeil, J.N.; Donly, C. The effect of diet on midgut and resulting changes in infectiousness of AcMNPV baculovirus in the cabbage looper, Trichoplusia ni. Front. Physiol. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Wilson, K.; Grzywacz, D.; Curcic, I.; Scoates, F.; Harper, K.; Rice, A.; Paul, N.; Dillon, A. A novel formulation technology for baculoviruses protects biopesticide from degradation by ultraviolet radiation. Sci. Rep. 2020, 10, 13301. [Google Scholar] [CrossRef]
- Cory, J.S.; Hirst, M.L.; Williams, T.; Hails, R.S.; Goulson, D.; Green, B.M.; Carty, T.M.; Possee, R.D.; Cayley, P.J.; Bishop, D.H.L. Field trial of a genetically improved baculovirus insecticide. Nature 1994, 370, 138–140. [Google Scholar] [CrossRef]
- Shim, H.J.; Choi, J.Y.; Wang, Y.; Tao, X.Y.; Liu, Q.; Roh, J.Y.; Kim, J.S.; Kim, W.J.; Woo, S.D.; Jin, B.R.; et al. Neurobactrus, a novel, highly effective, and environmentally friendly recombinant baculovirus insecticide. Appl. Environ. Microbiol. 2013, 79, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Goswami, A.; Debnath, N. Application of Baculoviruses as Biopesticides and the Possibilities of Nanoparticle Mediated Delivery in Nano-Biopesticides Today and Future Perspectives; Elsevier Inc.: Oxford, UK, 2019; ISBN 9780128158296. [Google Scholar]
- Collett, D. Modelling Binary Data; Chapman & Hall/CRC: Boca Raton, FL, USA, 2003. [Google Scholar]
- R Core Team. A Language and Environment for Statistical Computing; R Development Core Team: Wien, Austria, 2018. [Google Scholar]
- Plummer, M. JAGS: A program for analysis of bayesian graphical models using gibbs sampling. Proc. Dsc. 2003, 1–10. [Google Scholar]
- Su, Y.; Yajima, M. Package ‘R2jags’; CRAN: Vienna, Austria, 2015. [Google Scholar]
- Link, W.A.; Eaton, M.J. On thinning of chains in MCMC. Methods Ecol. Evol. 2012, 3, 112–115. [Google Scholar] [CrossRef]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Rubin, D.B. Bayesian Data Analysis; Taylor & Francis: Oxford, UK, 2014. [Google Scholar]
- Gelman, A.; Meng, X.L.; Stern, H. Posterior predictive assessment of model fitness via realized discrepancies. Stat. Sin. 1996, 6, 733–760. [Google Scholar]
- Kéry, M. Introduction to WinBUGS for Ecologists; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Granados, R.R.; Lawler, K.A. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 1981, 308, 297–308. [Google Scholar] [CrossRef]
- Fan, L.; Wang, G.; Hu, W.; Pantha, P.; Tran, K.-N.; Zhang, H.; An, L.; Dassanayake, M.; Qiu, Q.-S. Transcriptomic view of survival during early seedling growth of the extremophyte Haloxylon ammodendron. Plant. Physiol. Biochem. 2018, 132, 475–489. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Oh, D.; Barkla, B.J.; Vera-estrella, R.; Pantoja, O.; Lee, S.; Bohnert, H.J.; Dassanayake, M. Cell type-specific responses to salinity–the epidermal bladder cell transcriptome of Mesembryanthemum crystallinum. New Phytol. 2015, 207, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, A.; Bao, K.; Chen, Y.-R.; Chen, W.; Wang, P.; Fei, Z.; Blissard, G.W. Global analysis of Baculovirus Autographa californica Multiple Nucleopolyhedrovirus gene expression in the midgut of the lepidopteran host Trichoplusia ni. J. Virol. 2018, 92, e01277-18. [Google Scholar] [CrossRef] [Green Version]




| Transcriptome Assembly Features | S. frugiperda | T. ni |
|---|---|---|
| Coding a (Non-Coding b) | Coding a (Non-Coding b) | |
| Total assembled transcripts | 17,908 (101,169) | 19,472 (147,934) |
| Percent GC | 42.68 (35.28) | 41.16 (35.24) |
| Contig N50 (nt) | 2279 (532) | 2955 (656) |
| Average contig length (nt) | 1458 (495) | 1773 (549) |
| Smallest contig length (nt) | 297 (224) | 297 (201) |
| Longest contig length (nt) | 29,765 (14,343) | 30,823 (23,482) |
| Number of ORFs | 28,433 (-) | 31,292 (-) |
| Average ORF length (nt) | 920.28 (-) | 841.86 (-) |
| Smallest ORF length (nt) | 297 (-) | 297 (-) |
| Longest ORF length (nt) | 27,558 (-) | 27,408 (-) |
| Mapped sequenced read % to the reference assembly | 76 | 84 |
| Detected complete BUSCOs (%) c (Arthropoda reference) | 80.6 | 82.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantha, P.; Chalivendra, S.; Oh, D.-H.; Elderd, B.D.; Dassanayake, M. A Tale of Two Transcriptomic Responses in Agricultural Pests via Host Defenses and Viral Replication. Int. J. Mol. Sci. 2021, 22, 3568. https://doi.org/10.3390/ijms22073568
Pantha P, Chalivendra S, Oh D-H, Elderd BD, Dassanayake M. A Tale of Two Transcriptomic Responses in Agricultural Pests via Host Defenses and Viral Replication. International Journal of Molecular Sciences. 2021; 22(7):3568. https://doi.org/10.3390/ijms22073568
Chicago/Turabian StylePantha, Pramod, Subbaiah Chalivendra, Dong-Ha Oh, Bret D. Elderd, and Maheshi Dassanayake. 2021. "A Tale of Two Transcriptomic Responses in Agricultural Pests via Host Defenses and Viral Replication" International Journal of Molecular Sciences 22, no. 7: 3568. https://doi.org/10.3390/ijms22073568






